Abstract
The ABC gene family is recognized as one of the largest gene families in all kingdoms of life. Although many genes involved in the ABC superfamily have been annotated from several fish species, information on large sets of the ABC superfamily and their evolutionary characterization are still unclear. In the marine medaka Oryzias melastigma, 50 ABC transporters were identified with bioinformatics-aided in silico analyses, and their full-length cDNA sequences were characterized. Phylogenetic analysis revealed that they could be classified into the eight subfamilies (A–H) that include all members of all ABC subfamilies. Interestingly, several teleosts’ Abcg members were closely clustered with Abch members in a distinctive clade. The abch gene was also observed in the coelacanth and the spotted gar, suggesting that this gene was retained from a bilaterian ancestor and that a gene loss event recently occurred in the tetrapod lineage. In teleosts, the nomenclature of previously annotated abcg genes should be considered carefully, as they form a distinctive clade with the marine medaka abch subfamily and other teleost abch genes, but not with the members of the Abcg subfamily.
The ATP-binding cassette (ABC) transporters are active efflux pumps that use an ATP cleavage-driven energy source for transporting endogenous and exogenous substrates (e.g., amino acids, peptides, vitamins, sugars, lipids, sterols, hormones, endogenous metabolites, inorganic anions, drugs, and metal ions) from the cytosol to the intracellular and/or extracellular region1,2. In most ABC proteins, two highly conserved structural domains, nucleotide binding domain (NBD) and/or transmembrane domain (TMD), are included in a single polypeptide possessing different numbers of each domain. Full transporters are comprised of two NBDs and two TMDs, but half transporters require homo- or heterodimerization to form a functional unit as they contain only one of each type of domain (i.e. NBD and TMD). Specifically, the NBDs have a responsibility to bind and hydrolyze ATP for substrate translocation across biological membranes, while the TMDs provide substrate specificity with five to six membrane-spanning helices3,4. Based on sequence similarity and phylogenetic analysis using the NBDs, the ABC transporter superfamily has been divided into eight subfamilies, A to H. Of them, the Abch subfamily was first identified in the Drosophila melanogaster genome and is present in sea urchin and in all of the sequenced arthropod genomes but not in fungi, plants, and mammals5,6,7. Interestingly, in teleosts, the abch gene appears only in the zebrafish (Danio rerio) and the green spotted pufferfish (Tetraodon nigroviridis)8 while no member of the Abch subfamily has been identified in other teleosts.
In teleosts, the tissue distribution and molecular and biochemical functions of ABC transporters were extensively reviewed, with a focus on multidrug and/or multixenobiotic resistance (MDR/MXR) and physiological properties8,9. Also, the availability of genome information for several teleosts enables us to examine the comparative evolutionary distance of the entire set of ABC transporter genes10. Two successive whole genome duplication (WGD) events and the additional teleost-specific genome duplication (TGD) event may have important roles in the diversification of large gene superfamilies including the ABC transporter gene families11,12. As a result, comparative evolutionary relationships of the entire ABC superfamily have been intensively examined from tetrapods to teleosts (e.g. Danio rerio, Gadus morhua, Gasterosteus aculeatus, Ictalurus punctatus, Latimeria chalumnae, Oreochromis niloticus, Oryzias latipes, Takifugu rubripes, and Tetraodon nigroviridis)8,10.
The marine medaka Oryzias melastigma (also known as Indian medaka or brackish medaka) is phylogentically close to the freshwater counterpart Oryzias latipes as a sister species13. In detail, the salinity-tolerant O. melastigma and O. javanicus, showing high adaptability in response to acute changes of the ambient osmotic pressure from freshwater to seawater, have been clustered into the same clade on phylogenetic analysis13,14. The marine medaka has increasingly been recognized as a potential model for marine environmental research, as fertilization, sperm activity, and hatchability of O. melastigma are feasible in freshwater and in seawater14. The marine medaka has several promising advantages including its small size (~3–4 cm), daily spawning, short generation time (less than 3–4 months), sexual dimorphism, responsiveness to diverse chemicals, and ease of maintenance and breeding in the laboratory15. Also, most of the standard experimental procedures, establishing in model fishes such as zebrafish and Japanese medaka (e.g. developmental biology using transparent embryos, semen cryopreservation and in vitro fertilization, cell transplantation, microinjection with DNA or RNA, primary cell culture, mutant screening, in situ hybridization, immunohistochemistry, and stable fluorescent transgenic lines), can be applied with slight modifications in marine medaka. To establish the marine medaka as a model animal, we obtained draft genome information with Next Generation Sequencing (NGS) technologies (unpublished data), and the information was successfully employed for the evolutionary comparison of teleosts’ gene superfamilies such as the cytochrome P450 genes16. Subsequently, we tried to identify the marine medaka abch gene, which was not annotated in the genome database of Japanese medaka, although the whole genome information of O. latipes is already available17. Interestingly, one of identified ABC members showed high similarity with the abch genes of zebrafish and green spotted pufferfish in our preliminary BLASTX search. Thus, our next question has been raised on the controversial suggestions of absence or presence of abch gene in teleosts.
In this study, we characterized 50 putative ABC transporters in O. melastigma and identified one putative abch gene. In addition, a novel phylogenetic relationship between the Abcg and Abch subfamilies in fish was uncovered and is discussed to clarify the controversial annotation of the teleost Abch family.
Results and Discussion
Identification and annotation of O. melastigma ABC transporters
In this study, all NBD-containing reads were examined by BLASTX searches to the non-redundant (NR) database of NCBI, and subsequently, 50 ABC transporters were identified in the genome information of O. melastigma. Full-length sequences of all O. melastigma ABC genes were characterized and registered in the GenBank database (Table 1) after annotation of each gene using in silico analysis (i.e. BLAST search, amino acid similarity and identity comparison, and domain search). The length of these full-length ABC proteins ranged from 611 amino acids (aa) (Abcf2) to 4,758 aa (Abca12). Although most ABC genes were annotated in O. latipes in the GenBank database, in-depth nomenclature and phylogenetic analysis had not been conducted in the Genus Oryzias as yet. Thus, this is the first report on the characterization of the entire ABC superfamily in the Genus Oryzias. In this paper, description and previous relevant findings on each ABC subfamily are mostly omitted as these topics were discussed thoroughly in several recent valuable reviews on fish ABC transporters8,9,10.
Table 1. 50 ABC transporters identified in the marine medaka O. melastigma genome.
| Gene | Size (AA) | Topology | Accession No. | Matched gene | Matched species | E-value |
|---|---|---|---|---|---|---|
| abca1-1 | 2295 | TM-NBD-TM-ABC | KP725006 | abca1 (XP_004086583) | Oryzias latipes | 0.0 |
| abca1-2 | 2271 | TM-NBD-TM-ABC | KP725007 | abca1 (XP_004066316) | Oryzias latipes | 0.0 |
| abca1-like | 2073 | TM-NBD-TM-ABC | KP725008 | abca1 (XP_004554555) | Maylandia zebra | 0.0 |
| abca2 | 2514 | TM-NBD-TM-ABC | KP725010 | abca2 (XP_005447927) | Oreochromis niloticus | 0.0 |
| abca3 | 1707 | TM-NBD-TM-ABC | KP725009 | abca3 (XP_004071707) | Oryzias latipes | 0.0 |
| abca4-1 | 2275 | TM-NBD-TM-ABC | KP725013 | abca4 (XP_004080989) | Oryzias latipes | 0.0 |
| abca4-2 | 2312 | TM-NBD-TM-ABC | KP725014 | abca4 (XP_005476689) | Oreochromis niloticus | 0.0 |
| abca5 | 1687 | TM-NBD-TM-ABC | KP725005 | abca5 (XP_004080721) | Oryzias latipes | 0.0 |
| abca7 | 2353 | TM-NBD-TM-ABC | KP725011 | abca1 (XP_004068024) | Oryzias latipes | 0.0 |
| abca12 | 4758 | TM-NBD-TM-ABC | KP725012 | abca12 (XP_003445395) | Oreochromis niloticus | 0.0 |
| abcb2 | 732 | TM-NBD | KP725027 | abcb2 (XP_004078551) | Oryzias latipes | 0.0 |
| abcb3 | 713 | TM-NBD | KP725050 | abcb3 (XP_004080734) | Oryzias latipes | 0.0 |
| abcb4 | 1290 | TM-NBD-TM-ABC | KP725016 | abcb1 (ADQ20481) | Poeciliopsis lucida | 0.0 |
| abcb6 | 850 | TM-NBD | KP725020 | abcb6 (XP_004066768) | Oryzias latipes | 0.0 |
| abcb7 | 746 | TM-NBD | KP725018 | abcb7 (XP_004073395) | Oryzias latipes | 0.0 |
| abcb8 | 716 | TM-NBD | KP725026 | abcb8 (XP_004079446) | Oryzias latipes | 0.0 |
| abcb9 | 806 | TM-NBD | KP725040 | abcb9 (XP_005806663) | Xiphophorus maculatus | 0.0 |
| abcb10 | 701 | TM-NBD | KP725031 | abcb10 (XP_004067419) | Oryzias latipes | 0.0 |
| abcb11a | 1364 | TM-NBD-TM-ABC | KP725033 | abcb11 (XP_004066603) | Oryzias latipes | 0.0 |
| abcb11b | 1306 | TM-NBD-TM-ABC | KP725019 | abcb11 (XP_004081955) | Oryzias latipes | 0.0 |
| abcc1 | 1511 | TM-NBD-TM-ABC | KP725025 | abcc1 (XP_004573179) | Maylandia zebra | 0.0 |
| abcc2 | 1567 | TM-NBD-TM-ABC | KP725035 | abcc2 (XP_004077201) | Oryzias latipes | 0.0 |
| abcc3 | 1543 | TM-NBD-TM-ABC | KP725036 | abcc3 (XP_004086831) | Oryzias latipes | 0.0 |
| abcc4-1 | 1324 | TM-NBD-TM-ABC | KP725032 | abcc4 (XP_005744288) | Pundamilia nyererei | 0.0 |
| abcc4-2 | 1329 | TM-NBD-TM-ABC | KP725029 | abcc4 (XP_005945197) | Haplochromis burtoni | 0.0 |
| abcc5a | 1383 | TM-NBD-TM-ABC | KP725030 | abcc5 (XP_005751691) | Pundamilia nyererei | 0.0 |
| abcc5b | 1384 | TM-NBD-TM-ABC | KP725039 | abcc5 (XP_004073112) | Oryzias latipes | 0.0 |
| abcc6 | 1510 | TM-NBD-TM-ABC | KP725034 | abcc6 (XP_004066300) | Oryzias latipes | 0.0 |
| abcc7 | 1502 | TM-NBD-TM-ABC | KP725028 | abcc7 (AFV39711) | Oryzias dancena | 0.0 |
| abcc8 | 1587 | TM-NBD-TM-ABC | KP725041 | abcc8 (XP_005803356) | Xiphophorus maculatus | 0.0 |
| abcc9 | 1568 | TM-NBD-TM-ABC | KP725042 | abcc9 (XP_004083272) | Oryzias latipes | 0.0 |
| abcc10 | 1535 | TM-NBD-TM-ABC | KP725043 | abcc7 (XP_004065626) | Oryzias latipes | 0.0 |
| abcc12 | 1368 | TM-NBD-TM-ABC | KP725038 | abcc9 (XP_004067574) | Oryzias latipes | 0.0 |
| abcd1 | 772 | TM-NBD | KP725046 | abcd1 (XP_004086440) | Oryzias latipes | 0.0 |
| abcd2 | 734 | TM-NBD | KP725048 | abcd2 (XP_004082884) | Oryzias latipes | 0.0 |
| abcd3 | 657 | TM-NBD | KP725044 | abcd3 (XP_004080884) | Oryzias latipes | 0.0 |
| abcd4 | 616 | TM-NBD | KP725045 | abcd4 (XP_004083921) | Oryzias latipes | 0.0 |
| abce1 | 599 | NBD-NBD | KP725037 | abce1 (XP_004079653) | Oryzias latipes | 0.0 |
| abcf1 | 819 | NBD-NBD | KP725049 | abcf1 (XP_004074386) | Oryzias latipes | 0.0 |
| abcf2 | 611 | NBD-NBD | KP725051 | abcf2 (XP_004081198) | Oryzias latipes | 0.0 |
| abcf3 | 711 | NBD-NBD | KP725047 | abcf3 (XP_003457877) | Oreochromis niloticus | 0.0 |
| abcg1 | 670 | NBD-TM | KP725015 | abcg1 (XP_004086643) | Oryzias latipes | 0.0 |
| abcg2-1 | 652 | NBD-TM | KP725017 | abcg2 (XP_004079651) | Oryzias latipes | 0.0 |
| abcg2-2 | 664 | NBD-TM | KP725054 | abcg2 (XP_006787117) | Neolamprologus brichardi | 0.0 |
| abcg2-like | 616 | NBD-TM | KP725024 | abcg2 (XP_004066087) | Oryzias latipes | 0.0 |
| abcg4-1 | 648 | NBD-TM | KP725023 | abcg4 (XP_004076001) | Oryzias latipes | 0.0 |
| abcg4-2 | 642 | NBD-TM | KP725022 | abcg4 (XP_004076385) | Oryzias latipes | 0.0 |
| abcg5 | 638 | NBD-TM | KP725021 | abcg5 (XP_004077344) | Oryzias latipes | 0.0 |
| abcg8 | 666 | NBD-TM | KP725052 | abcg8 (XP_003438483) | Oreochromis niloticus | 0.0 |
| abch | 700 | NBD-TM | KP725053 | abcg23-like (XP_011482890) | Oryzias latipes | 0.0 |
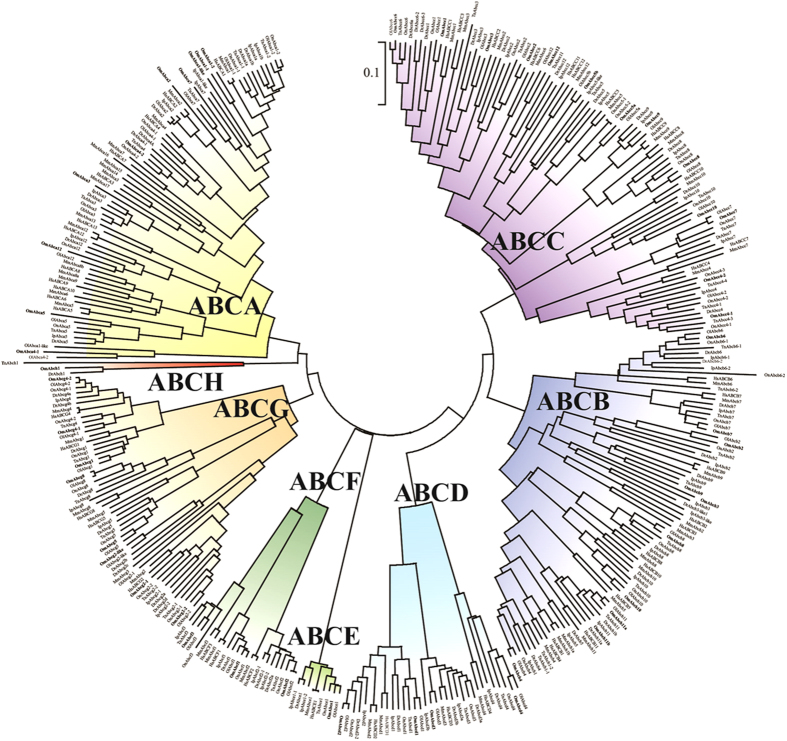
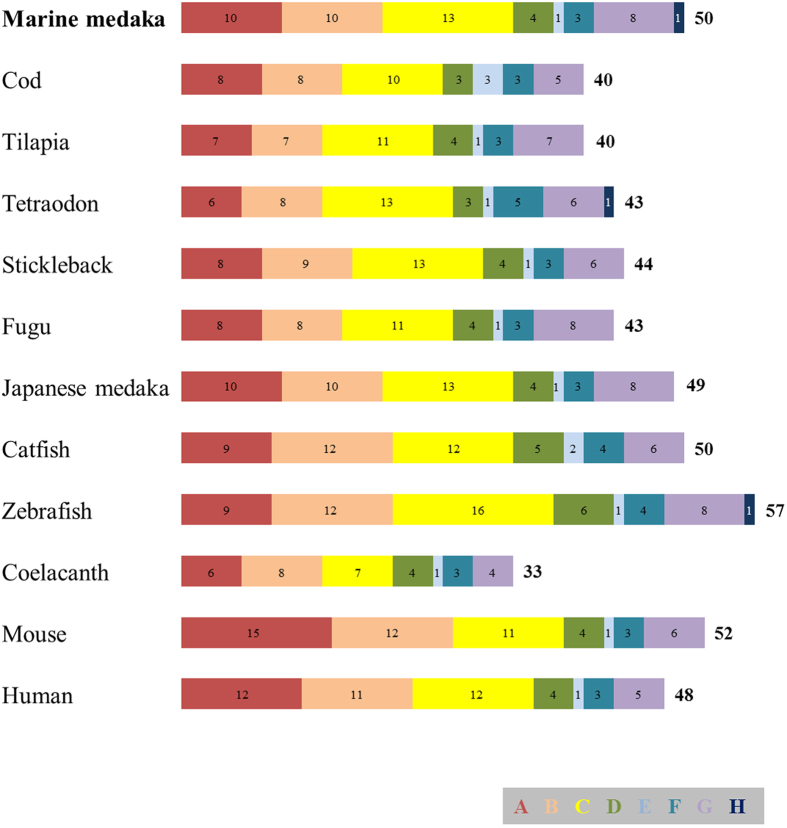
An ML phylogenetic analysis revealed that the 50 O. melastigma ABC transporters could be separated into the 8 subfamilies of A to H (Fig. 1). All O. melastigma ABC transporters possessed one or two conserved NBDs in their amino acid sequences, and the highest percentage of the ABC transporters belonged to the Abcc subfamily (26%), followed by the Abca and Abcb subfamilies (20% each) (Fig. 2). Overall composition rates and percentage rank of all O. melastigma ABC transporters were similar to those of other teleosts, while the absence or presence of some members belonging to each subfamily were slightly different between fish, suggesting that lineage-specific gene evolution has accumulated in teleosts (Table 2). Although overall composition of the genes seems to be conserved from teleosts to mammals, greater divergence was observed in the ABCA/Abca subfamily, as the mean number of ABCA/Abca members was higher in two mammals (i.e. mouse and human) compared to teleosts (Fig. 2). In this subfamily, the highest rate of gene duplication and/or loss events during evolution was suggested to be due to the absence of abca6, abca8, abca9, abca10, and abca13 genes in teleosts18.
Figure 1. Phylogenetic analysis of 50 O. melastigma ABC proteins.
Phylogenetic distance was calculated with combined NBD-TMD amino acid sequences from O. melastigma and other species. A best-fit substitution model was established using maximum likelihood (ML) analysis. Numbers at nodes represent the ML bootstrap support values and Bayesian posterior probabilities (=1.00). Details on the model test and parameters are explained in the Materials and Methods section. The tree is proportionally scaled and the scale bar indicates sequence distance in units of substitutions. Species abbreviations: Dr, Danio rerio; Hs, Homo sapiens; Ip, Ictalurus punctatus; Mm, Mus musculus; Ol, Oryzias latipes; Om, Oryzias melastigma; On, Oreochromis niloticus; Tn, Tetraodon nigroviridis.
Figure 2. Simplified comparison of the number of genes in each subfamily of ABC transporters between O. melastigma and other vertebrates.
Table 2. Number of ABC transporters in vertebrates and the composition of each ABC subfamily.
| Gene | Human | Mouse | Catfish | Zebrafish | Japanese Medaka | Marine Medaka | Fugu | Stickleback | Tetraodon | Tilapia | Cod | Coelacanth |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABCA1 | 1 | 1 | 3 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 1 |
| ABCA2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCA3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCA4 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 |
| ABCA5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCA6 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCA7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| ABCA8 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCA9 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCA10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCA12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| ABCA13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCA14 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCA15 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCA17 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCB1 | 1 | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| ABCB2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCB3 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCB4 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ABCB5 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ABCB6 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 |
| ABCB7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCB8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCB9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| ABCB10 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCB11 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 0 | 1 | 1 |
| ABCC1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| ABCC2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCC3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| ABCC4 | 1 | 1 | 1 | 1 | 2 | 2 | 3 | 4 | 4 | 3 | 3 | 1 |
| ABCC5 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 0 |
| ABCC6 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 |
| ABCC7 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ABCC8 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCC9 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ABCC10 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCC11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCC12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 |
| ABCC13 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCD1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| ABCD2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| ABCD3 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCD4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCE1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 |
| ABCF1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 |
| ABCF2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCF3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCG1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 |
| ABCG2 | 1 | 1 | 2 | 3 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 0 |
| ABCG3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABCG4 | 1 | 1 | 1 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 1 | 1 |
| ABCG5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCG8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCH | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ABCH (updated) | 0 | 0 | ? | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Total | 48 | 52 | 50 | 57 | 50 | 50 | 44 | 45 | 43 | 41 | 41 | 34 |
This table was updated from Liu et al.10. In detail, the abch gene was recently confirmed in the green spotted pufferfish (Tetraodon nigroviridis)8 . In this study, additional 12 ABC transporter members were newly identified in the genome database (http://genome.ucsc.edu) of Japanese medaka (Oryzias latipes), although Liu et al.10 reported 38 ABC transporters in the fish.
Several ABC transporters could also be divided into 26 full transporters (52%) and 20 half transporters (40%), based on the numbers of NBDs and TMDs (Fig. S1). Particularly, full transporters were observed in the Abca (10 proteins), Abcb (3 proteins), and Abcc (13 proteins) subfamilies, while half transporters were seen in the Abcb (7 proteins), Abcd (4 proteins), Abcg (8 proteins), and Abch (1 protein) subfamilies. In addition, four members involved in Abce and Abcf subfamilies that possess two linked NBDs but lack TMDs were identified in O. melastigma.
Phylogenetic analysis in each ABC subfamily
To analyze the evolutionary placement of the 50 O. melastigma ABC genes, in-depth phylogenetic analysis was conducted for each subfamily in comparison to those of zebrafish (D. rerio), channel catfish (I. punctatus), Japanese medaka (O. latipes), Nile tilapia (O. niloticus), green spotted pufferfish (T. nigroviridis), mouse (M. musculus), and human (H. sapiens) (Figs S2–S7). These species were chosen to create a representative set covering general evolutionary features within the vertebrate ABC superfamily.
Abca
Ten Abca proteins were identified in the O. melastigma genomic database and were examined using phylogenetic analysis to obtain their annotations (Fig. S1, Table 1). The members of the Abca subfamily of all teleosts, including marine medaka, were full transporters, as is seen in human2, suggesting it is a vertebrate-specific characteristic, since several insects and copepods are known to possess half transporters in this subfamily10,19. Overall phylogenetic analysis found that each member of the O. melastigma Abca subfamily was clustered with its counterpart from other animals. Particularly, evidence of the teleost-specific gene duplication after WGD was observed in abca1 (abca1-1 and abca1–2) and abca4 (abca4–1 and abca4–2) genes that, in mammals, are involved in cholesterol efflux of high-density lipoproteins (HDL) and the transport of retinoid-lipid complexes out of photoreceptor cells, respectively18,20,21. Interestingly, the abca1-like gene identified in catfish was also observed in both O. melastigma and O. latipes though a putative abca1-like gene has not been identified as yet in any other fish. Thus, the abca1-like gene appears to be derived from abca1 by further genome duplication after the abca7 duplication event in the abca gene lineage.
In O. melastigma, the mammalian-specific ABCA6, ABCA8, ABCA9, and ABCA10 genes are not present in the genome, just as they are absent in other teleosts (Fig. S2). In the human genome, all these genes are located in a single cluster together with the ABCA5 gene18 and in vertebrates, the ABCA5/Abca5 subfamily is highly conserved. Mammalian ABCA6/8/9/10 subfamilies form a sister clade with ABCA5 members but are not found in teleosts, indicating that the ABCA5/Abca5 lineage was split in a teleost-specific manner5,10.
Abcb
In O. melastigma, there are 3 Abcb full transporters and 7 NBD-containing half transporters (Table 1), and each gene forms robust clades with their corresponding counterparts (Fig. S3). The Abcb1, Abcb4, and Abcb5 members are known to share a common ancestor22, but there are controversial reports on the nomenclature for the putative abcb1/4 gene in teleosts due to a duplication event within a range of vertebrates10. In detail, mammalian ABCB1 and ABCB4 arose from a lineage-specific gene duplication (Fig. S3), and this phenomenon also observed in the chicken Gallus gallus8. However, teleost Abcb1 or Abcb4 are not one-to-one orthologues to either ABCB1 or ABCB4 but co-orthologues to both, resulting that all teleost possesses at least one transporter in the Abcb1/Abcb4 subfamily with different annotation names (i.e. Abcb1-like, p-glycoprotein, Abcb4-like, or Abcb4)8, as shown in Table S1. In this study, we identified one putative abcb4 gene in O. melastigma with its nomenclature. Although mammalian ABCB4 has a specific physiological function in the liver and transports certain fatty acids, a recent study showed that zebrafish Abcb4 (previously annotated as Abcb1b5) plays as a cellular toxicant transporter and provides protection for embryos in response to toxic chemicals23.
In O. melastigma, we could not identify the abcb5 gene that was observed in catfish and zebrafish10, and no homologous sequences were found in the transcriptomes and/or genome information of Japanese medaka, Nile tilapia, and green spotted pufferfish in our in silico analysis. Although incomplete genomic sequence databases of these fish need to be updated in order to conduct appropriate synteny analysis, we assume that species-specific gene duplication events in Abcb5 occurred in the Abcb1 lineage. In addition, two duplicated forms of abcb11 (abcb11a and abcb11b) appeared in marine medaka, as is also seen in Japanese medaka, zebrafish, and Atlantic cod (Ensembl Gene ID: abcb11a, ENSGMOG00000014190; abcb11b, ENSGMOG00000010088), while other teleosts have no duplicated members in the abcb11 gene.
Abcc
The Abcc subfamily consists of 13 full transporters in O. melastigma (Fig. S1, Table 1). This is the largest number of ABC transporters in all teleosts, including the marine medaka, when compared with those of mammals (Fig. 2). All members of Abcc formed distinct clades showing individual evolutionary branches (Fig. S4). In the case of abcc4, two types (abcc4a and abcc4b) were identified in O. melastigma, as is seen in other teleosts that have multiple copies of abcc4 (two members for O. latipes; three members for O. niloticus; four members for T. nigroviridis), while only a single gene was identified in mammals, indicating that teleost-specific genome duplications have occurred during evolution in fish.
An interesting feature of Abcc5 divergence was observed. For example, in O. melastigma, two members of abcc5 (abcc5a and abcc5b) were observed, as is seen in Japanese medaka (abcc5a and abcc5b) and catfish (abcc5 and abcc5-like), but no additional genes were observed in tetraodon, zebrafish, or mammals (Fig. S4). Although they diverged from a common ancestor, amino acid similarity of multiple Abcc5 members showed relatively low identity/similarity within the same group (Fig. S8, Table S2), suggesting a medaka- or catfish-specific duplication within teleosts.
A single member (abcc12) was incorporated into the combined clade of Abcc11/Abcc12, suggesting that the phylogenetic placement with diverse Abcc11/12 members from vertebrates requires revisiting. No member of abcc13 was observed in the O. melastigma genome, as is seen in most teleosts, while a single gene of abcc13 has been identified in zebrafish5.
Abcd
The ABCD subfamily generally harbors four highly conserved members in vertebrates, while several teleosts possess the following Abcd isoforms. In O. melastigma, all 4 abcd transporters were identified as half transporters (Fig. S1, Table 1), and each abcd member showed a clear homologous relationship to that of vertebrates (Fig. S5). Zebrafish has two isoforms of abcd2 and abcd3 and catfish contains two isoforms of abcd3, while only a single gene has been identified in the genus Oryzias (O. melastigma and O. latipes), implying that abcd genes expanded in a lineage-specific manner in teleosts.
Abce and Abcf
Since the subfamilies ABCE and ABCF lack TMDs and are comprised of a pair of linked NBDs, all ABC transporters are not restricted to having a function in ATP-dependent active transport. In the case of O. melastigma Abce and Abcf, all of the subfamily members are believed to be involved in biological processes other than transport5,18, as they possess two linked NBDs but lack TMDs (Fig. S1). Vertebrate ABCE1 was originally described as RNase L inhibitor24, and in recent studies novel evidences on ABCE1 roles were discovered in translation initiation, elongation, termination, and ribosome recycling25,26,27. The general and specific functions of vertebrate ABCF are still unclear as yet. Previously, human ABC50 (ABCF1) was identified as a tumor necrosis factor-α inducible gene in synoviocytes28. Subsequently, the protein was co-purified with eukaryotic translation initiation factor eIF229, suggesting that it plays a role in the translation initiation at an internal ribosome entry site (IRES) in vitro30. Phylogenetic analysis revealed that all Abce and Abcf transporters formed a unique cluster, similar to those seen in vertebrates (Fig. S6). As shown in most eukaryotes, a single Abce subfamily was found in marine medaka. The Abce protein is one of the most conserved proteins as most vertebrates and all invertebrates examined so far have a single member of this group, except for catfish (two genes) and cod (three genes)6,7,19,31 (Table 2). In the case of Abcf, all Abcf homologs (abcf1, abcf2, and abcf3) were identified in the marine medaka and Japanese medaka genomes, while abcf2 was duplicated in catfish and zebrafish10 (Fig. S6).
Abcg
Eight members of the Abcg subfamily were identified as half transporters in O. melastigma, and all the Abcg members were composed with a reverse-domain architecture (NBD-TMD) (Fig. S1). In most metazoan species, this unique structural feature is common except for fungi and plants32,33. The phylogenetic analysis of O. melastigma Abcg supported their annotations (Fig. S7). As shown in catfish (abcg2–1 and abcg2–2), green spotted pufferfish (abcg2–1 and abcg2–2), and zebrafish (abcg2a, abcgb, abcgc, and abcgd), the abcg2 gene was duplicated in the O. melastigma and O. latipes genomes (abcg2–1, abcg2–2, and abcg2-like). Likewise, stickleback and tilapia have 2 members and cod possesses 4 members in their genomes8. In particular, the O. melastigma abcg2-like gene was clustered with O. latipes abcg2-like and formed a distinct clade with zebrafish abcg2b and abcg2c. In detail, the O. melastigma abcg2-like gene showed low values in similarity and identity to O. melastigma abcg2–1 and abcg2–2 genes (Table S3). Likewise, the similarity and identity of O. latipes abcg2-like gene were high compared to those of the O. melastigma abcg2-like gene, while low similarity and identity were observed in comparison to the O. latipes abcg2–1 and abcg2–2 genes. Two zebrafish abcg2 genes (abcg2b and abcg2c), clearly separated from other abcg2 genes, show comparable patterns in similarity and identity. Thus, WGD and additional TGD may have occurred in the lineage of the teleost Abcg2 subfamily during evolution. Lineage-specific gene duplication was also observed in the Abcg4 group, as two isoforms were identified in O. melastigma, O. latipes, and O. niloticus, while only a single gene was observed in I. punctatus and T. nigroviridis.
Abch
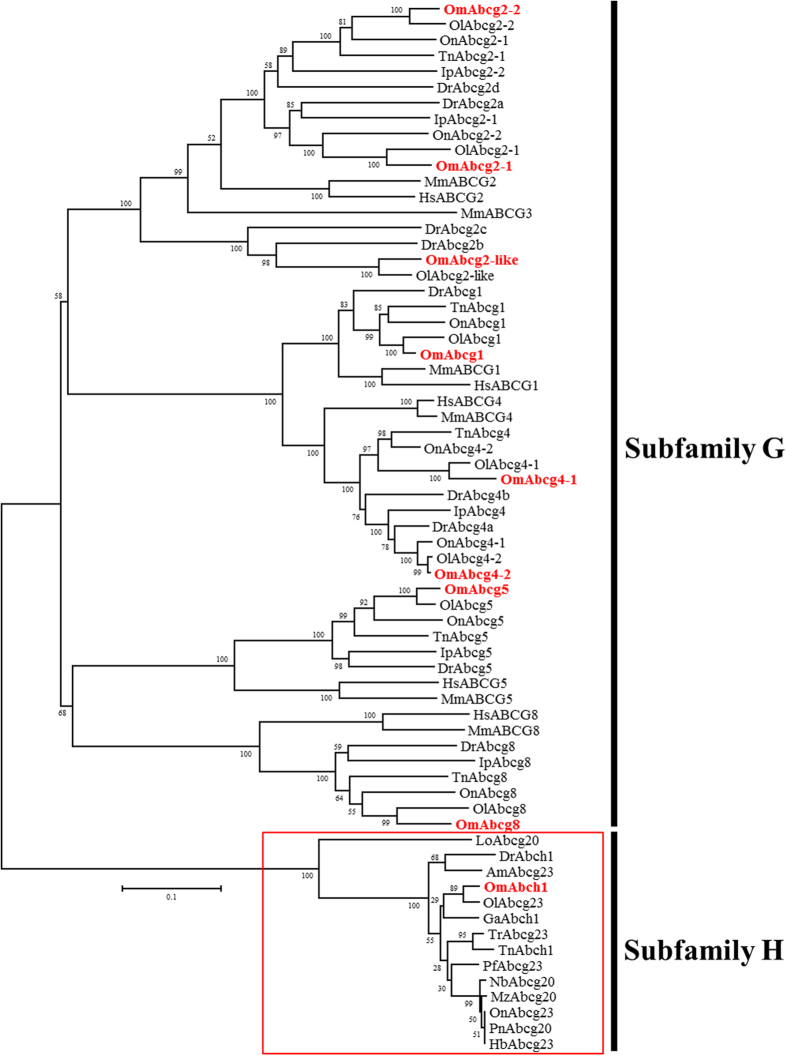
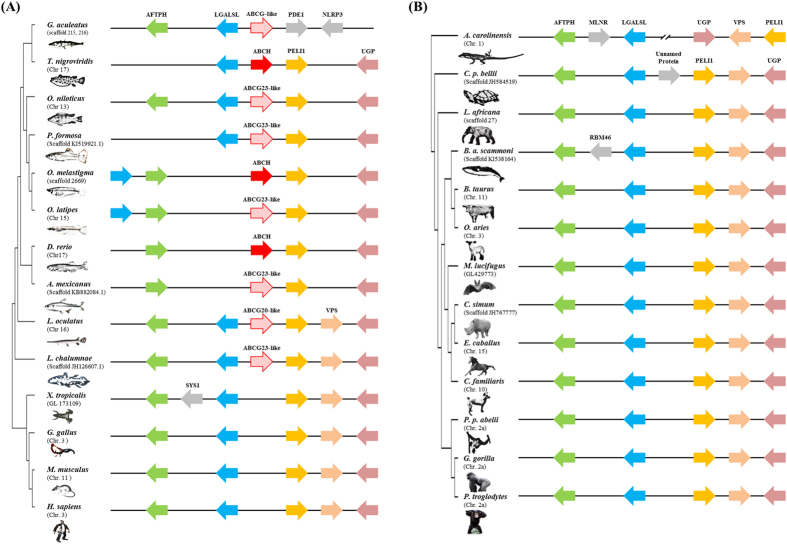
To date, the abch genes have only been annotated in sea urchin, arthropods, and two teleosts, zebrafish and green spotted pufferfish5,6,7,8,18,19. In fact, the presence of the Abch subfamily in fish is controversial. Previously, the abch gene was annotated only in zebrafish and a putative form was identified in green spotted pufferfish34, which has been confirmed in a recent review of ABC drug transporters8. The Abch subfamily has not been identified in other fishes such as catfish, Japanese medaka, fugu, stickleback, tetraodon, tilapia, cod, or coelacanths10. Interestingly, in O. melastigma, one transporter formed the same clade with the abch genes of zebrafish (abch1) and green spotted pufferfish (abch1), but this Abch clade seems to be an outgroup of the Abcg subfamily (Fig. S7). To clarify the identity of the Abch group, several Abcg members (abcg20 to abcg23) from other fish that contained only partial ABC information were employed in the phylogenetic analysis (Fig. 3). To date, no information on the annotation or phylogenetic distance of these Abcg members has been found. Regardless of the absence of the information for the Abch clade, our phylogenetic analysis strongly supported the formation of a unique clade for the three Abch members and additional Abcg20/Abcg23 members. In this study, we supposed that O. melastigma has the Abch subfamily, although the distinct clade containing Abch members and some Abcg members were duplicated from the Abcg subfamily. An independent lineage-specific expansion likely induced this kind of unique evolutionary event, as the Abch subfamily was not identified in coelacanths, the most primitive fish. A synteny comparison supported the orthologous relationship, as the genomic structures of the neighboring genes were very similar among teleosts (Fig. 4A). Interestingly, the abch gene was conserved in the coelacanth Latimeria chalumnae and the spotted gar Lepisosteous oculatus genomes, while there is no Abch orthologue in tetrapods (Fig. 4B). These results suggest that the abch gene was retained in the ancestor of teleosts and sarcopterygian. In fact, the abch genes are conserved in nematodes, copepods, and insects6,7,10,19,31 and observed in the Pacific oyster Crassostrea gigas (EKC37771), the acorn worm Saccoglossus kowalevskii (XP_006817503), and the sea urchin Strongylocentrotus purpuratus (XP_003731550; also registered in the Sea urchin genome database (http://www.echinobase.org); ID: SPU_026438) by GenBank database search, suggesting that the abch gene was retained from the bilaterian ancestor and a gene loss event recently occurred in the tetrapod lineage (Fig. 5). In the GenBank database, additional sea urchin ABC transporter H family member 2-like (S. purpuratus; LOC100889169), the dolphin ABC transporter H family member 2-like (Lipotes vexillifer; LOC103069646), and the Tibetan antelope ABC transporter H family member 2-like (Pantholops hodgsonii; LOC102331264) were registered as potential ABCH/Abch members, but they formed a distinct phylogenetic clade between Abcg and Abcf subfamilies apart from the Abch subfamily (Fig. S9, S11). Thus, the gene loss event of Abch subfamily in the tetrapod lineage is robust based on this result. At the moment, no putative abch gene has been identified in the Florida lancelet (Branchiostoma floridae) and Cnidarian. Identification and characterization of additional metazoan abch genes would be helpful for a better understanding of Abch evolution. Taken together, the nomenclature of several members of the Abcg subfamily that cluster with Abch should be reconsidered, as this may be an independent subfamily. This information provides a resource as a whole set from an essential gene family in teleosts, and their phylogenetic relationships with synteny analysis will be useful for a better understanding of evolutionary aspects of the ABC superfamily in teleosts.
Figure 3. Phylogenetic analysis of Abcg/Abch transporters with additional Abch candidates.
Phylogenetic distance was calculated with combined NBD-TMD amino acid sequences from O. melastigma and other species. A best-fit substitution model was established using maximum likelihood (ML) analysis. Numbers at nodes represent the ML bootstrap support values and Bayesian posterior probabilities. The tree is proportionally scaled, with the scale bar indicating sequence distance in units of substitutions. Species abbreviations: Am, Astyanax mexicanus; Dr, Danio rerio; Ga, Gasterosteus aculeatus; Hb, Haplochromis burtoni; Hs, Homo sapiens; Ip, Ictalurus punctatus; Lo, Lepisosteus oculatus; Mm, Mus musculus; Mz, Maylandia zebra; Nb, Neolamprologus brichardi; Ol, Oryzias latipes; Om, Oryzias melastigma; On, Oreochromis niloticus; Pf, Poecilia Formosa; Pn, Pundamilia nyererei; Tn, Tetraodon nigroviridis; Tr, Takifugu rubripes.
Figure 4.
(A) Syntenic analysis of the genomic structure harboring the abch gene in teleosts. Genes are represented by colored arrows. Distances within genes are not represented to scale. The different species were drawn by author Chang-Bum Jeong using Photoshop. GenBank accession numbers of ABCG/Abcg members used in the syntenic analysis are as follows: Astyanax mexicanus Abcg23-like (cavefish; XP_007244688), Gasterosteus aculeatus Abcg-like (Stickleback; ENSGACG00000000159), Latimeria chalumnae Abcg23-like (coelacanth; XP_005989631), Lepisosteus oculatus Abcg20-like (spotted gar; XP_006638801), Oreochromis niloticus Abcg23-like (tilapia; XP_005473794), Oryzias latipes Abcg23-like (Japanese medaka; XP_011482890), and Poecilia formosa Abcg23-like (Amazon molly; XP_007572984). Gene name abbreviations: aftiphilin, AFTPH; galectin-related protein, LGALSL; NACHT, LRR and PYD domain-containing protein, NLRP3; E3 ubiquitin-protein ligase, PELI1; calmodulin-dependent phosphodiesterase 1C, PDE1; golgi-localized integral membrane protein, SYS1; UTP-glucose-1-phosphate uridylyltransferase, UGP; vacuolar protein sorting-associated protein, VPS. (B) Further syntenic analysis of the genomic structure of abch gene in tetrapods. Genes are represented by colored arrows. Distances within genes are not represented to scale. Entire images were drawn by the author Chang-Bum Jeong. Gene name abbreviations: aftiphilin, AFTPH; galectin-related protein, galectin-related protein, LGALSL; E3 ubiquitin-protein ligase, PELI1; motilin receptor, MLNR; probable RNA-binding protein 46 isoform 2, RBM46; UTP-glucose-1-phosphate uridylyltransferase, UGP; vacuolar protein sorting-associated protein, VPS.
Figure 5. Schematic diagram for evolutionary scenario of the abch gene in eumetazoans.
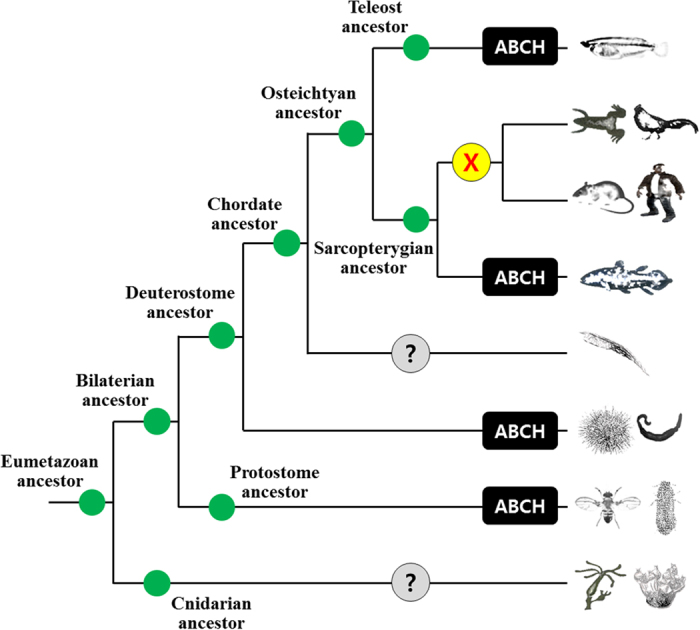
The diagram depicts a phylogenetic tree of metazoan species. Gene loss event is marked with an “X” in a yellow circle. Unknown information on the existence of the abch gene is marked with a “?” in a gray circle. The different species were drawn by the author Chang-Bum Jeong using Photoshop. GenBank accession numbers of ABCG genes used in the syntenic analysis are as follows: Drosophila melanogaster CG9990, CG11147, and CG33970 (fruitfly; AAF56807, AAF52284, and ABC66191, respectively); Latimeria chalumnae abch (coelacanth; ENSLACG00000015557); Oryzias melastigma abch (marine medaka; KP725053); Saccoglossus kowalevskii abch (acorn worm; XP_006817503); Tribolium castaneum abch1, abch2, and abch3 (red flour beetle; XP_973444, XP_967359, and XP_974932, respectively).
Materials And Methods
Ethics in experiments
All animal handling and experimental procedures were approved by the Animal Welfare Ethical Committee and the Animal Experimental Ethics Committee of the Sungkyunkwan University (Suwon, South Korea).
Fish
The marine medaka O. melastigma were kindly provided by Dr. Doris W.T. Au (City University of Hong Kong, Hong Kong SAR, China) and were maintained at the aquarium facility of the Department of Biological Science, Sungkyunkwan University (Suwon, South Korea). The fish were reared in accordance with the Animal Welfare Ethical Committee of the Sungkyunkwan University. Briefly, the fish were maintained in automatically controlled conditions at 26°C with a light/dark ratio of 12L:12D and artificial seawater (TetraMarine Salt Pro, Tetra™, Cincinnati, OH, USA; 5.71 ± 0.19 mgO2/L) adjusted to 12 practical salinity units (psu). The automated water-changing system was set for constant flow-through, and water quality (pH, salinity, and temperature) was recorded using various instruments. Fish were maintained in glass aquaria (60 L capacity) and each aquarium accommodated up to 30 adult fish (both sexes). They were fed Artemia salina (<24 h after hatching) once a day until satiation.
Retrieval of ABC transporter genes
An O. melastigma genomic DNA database was constructed using next generation sequencing (NGS) technologies and bioinformatics (unpublished data; scaffold no.: 24,820; total length: 671,972,662 bp; longest length: 1,068,498 bp; average length: 27,074 bp; N50 value: 115,707 bp). Briefly, the genomic DNA of O. melastigma was mechanically sheared into fragments, and a genomic DNA library was created according to the manufacturer’s instructions (Roche Applied Science, Genome Sequencer System, Pleasanton, CA, USA). Genomic DNA of O. melastigma was sequenced using a GS-FLX-Titanium genomic DNA sequencer (Roche Diagnostics, Mannheim, Germany) and SOLEXA sequencer (Illumina, San Diego, CA, USA), and then properly assembled with the software NGS Cell (Ver. 4.06 beta 67189, CLC Bio, Boston, MA, USA) and Velvet (EMBL-EBI, Wellcome Trust Genome Campus, Cambridge, UK). Currently, the number of unidentified N in the consensus sequence is 50,688,736 bp, indicating that approximately 7.5% of nucleotides are unidentified. To date, we have obtained 46,461 genes (unpublished data; total length: 27,691,766 bp; longest length: 7,011 bp; N50 value: 2,265 bp) after de novo genome assembly and RNA-sequencing (contig no.: 51,014; total length of contigs: 135,850,078 bp; longest length among contigs: 21,254 bp) with bioinformatics-aided gene annotation.
To obtain the sequence information for all ABC transporters after assembly, all contigs containing NBDs and/or TMDs in O. melastigma genomic DNA and transcriptome databases were subjected to BLAST analysis using the non-redundant (NR) database at GenBank (http://www.ncbi.nlm.nih.gov/genome/seq/database.html).
Nucleotide sequence validation
O. melastigma pooled tissues were homogenized in three volumes of TRIZOL® reagent (Invitrogen, Paisley, Scotland) with a tissue grinder and stored at −80 °C until use. Total RNA was extracted according to the manufacturers’ instructions and stored at −80 °C until use. DNA digestion was performed using DNase I (Sigma, St. Louis, MO, USA). Total RNA was quantified by absorption of light at 230, 260, and 280 nm (A230/260, A260/280) using a spectrophotometer (Qiaxpert®, Qiagen, Hilden, Germany). To verify that there was no genomic DNA contamination, total RNAs were loaded in a 1% agarose gel that contained ethidium bromide (EtBr) and visualized on a UV transilluminator (Wealtec Corp., Sparks, NV, USA). Subsequently, total RNAs were loaded in a 1% formaldehyde/agarose gel with EtBr staining in order to verify total RNA quality and verify 18/28S ribosomal RNA integrity. After RNA quality was determined, single-stranded cDNA was synthesized from 2 μg of total RNA from each sample using oligo (dT)20 primers for reverse transcription in 20 μl reactions (SuperScriptTM III RT kit, Invitrogen).
To confirm exon/intron boundaries and start/stop codons of O. melastigma ABC transporter genes, genomic structures of the obtained genes were compared between genomic clones and the transcripts for each gene. Some incomplete ABC transporter sequences were subjected to 5′- and 3′-RACE according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). To validate cDNA sequences of entire ABC genes identified in O. melastigma, RT-PCR was employed with two primers: a forward primer containing a start codon, and a reverse primer containing a stop codon. RT-PCR was conducted in a reaction mixture comprising 1 μl of first strand cDNA, 5 μl of 10× PCR reaction buffer, 1 μl of 10 mM dNTPs, 10 pM concentrations of each primer, and 0.5 μl of NeoTherm™ Taq polymerase (GeneCraft, Köln, Germany). Reaction mixtures were subjected to amplification (1 cycle, 95 °C, 5 min; 30 cycles, 94 °C, 30 sec, 55 °C, 30 sec, and 72 °C, 30 sec; 1 cycle, 72 °C, 7 min) using an iCycler (Bio-Rad, Hercules, CA, USA). The final PCR products were isolated from 1% agarose/Tris-Borate-EDTA (TBE) gels, cloned into pCR2.1 TA vectors (Invitrogen), and sequenced using an ABI PRISM 3700 DNA analyzer (Bionics Co., Seoul, South Korea).
Annotation of whole ABC transporter genes
Annotation and nomenclature of all the O. melastigma ABC genes followed amino acid sequence similarities compared to ABC superfamilies of other animals in terms of in silico domain analysis [Pfam HMM search, http://pfam.sanger.ac.uk; Motif Scan, http://myhits.isb-sib.ch/cgi-bin/motif_scan; National Center for Biotechnology Information (NCBI)’s Conserved Domain Database (CDD)]. All gene information was registered with the GenBank database, and the accession numbers of each gene are appended in Table 1.
Phylogenetic analysis
To investigate evolutionary relationships and the nomenclature of O. melastigma putative ABC transporters, amino acid sequences of ABC transporters identified in O. melastigma were subjected to phylogenetic analysis and compared with other species (Danio rerio, Ictalurus punctatus, Oreochromis niloticus, Oryzias latipes, Tetraodon nigroviridis, Mus musculus, and Homo sapiens) in the NCBI GenBank database by performing BLASTX searches. Combined NBD-TMD amino acid sequences from O. melastigma and other species were aligned using MEGA software (ver. 6.0; Center for Evolutionary Medicine and Informatics, Tempe, AZ, USA) with the ClustalW alignment algorithm. To set a best-fit substitution model for phylogenetic analysis, a model showing the lowest score in the Bayesian Information Criterion (BIC)35 and the Akaike Information Criterion (AICc)36,37 was determined with Maximum Likelihood (ML) analysis. According to the results of the model test, the WAG + G + I + F model was chosen to generate a phylogenetic tree. MrBayes (ver. 3.1.2) was used to reconstruct phylogenetic trees based on Bayesian inference38. The Markov chain Monte Carlo (MCMC) process was conducted with four chains and run for 5,000,000 generations. Sampling frequency was every 100 generations. After analysis, the first 5,000 trees were deleted as part of the burn-in process. A consensus tree was constructed and was visualized using SeaView ver.4.2.139. Nodal support was reported as Bayesian posterior probabilities (=1.00).
Additional Information
How to cite this article: Jeong, C.-B. et al. Marine medaka ATP-binding cassette (ABC) superfamily and new insight into teleost Abch nomenclature. Sci. Rep. 5, 15409; doi: 10.1038/srep15409 (2015).
Supplementary Material
Acknowledgments
This work was supported by a grant of the Basic Science Research Program through the National Research Foundation (NRF) of Korea (NRF-2012R1A1A2002806) funded to Ik-Young Choi and was also supported by a grant of the Marine Biotechnology Program (PJT200620; Genome analysis of marine organisms and development of functional application) funded by the Ministry of Oceans and Fisheries, Korea to Jae-Seong Lee.
Footnotes
Author Contributions J.-S.R. and J.-S.L. designed the research study. C.-B.J., B.-M.K., H.-M.K., and I.-Y.C. analyzed the data and prepared entire figures. J.-S.R. and J.-S.L. wrote the manuscript with input from C.-B.J. and B.-M.K. All authors reviewed and approved the final manuscript.
References
- Roninson I. B., Abelson A. T., Housman D., Howell N. & Varshavsky A. Amplification of specific DNA sequences correlates with multi-drug resistance in Chinese hamster cells. Nature 309, 626–628 (1984). [DOI] [PubMed] [Google Scholar]
- Dean M., Rzhetsky A. & Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11, 1156–1166 (2001). [DOI] [PubMed] [Google Scholar]
- Avidson A. L., Dassa E., Orelle C. & Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J. & Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annilo T. et al. Evolution of the vertebrate ABC gene family: Analysis of gene birth and death. Genomics 88, 1–11 (2006). [DOI] [PubMed] [Google Scholar]
- Sturm A., Cunningham P. & Dean M. The ABC transporter gene family of Daphnia pulex. BMC Genomics 10, 170 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermauw W. & Van Leeuwen T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 45, 89–110 (2014). [DOI] [PubMed] [Google Scholar]
- Luckenbach T., Fischer S. & Sturm A. Current advances on ABC drug transporters in fish. Comp. Biochem. Physiol. C. 165, 28–52 (2014). [DOI] [PubMed] [Google Scholar]
- Ferreira M., Costa J. & Reis-Henriques M. A. ABC transporters in fish species: a review. Front. Physiol. 5, 266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Li Q. & Liu Z. Genome-wide identification, characterization and phylogenetic analysis of 50 catfish ATP-binding cassette (ABC) transporter genes. PLoS ONE 8, e63895 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. & Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11, 699–704 (1999). [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication, Heidelberg, Germany: Springer-Verlag; Springer (1970).
- Parenti L. R. A phylogenetic analysis and taxonomic revision of ricefishes, Oryzias and relatives (Beloniformes: Adrianichthyidae). Zool. J. Linn. Soc. 154, 494–610 (2008). [Google Scholar]
- Inoue K. & Takei Y. Diverse adaptability in Oryzias species to high environmental salinity. Zool. Sci. 19, 727–734 (2002). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. Molecular staging of marine medaka: a model organism for marine ecotoxicity study. Mar. Pollut. Bull. 63, 309–317 (2011). [DOI] [PubMed] [Google Scholar]
- Rhee J.-S. et al. Whole spectrum of cytochrome P450 genes and molecular responses to water-accommodated fractions exposure in the marine medaka. Environ. Sci. Technol. 47, 4804–4812 (2013). [DOI] [PubMed] [Google Scholar]
- Kasahara M. et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 477, 714–719 (2007). [DOI] [PubMed] [Google Scholar]
- Dean M. & Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 6, 123–142 (2005). [DOI] [PubMed] [Google Scholar]
- Jeong C.-B., Kim B.-M., Lee J.-S. & Rhee J.-S. Genome-wide identification of whole ATP-binding cassette (ABC) transporters in the intertidal copepod Tigriopus japonicus. BMC Genomics 15, 651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R. et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat. Genet. 15, 236–246 (1997). [DOI] [PubMed] [Google Scholar]
- Oram J. F. & Lawn R. M. ABCA1: the gatekeeper for eliminating excess cholesterol. J. Lipid Res. 42, 1173–1179 (2001). [PubMed] [Google Scholar]
- Moitra K. et al. Molecular Evolutionary Analysis of ABCB5: The Ancestral Gene Is a Full Transporter with Potentially Deleterious Single Nucleotide Polymorphisms. PLoS ONE 6, e16318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S. et al. Abcb4 acts as multixenobiotic transporter and active barrier against chemical uptake in zebrafish (Danio rerio) embryos. BMC Biol. 11, 69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbal C., Martinand C., Silhol M., Lebleu B. & Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J. Biol. Chem. 270, 13308–13317 (1995). [DOI] [PubMed] [Google Scholar]
- Chen Z. Q. et al. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J. Biol. Chem. 281, 7452–7457 (2006). [DOI] [PubMed] [Google Scholar]
- Barthelme D. et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc. Natl. Acad. Sci. USA 108, 3228–3233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva V. P., Skabkin M. A., Hellen C. U., Pestova T. V. & Pisarev A. V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30, 1804–1817 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Drouin R. & Beaulieu A. D. ABC50, a novel human ATP-binding cassette protein found in tumor necrosis factor-alpha-stimulated synoviocytes. Genomics 53, 137–145 (1998). [DOI] [PubMed] [Google Scholar]
- Tyzack J. K., Wang X., Belsham G. J. & Proud C. G. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem. 275, 34131–34139 (2000). [DOI] [PubMed] [Google Scholar]
- Paytubi S. et al. ABC50 promotes translation initiation in mammalian cells. J. Biol. Chem. 284, 24061–24073 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheps J. A., Ralph S., Zhao Z., Baille D. L. & Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 5, R15 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk A. & Driessen A. J. Phylogenetic analysis of fungal ABC transporters. BMC Genomics 11, 177 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier P. J. et al. Plant ABC proteins - a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159 (2008). [DOI] [PubMed] [Google Scholar]
- Popovic M., Zaja R., Loncar J. & Smital T. A novel ABC transporter: the first insight into zebrafish (Danio rerio) ABCH1. Mar. Environ. Res. 69, S11–S13 (2010). [DOI] [PubMed] [Google Scholar]
- Schwarz G. Estimating the dimension of a model. Annal. Statistics 6, 461–464 (1978). [Google Scholar]
- Hurvich C. M. & Tsai C.-L. Regression and time series model selection in small samples. Biometrika 76, 297–307 (1989). [Google Scholar]
- Posada D. & Buckley T. R. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches Over Likelihood Ratio Tests. Systematic Biology 53, 793–808 (2004). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S. & Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.