Abstract
We present an integrated and low-cost microfluidic platform capable of extraction of nucleic acids from real biological samples. We demonstrate the application of this platform in pathogen detection and cancer screening. The integrated platform consists of three units including a pretreatment unit for separation of nucleic acids from lysates, a preconcentration unit for concentration of isolated nucleic acids and a sensing unit localized at a designated position on the chip for specific detection of the target nucleic acid. The platform is based on various electrokinetic phenomena exhibited by ion exchange membranes in a DC electrical field that allow them to serve as molecular filters, analyte preconcentrators and sensors. In this manuscript, we describe each unit of the integrated chip separately and show specific detection of a microRNA (miRNA 146a) biomarker associated with oral cancer as a proof-of-concept experiment. This platform technology can easily be extended to other targets of interest by optimizing the properties of the ion exchange membranes and the specific probes functionalized onto the sensors.
Keywords: point-of-care diagnostics, ion exchange membranes, oral cancer, electrokinetics, integrated platform technology, sample-to-answer detection
Graphical abstract
Introduction
The development of an integrated microfluidic diagnostic platform has been of considerable interest in recent years as it enables molecular detection with miniature devices that are amenable to use in any setting without the need for extensive resources. Such settings may include the patient's bedside at a modern hospital, or a clinic in a low-resource setting. The device may be used by first responders investigating a biological event or a medic at a military base. Eventually, such an integrated platform can reduce the disease-associated morbidity and mortality rate, particularly for infectious or cancer-related diseases [1]. The lack of availability of good diagnostic tests significantly contributes to health problems in developing countries [2]. In low-resource settings, the diagnosis is often based on the physician's experience rather than results from laboratory tests. Thus an integrated platform capable of providing sample-to-answer solution in low-resource settings as well as in a timely fashion could significantly impact disease control and advance the benefits of treatment in the developing world [3,4].
Conventional and standard laboratory techniques for pathogen detection generally use cell culture method, enzyme-linked immunosorbent assay (ELISA) or polymerase chain reaction (PCR). However, these tests cannot be employed in low-resource settings due to their long assay times, high costs per test and their need for trained personnel and infrastructure resources [5,6]. Compared to current laboratory-based detection techniques, fully automated push-button integrated devices would need to be rapid and low-cost with simple assay protocol to be useful in such settings [7].
In recent years, there have been significant efforts to develop sample-to-answer, low-cost, integrated microfluidic platforms capable of analyzing biological samples like oral rinse [8-10], nasal fluids [11], urine [12], serum [13], and whole blood [14-16]. The use of microfluidic architecture to develop point-of-care (POC) diagnostic devices bring some inherent advantages such as portability, short assay time, lower sample/reagent requirement, lower power consumption, disposability and automation. This significantly reduces the cost, time, complexity and contamination of assay without compromising the sensitivity and specificity of the test [5]. Researchers have developed different promising microfluidic technologies such as droplet-based devices [17,18], centrifugal devices [19,20], capillary devices [21,22], paper-based devices [5,6,23], and lateral flow assays [1,8,24,25] to realize such integrated diagnostic platforms. For instance, Sauer-Budge et al. used silica and polymeric monolith in the lateral flow assay strip to detect different types of pathogens from fluid samples [24]. These microfluidic cassettes can perform cell lysis, mixing, target isolation, PCR, and detection of amplified products in the integrated chip [25]. Commercially-available diagnostic devices based on lateral flow include GenoType (Hain Lifescience GmbH) and INNO-LiPA (Innogenetics NV) [26]. Yang et al. used an affinity column and capillary electrophoresis channels in an integrated microdevice to detect multiple biomarkers from human blood serum [27]. Such systems used the solid phase extraction principle to capture the target on the microchip column functionalized with antibody and later elute the target for downstream analysis. Lee et al. developed a portable, fully-automated immunoassay platform to test for infectious disease from whole blood sample using centrifugal microfluidic compact disk technology [14,15]. However, most of these platforms employ a high level of integration and functionality to perform an assay. The use of a multilayer design, different types of materials, and a number of assembly steps for all these platforms renders their mass production expensive. Although some commercially available medical diagnostics products are FDA-approved (e.g. Focus Dx (Quest), Handy Lab (BD), i-Stat Corp (Abbott), IQuum, Idaho Technologies, and Cepheid GeneXpert), they are still not amenable to low-resource settings [4,21,28,29].
As an extension of the current effort, we report a simple, low-cost, label-free, PCR-free, integrated microfluidic biosensing platform capable of analyzing real samples using commercially available heterogeneous ion-selective nanoporous membranes. The integrated design has been conceived in a way to allow for easy automation and would only require the user for sample loading, thus fully exploiting the simplicity of our integrated platform. The device consists of three units: pretreatment, preconcentration and sensing. All units use ion-exchange membranes for various purposes: to isolate nucleic acids from the lysed cell sample (pretreatment unit), to concentrate the isolated nucleic acid molecules at a precise location (pre-concentration unit), and to specifically detect target nucleic acid biomarkers (sensing unit). The three units of the integrated platform utilize the inherent negative charge of the nucleic acid molecules and the ion-selective properties of cation- and anion-exchange membranes to transport, concentrate and detect target nucleic acid biomarkers in the integrated microfluidic platform, whereas electric field is used to extract the nucleic acid molecules from the sample through an agarose gel.
Recently, several sample pretreatment lab-on-a-chip techniques have been developed to process real biological samples including hydrodynamic, mechanical, acoustic, electric, magnetic bead, and ultrasonic approaches [4,26,29,30]. Some of these lysing methods require complicated metal electrode fabrication, expensive instrumentation and multistep protocols [30]. Here, we demonstrate an easy-to-integrate, on-chip sample pretreatment process to extract nucleic acids from real samples by integrating gel electrophoresis into our microfluidic platform. By applying a DC field, we separated and extracted the negatively-charged nucleic acid molecules from cell debris and other positively charged biomolecules like proteins below their isoelectric point, thus reducing contamination for downstream functionalities.
To accomplish rapid detection of target, we used the cation exchange membrane under DC field to create an electrokinetic filter, which does not create any hydrodynamic resistance, to concentrate the target molecules close to the sensor. We used the ion depletion feature of the membrane on one side to exclude all charged molecules, such that subsequent application of flow opposite to the depletion front concentrates all the molecules at a precise location in the microfluidic channel. This simple analyte concentration design reduces diffusion time to the sensor and allows for rapid target detection.
Several sensing technologies exist for molecular targets, such as colorimetric, fluorescence-based, and electrochemical detection [1,4,21,25]. However, colorimetric assay generally lacks the necessary detection limit. Optical detection methods provide excellent detection limit down to an impressive single-molecule level, but require costly and bulky optical systems. Several low-cost, label-free molecular sensing technologies like electrochemical sensors which amplify signals using redox reporters can enhance detection sensitivity, yet are hampered by instability of the electrochemical signal and calibration difficulties [31, 32]. Capacitance, conductance and field-effect transistor (FET) electrode sensors are typically insensitive as the ionic strength within the electrical Debye layer is about 2 to 3 times higher than the bulk and the presence of the nucleic acid molecules would not significantly affect the change of local conductance. Additionally, the screening effect of the thin Debye layer would allow the detection of only the smallest charged molecules [33, 34]. Importantly, they all require an expensive micro-fabricated electrode. The largest drawback of all electrode sensors is their long assay time due to diffusion-based transport of large nucleic acid molecules to the electrode surface [35]. Several techniques have been suggested to remove the need for transport. One involves the activation of high voltage at the electrode sensor to electrophoretically attract nearby DNAs [36]. However, this electrophoretic concentration technique is non-specific and the elevated voltage can produce undesirable Faradaic reactions for high-ionic strength buffers resulting in false current or voltage signals. Further, all the existing nucleic acid detection technologies require purified samples, rendering them ineffective for field applications. Recently, we demonstrated the detection of target RNA molecules from different pathogens including dengue virus, brucellosis, and E.coli using a commercially available anion exchange membrane [37-40]. The membrane sensor is highly stable over time and the sensor signal is reproducible [37]. This is due to the absence of any electron transfer reaction on the membrane surface, unlike the spurious side reactions in unstable electrochemical sensors [41]. We used the probe-functionalized anion exchange membrane sensor in this integrated platform for rapid and sensitive detection of target molecules. To successfully demonstrate the performance of our integrated platform, we have chosen oral cancer as our model target. Head and neck squamous cell carcinoma (HNSCC), which includes tumors of the oral cavity, larynx and oropharynx, is the 6th most prevalent cancer worldwide, representing a major global health problem, with >400,000 cases/year and >200,000 deaths annually [42,43]. Thus, development of an integrated platform to detect the upregulated cancer-relevant microRNA is highly desirable. In the future, a fully developed instrument along with the integrated platform described in this manuscript can be used in dental clinics for early diagnosis of oral cancer. As the integrated platform uses the inherent negative charge of the target nucleic acids, this platform can be used for detection of any target nucleic acid provided their sequences are known so the complementary oligoprobes can be employed.
Materials and methods
2. Experimental procedure
2.1. Reagents and chemicals
Commercially-available heterogeneous anion and cation exchange nanomembranes were obtained from Mega a.s., Czech Republic (http://www.mega.cz/hetero genous-ion-exchange-membranes-ralex.html). A two-component silicon RTV system and Quick cast polyurethane resin were purchased from TAP Plastic Inc. (San Leandro, CA, USA). Phosphate Buffer Saline (PBS) 10× solution containing 1.37M sodium chloride, 27mM potassium chloride, 100mM disodium hydrogen phosphate, and 18mM potassium dihydrogen phosphate was purchased from Fisher Scientific (Waltham, MA, USA). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), benzophenone-3,3,4,4-tetracarboxylic acid, and sodium hydroxide were purchased from Sigma Aldrich (St. Louis, MO, USA). TRIzol, fluorophore FAM and amine-coupled oligoprobes were obtained from Life Technologies (Grand Island, NY, USA). RNeasy Mini Kit was obtained from Qiagen (Valencia, CA, USA). The oral cancer cell line OSCC1 engineered to re-express miR-146a or housekeeping microRNAs were prepared as previously described [37] and obtained from Prof. Stack's laboratory at the Harper Cancer Research Institute, University of Notre Dame.
2.2. Attachment of oligoprobe on the sensing surface
The oligoprobe was covalently attached to the anion exchange nanomembrane as previously described [37,44]. Briefly, COOH groups were created on the surface of the anion exchange nanomembrane by adsorbing 10 μl of photoreactive benzophenone-3,3,4,4-tetracarboxylic acid (1 mg in 10 μl water at pH 6-7) and exposing it to UV light for 20 min. Next, the membrane was incubated with 0.4 M EDC in MES buffer for 30 minutes followed by an overnight incubation with 10 μM of the amine-coupled oligoprobe in 0.1× PBS solution to covalently attach the probe to the sensing surface.
2.3. Fabrication of Integrated Chip
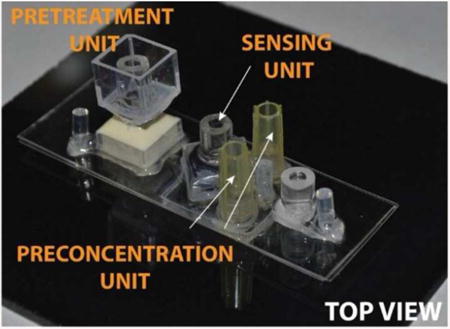
The sample pretreatment, preconcentration and sensing units were integrated on to a single chip with dimensions of approximately 25mm × 75mm. The schematic of the integrated chip with the three units is depicted in Figure 1d and the actual microfluidic chip is shown in Figure 1e.
Figure 1.
Schematics of a (a) pretreatment unit, (b) preconcentration unit (c) flow-through sensing unit, (d) integrated platform with all electrical connections. (e) Top view of an actual integrated device.
The sample pretreatment unit was fabricated by gluing a plastic cuvette to a perforated tube using a UV curing Acrifix 192. The inner tube acts as a reservoir for target sample whereas cuvette acts as a buffer reservoir. Prior to placing the pretreatment unit into the main integrated platform, the interface between the sample reservoir and the main microfluidic channel is plugged with 2% agarose gel. A diagrammatic representation of the sample pretreatment unit is shown in Figure 1a.
The preconcentration unit, shown in Figure 1b, was made by placing two small strips of a cation exchange membrane at designated spots in a silicone mold and embedding in a UV curable Acrifix 192 glue capped with a small PMMA plate. The preconcentration unit was fixed to the main body of the integrated chip with UV curable glues Loctite 3492 and Acrifix 192.
We fabricated the sensor by embedding a small piece of anion exchange membrane in a two-component fast curing polyurethane resin (1:1 ratio) by using a specifically developed molding protocol. Release of the silicone mold yielded a polyurethane-based microfluidic biochip disk with an exposed sensing membrane. Later, a PMMA tube was glued to the biochip disk. This PMMA tube served for connecting electrodes needed to measure signals from the sensor. A depiction of the sensing unit is shown in Figure 1c.
The main body of the integrated chip consisted of three layers of polycarbonate sheets structured by cutting on a plotter Graphtec 7000. The three structured polycarbonate sheets were aligned and thermally laminated together. The top polycarbonate sheet contained openings for the modular designs of the pretreatment and sensing units, the middle sheet for the main microfluidic channel and the bottom sheet cutouts for fixing the preconcentration unit. Tygon tubing as fluidic inlets and outlets and cut pipet tips as buffer reservoirs were glued into their designated places on the integrated chip.
In integrating the units on a single chip, we simplified the system by using one reservoir with a cation exchange membrane for both pretreatment and preconcentration and also placed the cation exchange membranes for preconcentration unit at the bottom of the channel. This allowed us to independently change the position of the sensing and preconcentration unit based on the preliminary experimental results and spatial requirements of each unit. On the other hand, the pretreatment and sensing units were designed in a modular fashion to allow easy replacements of these units in the integrated platform.
2.4. Electrical measurements
The Gamry 500 potentiostat (Gamry Instruments, Warminster, PA, USA) was used to perform the electrical measurement of the nanomembrane sensor in a four-electrode setup. Two reference Ag-AgCl electrodes were used for the measurement of the voltage across the membrane whereas two platinum electrodes were used to apply the current load. The current voltage characteristics (CVC) was obtained by applying current load from 0 to 40 μA at the step rate of 1 μA/s and measuring voltage drop across the membrane. The sensing reservoir (side of the anion exchange membrane functionalized with the oligoprobe) was connected to the ground, whereas a positive potential was applied in the other reservoir. In a typical experiment, a first CVC was measured for the oligoprobe functionalized sensor in a 0.1× PBS solution, which acted as the baseline signal. 10 μl of lysing buffer and 50 μl cancer cell lines were mixed with 50 μl of 1% agarose gel and transferred into the sample loading chamber on top of the 2% agarose gel. 10× TAE buffer (pH 7.4) was used as a sample loading buffer. A constant DC current (800 mV) was applied for 15 minutes that allowed all the negatively charged molecules including microRNA to move to the microfluidic channel. Extracted nucleic acids were concentrated by applying 150 V to the preconcentration unit. The location of the concentrated slug was adjusted to align with the location of the sensing unit by optimizing the electric DC field and flow rate. The concentrated nucleic acid slug was then incubated for 15 minutes to allow for hybridization of target molecules with the probes functionalized on the sensing surface. Non-specifically bound DNA/RNA molecules on membrane surface were washed three times using a high-salt 4× PBS solution. A final CVC was measured after equilibrating the reservoirs in 0.1× PBS solution. Any shift in CVCs in the overlimiting region before and after hybridization indicated the successful detection of the target nucleic acids.
Results and Discussion
The work presented in this paper focuses on the fundamental aspects of developing a standalone platform that can process real biological samples, prepare them for further processing and finally perform specific detection of targeted nucleic acids. The concept of the integrated platform for detection of chosen nucleic acid biomarkers in complex samples has been built on the following requirements: (i) the platform must release the nucleic acid biomarkers from their intracellular environment and separate them from cell debris and other biomolecules that may interfere with the sensing; (ii) the platform must actively preconcentrate and localize the target molecules at the sensor to decrease the time associated with the transport of the molecules to the sensor and increase the limit of detection; and (iii) the platform must realize the actual sensing that is based on hybridization on the ssDNA oligoprobes complementary to the single-stranded targets. The platform is divided into three main functional units (pretreatment, preconcentration and sensing) which provide the three different functions described above. The entire platform is based on the use of commercially available heterogeneous ion exchange membranes. The heterogeneous ion exchange membranes are used for a number of different purposes, namely as molecular filters, molecular preconcentrators and even sensors. These membranes in conjunction with microfluidics and external DC voltage exhibit phenomena stemming from their unique property of ion selectivity that offer unprecedented possibilities in both control of the transport and manipulation of nucleic acids and their specific detection. The other important aspect of the platform is its ability to exploit the omnipresent negative charge of the nucleic acids. This negative charge is used to transport the nucleic acid in the DC electrical field and also to sense the presence of the target nucleic acids after hybridizing to the complementary probes attached on the sensor. In the next section we describe each unit of the platform in detail with the emphasis on the role of ion exchange membranes and eventually show the sensing of a miRNA target from oral cancer cells using the fully integrated platform.
Pretreatment unit
The operation of the pretreatment unit is based on chemical lysis of biological samples and allows separation of negatively charged nucleic acids while leaving behind large cellular debris, uncharged molecules and positively charged molecules. The separation is based on electrophoretic extraction from the lysate through an agarose gel layer and it significantly purifies the actual sample for downstream processing. The schematics of the pretreatment unit with its major components is depicted in Figure 1a, and the actual chip in shown in Figure 2a. The unit consists of three major fluidic components: (i) a sample reservoir for introduction of biological samples mixed with chemical lysis buffers, (ii) a counter reservoir with a cation exchange membrane and (iii) a fluidic channel connecting the two reservoirs. The process of electrophoretic separation of nucleic acids is based on the movement of the molecules towards the positively biased electrode through a layer of agarose gel. The agarose gel separates the sample reservoir from the main channel and thus prevents any convective mixing of the fluids in those spaces. Additionally, the gel blocks any molecules or cell debris larger than the pore size of the gel from entering the main channel. Typically, the pore size of 1% agarose gel used for our study was around 500nm [45, 46]. While all positively charged molecules will migrate towards the cathode (away from the microfluidic channel) and neutral molecules will remain in the sample reservoir this technique offers an easy way to purify and separate nucleic acids from other molecules that can interfere with the sensing. The cation exchange membrane in the counter reservoir acts as a nucleic acid filter that prevents the nucleic acid from escaping the main channel and migrating to the positive electrode. This molecular filtering is given by synergistic effects of very strong electrostatic repulsion that occurs between the negatively charged cation exchange membrane and the negative nucleic acids and steric restrictions of the membrane nanopores that simply do not allow these relatively large molecules to penetrate. The separated nucleic acid molecules thus accumulate underneath the cation exchange membrane.
Figure 2.
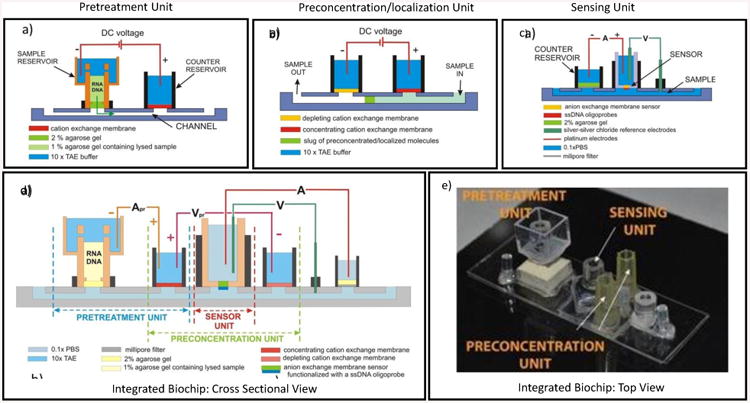
(a) Picture of the actual pretreatment unit. (b) Separation of a fluorescently labeled ssDNA from an oral cancer cell lysate,
The processing starts with pipetting a thin layer of 2% agarose gel at the bottom of the sample reservoir. Next, the biological sample is lysed with a chemical agent and the lysate is mixed with a warm 2% agarose gel yielding lysate dispersed in the agarose gel at final concentration of 1%. This mixture is quickly pipetted on top of the 2% agarose layer and refrigerated for 5-10 minutes to solidify the gel. This combination of two gels creates a layer that is mechanically stable enough to withstand the DC electrical field applied during electrophoretic extraction. The reservoirs are then filled with 10× TAE buffer that keeps the pH in the electrode compartments constant. We apply a constant DC electrical current and limit the voltage imposed on the system thus preventing the system from overheating due to Joule heating. This electrophoretic separation is performed in approximately 15 minutes.
Figure 2b shows our results from our separation experiment. In this case, the sample of lysed oral cancer cells was spiked with fluorescently labeled short ssDNA mimicking the real miRNA molecules in the sample so that we could observe the separation in real time. At the beginning of the experiment all the fluorescence comes from the sample reservoir. After applying the DC voltage, the fluorescently labeled ssDNA starts moving across the gel layer into the channel where it accumulates at the cation exchange membrane. The fluorescence in the sample reservoir slowly disappears over time whereas it increases at the membrane. The separation is completed in about 15 minutes and the extracted nucleic acid molecules are ready for further processing. Currently, we are performing studies aimed at analyzing the contents of the extracted nucleic acids using the basic tools of molecular biology.
Preconcentration/localization unit
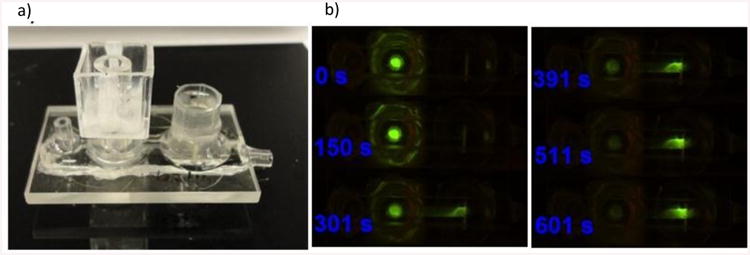
The main use of this unit is to concentrate the previously extracted nucleic acid molecules and localize them at a specific location in the channel. The preconcentration unit is based on balancing electrophoretic and convective transport of separated molecules in order to localize and concentrate the nucleic acid molecules at the position of the sensor. While the cation exchange membrane in the pretreatment unit acts as a “mechanical” filter of nucleic acids (the molecules cannot penetrate the membrane), the cation exchange membranes in the preconcentration unit develop a condition in the channel that works as a non-mechanical ionic plug. This ionic plug, depending on the conditions applied, desalts the solution pumped through the channel. The ionic plug is based on the effect of concentration polarization that takes place around the cation exchange membrane when DC voltage is applied. This concentration polarization creates zones in the electrolyte adjacent to the membrane in which the concentration of ions is either lower or higher than that of the electrolyte bulk. These zones are called depletion and enrichment zones, respectively. In our unit, we use two cation exchange membranes that are placed on top of the microfluidic channel and separate the electrode compartments from the main channel as shown in Figure 1b. When a DC voltage is applied on the system, the two membranes connected in series produce localized depletion and enrichment zones in the main channel simultaneously. The formation of the depletion and enrichment zones is reflected in the voltage drops that occur in different parts of the channel and thus in the local electrical field. Figure 3a depicts experimental voltage profile established between the two membranes at the applied voltage of 100 V and the flow rate of 5 μl/min. The voltage profile clearly shows two regions with different voltage drops, a large one closer to the depleting membrane and a smaller one at the concentrating membrane. This is reflected in the electrical field profile (Figure 3b) that was numerically calculated from the voltage profile. The electrical field is strongest under the depleting membrane and weakest under the concentrating membrane. The difference between the strongest and the weakest electrical field can be up to two orders of magnitude. The nonlinear profile of the electrical field results in a spatially dependent electrical force that acts upon charged molecules. In case of nucleic acids, this force acts against the direction of the electrical field (towards the positively biased electrode). By applying a convective flow through the channel, we can counterbalance the electrical force acting on the nucleic acids and localize them at a position along the channel where the resultant force on nucleic acids is essentially zero. By tuning the voltage and flow rate we can adjust the final position of the preconcentration slug. While we demonstrate that the flow rate and the voltage are major parameters affecting the position of the slug in the channel, other electrokinetic phenomena, like vortex instabilities, can also contribute to the overall behavior of the system.
Figure 3.
(a)Experimentally obtained voltage profile that develops between the depleting and concentrating membrane at the applied voltage of 100 V and the flow rate of 5 μl/min and b) the corresponding electric field profile (absolute values) numerically calculated from the voltage profile.
Figure 4a shows dynamics of the development of the preconcentration slug. In this case fluorescently labeled ssDNA diluted in 0.01× PBS is pumped through the channel while a voltage of 150V is applied simultaneously. The preconcentration slug develops immediately after starting the experiment, settling into its final position which does not change in time. However, the fluorescent intensity of the slug increases over time as more and more nucleic acid molecules accumulate at the region.
Figure 4.
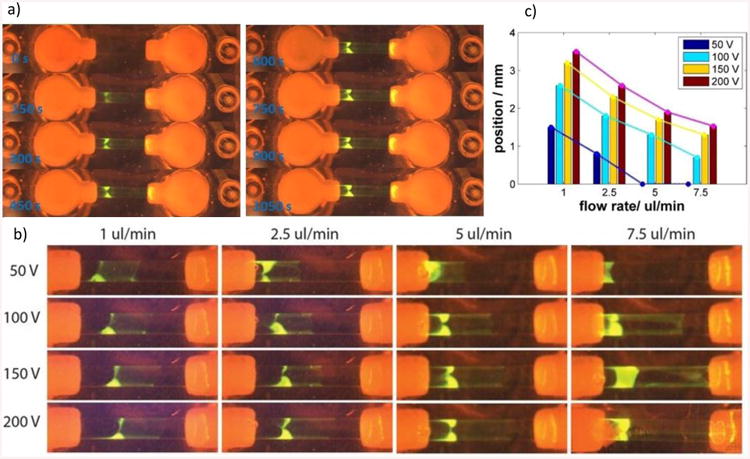
a) Dynamics of the preconcentration of a fluorescently labeled ssDNA, (b) Experimental study showing the effect of the voltage and the flow rate on the position of the preconcentration slug, (c) Graph showing the positions of the preconcentration slug related to the flow rate and voltage applied
We studied the effect of the voltage and the flow rate on the position of the preconcentration slug. The results are depicted in Figure 4b in which a set of images shows the fluorescence of the preconcentration slug obtained under different conditions in experiments that were run for 10 minutes. Each row shows the position of the slug at constant voltage but varying flow rate and each column at constant flow rate and varying voltage. The set of images clearly shows the effect of both parameters. While increasing voltage pushes the slug further away from the depleting membrane, the increasing flow rate acts in the opposite direction. Figure 4c shows the positions of the preconcentration slugs that have been evaluated as a distance of the front edge of the slug from the depleting membrane in the center of the channel. The trend of all curves confirms our hypotheses that the flow rate and electric field influence the location of the concentrated slug. This study also revealed the limits of the parameters. In case of low voltages and large flow rates, the electrical field is not strong enough to keep the molecules in the intermembrane region of the channel and the molecules are convected out of the channel (see Figure 4c for 50 V and flow rate of 5 and 7.5 μl/min).
Sensing unit
The sensing of nucleic acids using an anion exchange membrane is based on a phenomenon denoted as surface charge inversion as described earlier [36-40]. The anion exchange membrane is comprised of molecules that bear a fixed positive charge at a very high concentration. This fixed charge affects the transport of ions through the membrane that in turn gives rise to peculiar phenomena such as concentration polarization, electro-convection and water splitting. These behaviors are reflected in the non-linear current voltage characteristics of the membrane. When an oppositely charged, large poly-ion (e.g. DNA, RNA) is added on the appropriate side of the membrane, the polyanions tend to adsorb on the membrane by strong electrostatic charge-charge interactions. This process creates a thin layer of immobile opposite charge on the membrane that is reflected in the measurement of current voltage characteristics as a characteristic shift. By functionalizing the membrane with specific ssDNA oligoprobes complementary to the detected targets, the original nonspecific signal coming from any adsorbed polyanions is turned into specific detection of target nucleic acids hybridized on the complementary probes. The specific detection requires use of optimized washing buffers to remove all non-specifically bound molecules. The chip consists of a single channel, a counter reservoir, the actual sensor and a fitting for inserting a reference electrode (Figure 5a). As described earlier, the sensor is made by embedding a small piece of anion exchange membrane into a polyurethane resin and attaching it to a short piece of a plexiglass tubing. First, the probe is functionalized to the sensor as described earlier. The measurement of the current voltage characteristics is done in a four electrode setup. Two platinum source electrodes connect a given DC electrical signal to the membrane (A in Figure 1c) and two silver-silver chloride electrodes measure the voltage response of the system (V in Figure 1c). Figure 5b shows a representative result of a specific detection obtained in our flow-through sensor, in this case the target is ssDNA sequence corresponding to miRNA 146a. An artificial ssDNA nonspecific sequence was used as a negative control. The red curve in the figure represents the signal of the ssDNA oligoprobe covalently linked to the membrane. The black curve represents the signal of the non-target which clearly indicates the sensors specificity, as no shift was detected. Unlike the non-target, the signal for the target represented by the blue curve in Figure 5b provided a significant deviation from the baseline. The signal can be quantified by evaluating the voltage shift between the baseline and the signal for the target that occurs at the highest current applied to the system. For the results presented in Figure 5b this shift is approximately 0.8V (measured at 25 μA). This demonstrates our ability to selectively detect short single-stranded nucleic acids using changes in current-voltage characteristics. Changes in voltage shift can be used to not only gather information about the selectivity of the sensor but also the sensitivity of the sensor by measuring the amount of shift. An extensive study on the selectivity and sensitivity of the sensing unit was demonstrated previously by our group [37].
Figure 5.
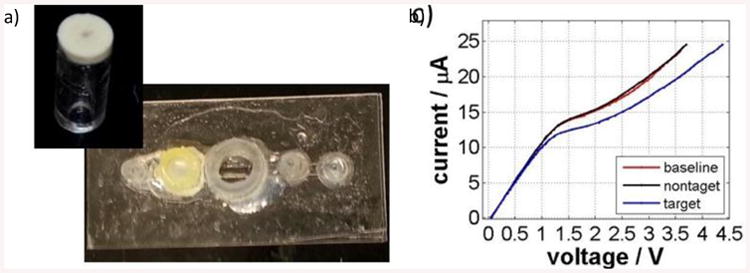
(a) Picture of an actual flow-through sensing unit and (b) Representative experiment showing specific detection of a DNA with a sequence corresponding to miRNA146a.
Integrated platform
Figure 6 shows some of the results obtained with the integrated chip. Because the platform aims to perform detection in a PCR-free manner, this limits the use of the platform for single stranded nucleic acids, e. g. microRNA, rRNA, ssDNA (in some viruses). MicroRNAs are a convenient target since they are short (20-24 nucleotides long) and are more stable than mRNAs. At the same time, miRNA profiles are being considered as novel biomarkers carrying a diagnostic value, e.g. for cancer. Here we present our results for detection of miRNA 146a in samples containing oral cancer cells OSCC1 in 30 minutes. The protocol for all experiments was as follows. Roughly 500,000 cancer cells of OSSC1 cell line were chemically lysed and the lysate was embedded into a 1% agarose gel in the pretreatment unit that was previously capped with a 2% agarose gel plug. This pretreatment unit and a freshly functionalized sensor were inserted in the appropriate fittings on the integrated chip and the chip was filled with 0.1× PBS and 10× TAE in appropriate reservoirs. All tubing and electrode connections were then established. The baseline corresponding to the probe functionalized on the membrane is measured. Then 800 mA are applied on the pretreatment unit (Apr in Figure 1d) and this current is connected for 15 minutes to separate targeted miRNA from the lysate. The target is then allowed to hybridize with the probes functionalized on the sensor for 5 minutes. The channel is then washed with 4× PBS buffer to remove nonspecifically bound molecules from the membrane and the channel is again filled with 0.1× PBS to equilibrate the sensor. The current voltage curve was measured again and compared to the baseline.
Figure 6.
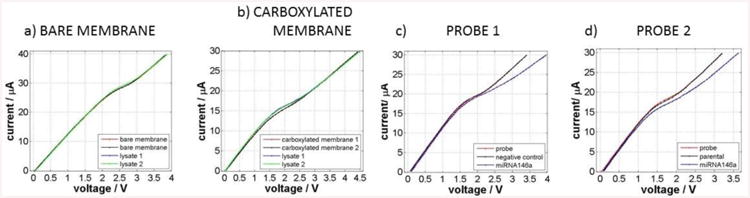
(a) Control experiment performed with a sensor that was not functionalized with a probe, (b) Control experiment performed with a sensor that was carboxylated but not functionalized with a probe, (c) Specific detection of ssDNA from clean samples, and (d) Detection of miRNA146a from oral cancer cell lysate.
Figure 6a shows the results for a system where a bare membrane without probe (negative control) was used as a sensor. As expected, no change in CVC was observed after performing the experiment thus clearly indicating no detection of target. This result implies that the nonspecific binding on the membrane can be removed with our washing protocol and that the membrane itself in not affected by the preceding processes done on the chip. Figure 6b shows the results for a system where the membrane creating the sensor was functionalized with carboxyl groups used for attachment of the ssDNA oligoprobe but no probe was attached. Similar to Figure 6a, when the target molecules were flowed through this setup followed by a washing step, no signal was detected indicating sensor selectivity. The results in Figure 6c show the detection of the actual target. We used solution of ssDNA complementary to the probe functionalized on the membrane as a target and a short ssDNA non-complementary as a negative control both at a concentration of 1 μM. In this case, the negative control experiment was run first. The CVCs in this graph show that the sensor specifically detected the target DNA sequence (shift of 0.7 V obtained at 30 μA) while no shift was observed when non-target was run through the system. Finally, the plot in Figure 6d shows the results for actual cell samples from the oral cancer cell line. We used the parental oral cancer cell line as a negative control and a genetically modified line that overexpresses miRNA 146a as a positive control. Again the experiments with the parental cell line were performed first and we did not detect any signal as demonstrated by no shift in voltage. However, the derived oral cancer cell line overexpressing the microRNA of interest provided a signal amounting to about 0.5 V at 30 μA thus showcasing our ability to selectively extract and detect microRNA biomarkers using our integrated platform. Successful discrimination of two oral cancer cell lines based on the presence of miRNA 146a provides a proof-of-concept data for further development of the detection platform.
Conclusion
We have successfully developed an integrated, sample-to-answer diagnostic platform for rapid detection of microRNA biomarkers from cancer cell lines in approximately 30 minutes. We used low cost polymeric materials to design a disposable integrated chip that can be easily manufactured. The integrated platform consisted of three units: a pretreatment unit to isolate nucleic acids from lysed cell sample, a preconcentration unit to concentrate the isolated nucleic acid molecules at a precise location where the sensing unit is placed to detect target nucleic acids. The ion-selective properties of the cation- and anion-exchange membranes and the negative charge of the nucleic acids were used to manipulate, concentrate and detect target nucleic acid biomarkers. The sample pretreatment is designed based on the principle of gel electrophoresis where an electric field is used to extract the nucleic acid molecules from the sample chamber through an agarose gel. The sensing is based on the charge inversion phenomenon when negatively charged target nucleic acids hybridize with oligoprobes attached on the surface of the positively charged membrane. Changes in current-voltage characteristics are used to selectively identify targets microRNA 146a biomarkers. As the sensor uses the inherent negative charge for detection, this platform can be used to detect any pathogen nucleic acids not only in medical diagnostics but also in food and environmental diagnostics provided probes of interest are known. This low-cost, integrated platform has the potential to reach low resource settings and conduct testing at the location of an outbreak, at dental clinics or at the point-of-care application thus significantly improving safety and reducing disease outbreaks.
Highlights.
-Integrated platform for sample-to-answer nucleic acid detection
-Sample pretreatment by electrophoretic separation
-Concentration of target nucleic acids using cation exchange membrane
-Specific detection of nucleic acids using anion exchange membrane
-Testing of microRNA (miRNA 146a) biomarker associated with oral cancer
Acknowledgments
This work has been supported by the Walther Cancer Foundation Grant 0120, NIH 1R21AI105361-01A1, USDA 2012-67005-19589, NSF-CBET 1065652 and NIH RO1CA085870 (M.S.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yager P, Domingo GJ, Gerdes J. Annu Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 2.Jones G, Steketee RW, Black RE, Bhutta ZA, Morris SS. Lancet. 2003;362:65–71. doi: 10.1016/S0140-6736(03)13811-1. [DOI] [PubMed] [Google Scholar]
- 3.Bissonnette L, Bergeron MG. Clin Microbiol Infect. 2010;16:1044–1053. doi: 10.1111/j.1469-0691.2010.03282.x. [DOI] [PubMed] [Google Scholar]
- 4.Su W, Gao X, Jiang L, Qin J. J Chromatogr A. 2015;1377:13–26. doi: 10.1016/j.chroma.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 5.Hu J, Wang S, Wang L, Li F, Belinda PM, Lu TJ, Xu F. Biosens Bioelectron. 2014;54:585–597. doi: 10.1016/j.bios.2013.10.075. [DOI] [PubMed] [Google Scholar]
- 6.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Anal Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 7.Hart RW, Mauk MG, Liu C, Qiu X, Thompson JA, Chen D, Malamud D, Abrams WR, Bau HH. Oral Dis. 2011;17:745–752. doi: 10.1111/j.1601-0825.2011.01808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Mauk M, Qiu X, Liu C, Kim J, Ramprasad S, Ongagna S, Abrams WR, Malamud D, Corstjens PL. Biomed Microdevices. 2010;12:705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson BS, Buchsbaum SF, Wu TT, Hsieh K, Xiao Y, Sun R, Soh HT. J Am Chem Soc. 2011;133:9129–9135. doi: 10.1021/ja203981w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul L. Lab Chip. 2009;9:768–776. [Google Scholar]
- 11.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feld-man SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP. Proc Natl Acad Sci. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaigala GV, Huskins RJ, Preiksaitis J, Pang XL, Pilarski LM, Backhouse CJ. Electrophoresis. 2006;27:3753–3763. doi: 10.1002/elps.200600061. [DOI] [PubMed] [Google Scholar]
- 13.Lee YF, Lien KY, Lei HY, Lee GB. Biosens Bioelectron. 2009;25:745–752. doi: 10.1016/j.bios.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee BS, Lee YU, Kim HS, Kim TH, Park J, Lee JG, Kim J, Kim H, Lee WG, Cho YK. Lab Chip. 2011;11:70–78. doi: 10.1039/c0lc00205d. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Weng X, Li D. Microfluid Nanofluid. 2011;10:941–964. [Google Scholar]
- 16.Lafleur L, Stevens D, McKenzie K, Ramachandran S, Spicar-Mihalic P, Singhal M, Arjyal A, Osborn J, Kauffman P, Yager P. Lab Chip. 2012;12:1119–1127. doi: 10.1039/c2lc20751f. [DOI] [PubMed] [Google Scholar]
- 17.Guo MT, Rotem A, Heyman JA, Weitz DA. Lab Chip. 2012;12:2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 18.Harshman DK, Reyes R, Park TS, You DJ, Song JY, Yoon JY. Biosens Bioelectron. 2014;53:167–174. doi: 10.1016/j.bios.2013.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho YK, Lee JG, Park JM, Lee BS, Lee Y, Ko C. Lab Chip. 2007;7:565–573. doi: 10.1039/b616115d. [DOI] [PubMed] [Google Scholar]
- 20.Lutz S, Weber P, Focke M, Faltin B, Hoffmann J, Muller C, Mark D, Roth G, Munday P, Armes N. Lab Chip. 2010;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- 21.Yager P, Domingo GJ, Gerdes J. Annu Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 22.Gervais L, Delamarche E. Lab Chip. 2009;9:3330–3337. doi: 10.1039/b906523g. [DOI] [PubMed] [Google Scholar]
- 23.Park TS, Li W, McCracken KE, Yoon JY. Lab Chip. 2013;13:4832–4840. doi: 10.1039/c3lc50976a. [DOI] [PubMed] [Google Scholar]
- 24.Sauer-Budge AF, Mirer P, Chatterjee A, Klapperich CM, Chargina D, Sharon A. Lab Chip. 2009;9:2803–2810. doi: 10.1039/b904854e. [DOI] [PubMed] [Google Scholar]
- 25.Mark D, Haeberle S, Roth G, Stetten Fv, Zengerle R. Chem Soc Rev. 2010;39:1153–1182. doi: 10.1039/b820557b. [DOI] [PubMed] [Google Scholar]
- 26.Chin CD, Chin SY, Laksanasopin T, Sia SK. Biol Med Phys Biomed. 2013:3–21. [Google Scholar]
- 27.Yang W, Yu M, Sun X, Woolley AT. Lab Chip. 2010;10:2527–2533. doi: 10.1039/c005288d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You DJ, Geshell KJ, Yoon Jy. Biosens Bioelectron. 2011;28:399–406. doi: 10.1016/j.bios.2011.07.055. [DOI] [PubMed] [Google Scholar]
- 29.Chin CD, Linder V, Sia SK. Lab Chip. 2012;12:2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 30.Wang JH, Wang CH, Lee GB. Ann Biomed Eng. 2012;40:1367–1383. doi: 10.1007/s10439-011-0473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakker E, Qin Y. Anal Chem. 2006;78:3965–3984. doi: 10.1021/ac060637m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umezawa Y, Aoki H. Anal Chem. 2004;76:321–326. [PubMed] [Google Scholar]
- 33.Stern E, Wagner R, Sigworth FJ, Breaker R, Fahmy TM, Reed MA. Nano Lett. 2007;7:3405–3409. doi: 10.1021/nl071792z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suni II. TrAC-Trend Anal Chem. 2008;27:604–611. [Google Scholar]
- 35.Nkodo AE, Garnier JM, Tinland B, Ren H, Desruisseaux C, McCormick LC, Drouin G, Slater GW. Electrophoresis. 2001;22:2424–2432. doi: 10.1002/1522-2683(200107)22:12<2424::AID-ELPS2424>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 36.Sosnowski R, Tu E, Butler WF, O'Conell JP, Heller MJ. Proc Natl Acad Sci USA. 1997;94:1119–1123. doi: 10.1073/pnas.94.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senapati S, Slouka Z, Shah S, Behura S, Stack S, Severson D, Chang HC. Biosens Bioelectron. 2014;60:92–100. doi: 10.1016/j.bios.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slouka Z, Senapati S, Chang HC. Ann Rev Anal Chem. 2014;7:317–335. doi: 10.1146/annurev-anchem-071213-020155. [DOI] [PubMed] [Google Scholar]
- 39.Slouka Z, Senapati S, Yan Y, Chang HC. Langmuir. 2013;29:8275–8283. doi: 10.1021/la4007179. [DOI] [PubMed] [Google Scholar]
- 40.Senapati S, Basuray S, Slouka Z, Cheng LJ, Chang HC. Tor Curr Chem. 2011:1–17. doi: 10.1007/128_2011_142. [DOI] [PubMed] [Google Scholar]
- 41.Bogomolova A, Komarova E, Reber K, Gerasimov T, Yavuz O, Bhatt S, Aldissi M. Anal Chem. 2009;81:3944–3949. doi: 10.1021/ac9002358. [DOI] [PubMed] [Google Scholar]
- 42.Miller DL, Puricelli MD, Stack MS. Biochem J. 2012;443:339–353. doi: 10.1042/BJ20112017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duvvuri U, Myers JN. Curr Probl Surg. 2009;46:114–117. doi: 10.1067/j.cpsurg.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Berens C, Courtoy PJ, Sonveaux E. Bioconjugate Chem. 1999;10:56–61. doi: 10.1021/bc980065f. [DOI] [PubMed] [Google Scholar]
- 45.Pernodet N, Maaloum M, Tinland B. Electrophoresis. 1997;18:55–58. doi: 10.1002/elps.1150180111. [DOI] [PubMed] [Google Scholar]
- 46.Narayanan J, Xiong JY, Liu XY. J Phys: Conf Series. 2006;28:83–86. [Google Scholar]


