SUMMARY
The vertebrate and neural-specific SR-related protein nSR100/SRRM4 regulates an extensive program of alternative splicing with critical roles in nervous system development. However, the mechanism by which nSR100 controls its target exons is poorly understood. We demonstrate that nSR100-dependent neural exons are associated with a unique configuration of intronic cis-elements that promote rapid switch-like regulation during neurogenesis. A key feature of this configuration is the insertion of specialized intronic enhancers between polypyrimidine tracts and acceptor sites that bind nSR100 to potently activate exon inclusion in neural cells, while weakening 3′ splice site recognition and contributing to exon skipping in non-neural cells. nSR100 further operates by forming multiple interactions with early spliceosome components bound proximal to 3′ splice sites. These multifaceted interactions achieve dominance over neural exon silencing mediated by the splicing regulator PTBP1. The results thus illuminate a widespread mechanism by which a critical neural exon network is activated during neurogenesis.
INTRODUCTION
Alternative splicing (AS) is the process by which different pairs of splice sites are selected in pre-mRNA to produce distinct mRNA and protein variants. Transcriptome-wide profiling has uncovered vast repertoires of splice variants in metazoan species, many of which are highly conserved and regulated in a cell- and tissue-specific manner (Irimia and Blencowe, 2012; Licatalosi and Darnell, 2010; Nilsen and Graveley, 2010). Important current challenges are to systematically determine the functions of individual tissue-regulated AS events and establish how they are regulated. These questions are especially important in the context of the vertebrate nervous system, which possesses among the most complex yet highly conserved repertoires of regulated AS events (Barbosa-Morais et al., 2012; Merkin et al., 2012).
Tissue-specific regulation of AS involves the complex interplay of numerous cis-acting elements and trans-acting factors. Experimental and computational studies have provided evidence that recognition of splice sites flanking neural and other tissue-regulated exons involves tissue-specific AS regulators that, depending on their binding location on pre-mRNA, can function to positively or negatively regulate the assembly of core components of the splicing machinery (spliceosome) at proximal or overlapping splice sites (Witten and Ule, 2011). For example, Nova proteins, which regulate a discrete network of exons enriched in synaptic and axon guidance genes, bind clusters of YCAY motifs concentrated near 5′ splice sites to promote exon inclusion, and to YCAY clusters proximal to 3′ splice sites to promote exon skipping (Licatalosi et al., 2008; Ule et al., 2006).
Ptbp1, and its paralog Ptbp2, bind intronic C/U-rich elements upstream of regulated exons to repress the inclusion of neural exons, yet Ptbp1 also promotes exon inclusion when binding to C/U-rich elements downstream of regulated exons (Licatalosi et al., 2012; Llorian et al., 2010; Xue et al., 2009). Ptbp1 and Ptbp2 have mutually exclusive patterns of expression in the developing nervous system. Expression of the microRNA miR-124 silences Ptbp1 in developing neurons (Makeyev et al., 2007). Loss of Ptbp1 facilitates the inclusion of a neural-specific exon (exon 10) in Ptbp2 transcripts that promotes Ptbp2 expression by preventing turnover of its transcripts by the nonsense-mediated mRNA decay pathway (Boutz et al., 2007; Makeyev et al., 2007; Spellman et al., 2007). The relative expression levels of Ptbp1 and Ptbp2 governed by this regulatory circuit contribute to establishing neural-specific AS patterns (Boutz et al., 2007; Li et al., 2014; Licatalosi et al., 2012; Zheng et al., 2012). However, the mechanisms by which Ptbp1/2-repressed and other neural-specific exons are activated during neurogenesis are not well understood.
Alternative splicing is additionally regulated by members of a large class of proteins harboring Ser/Arg (SR)-repeat regions (Änkö, 2014). The vertebrate- and neural-specific SR-related protein of 100 kDa (nSR100/SRRM4) regulates a network of brain-specific alternative exons concentrated in genes that function in various aspects of neurogenesis (Calarco et al., 2009; Raj et al., 2011). Among this network is exon 10 of Ptbp2, and a neural-specific “switch” exon in REST/NRSF, a transcriptional repressor of neurogenesis genes. Skipping of the switch exon in non-neural cells produces a repressive form of REST, whereas nSR100-dependent inclusion of the exon in differentiating neurons produces a truncated isoform that lacks repressive activity, thereby allowing neurogenesis to proceed (Raj et al., 2011). nSR100 regulates many additional brain-specific exons that are enriched in disordered regions of proteins, which function in modulating protein-protein interactions that contribute to neural-specific functions (Ellis et al., 2012). Recently, a mutation in the Bronx waltzer (bv) mouse, which causes deafness and balance defects, was mapped to the nSR100/Srrm4 locus and shown to disrupt AS of genes linked to secretion and neurotransmission that are expressed in the inner ear (Nakano et al., 2012). However, the molecular mechanisms that underlie nSR100-dependent neural-specific AS remain poorly understood.
In this study, we employed high-throughput RNA sequencing (RNA-Seq) and crosslinking and immunoprecipitation coupled to sequencing (CLIP-Seq) to identify an expanded network of neural-specific AS events regulated by nSR100. In addition to revealing new nSR100 target exons with important functions in neurogenesis, analyses of these data together with functional studies, demonstrate that nSR100 activates neural exon inclusion by binding to intronic UGC-containing motifs proximal to suboptimal 3′ splice sites. The same motifs also serve to weaken 3′ splice site recognition and contribute to skipping of neural exons in non-neural cells. We further provide evidence that this nSR100-dependent regulatory mechanism involves multiple interactions with early spliceosomal assembly components, and that it directly outcompetes widespread neural exon repression by PTBP1 during early stages of neurogenesis. These results thus reveal a novel mechanism underlying the activation of a critical network of neural-specific AS events.
RESULTS
Identification and characterization of a conserved exon network regulated by nSR100
To investigate the mechanism by which nSR100 controls neural-specific AS, we sought to computationally identify cis-features that mediate its activity in the regulation of conserved target alternative exons in human and mouse cells. RNA-Seq profiling was performed following knockdown of nSR100 in mouse neural N2A cells, and doxycycline (dox)-induced expression of nSR100 in human 293T cells. Consistent with our previous results from profiling a smaller set of exons using a focused AS microarray (Calarco et al., 2009), knockdown of nSR100 in undifferentiated N2A cells predominantly resulted in skipping of cassette exons that are specifically included in neural tissues (Figure 1A). Of 503 responsive exons (i.e. displaying an absolute Percent Spliced In change, |ΔPSI| ≥15%), 405 exhibited increased skipping, whereas 98 displayed increased inclusion. Remarkably, although nSR100 is not normally expressed in 293T cells, its dox-induced expression in this cell line resulted in widespread changes in the inclusion levels of exons, many of which are differentially regulated between neural and non-neural tissues (Figure 1B).
Figure 1. RNA-Seq identifies an extensive network of conserved neural-enriched exons regulated by nSR100.
(A and B) Top panels, scatterplots comparing changes in inclusion levels (|ΔPSI| ≥ 15%) of AS events upon altering nSR100 levels versus differences in inclusion levels between neural and non-neural tissues. ΔPSI nSR100, change in PSI levels of alternative exons following: (A) nSR100 knockdown in N2A cells, and (B) dox-induced nSR100 expression in 293T cells. ΔPSI Neural, difference in mean PSI level of regulated exons between neural and non-neural tissues (Table S1; Supplemental Experimental Procedures). Exons with ΔPSI Neural ≥ 20% (dotted line) were considered neural-enriched. Orthologous exons that show nSR100-dependent regulation in both N2A and 293T cells are circled in red (inclusion, 157 exons) or black (skipping, 5 exons). Bottom panels, RT-PCR assays and western blots confirming changes in nSR100 protein levels. Tubulin, gapdh and β-actin detection was used as loading controls.
(C) Venn diagrams indicating overlap between nSR100-promoted (left) and nSR100-inhibited (right) orthologous alternative exons in N2A and 293T cells.
(D and E) Examples of RT-PCR validations of conserved nSR100-regulated AS events upon: (D) nSR100 knockdown in N2A cells, and (E) dox-induced nSR100 expression in 293T cells. See also Figure S1 and Table S1.
To define a high-confidence set of conserved exons that are regulated by nSR100, we focused our analyses on all (157) orthologous exons that undergo skipping upon knockdown of nSR100 in N2A cells and that exhibit increased inclusion in 293T cells upon nSR100 expression (Figures 1A and 1B, red circles; Figure 1C). This set of 157 orthologous exons represents 53% and 62% of all conserved exons that are dependent on nSR100 for inclusion in 293T and N2A cells, respectively (p < 0.0001, Chi-square test). Further underscoring the physiological relevance of this set of AS events, the vast majority (95%) correspond to exons that are more included in neural versus non-neural tissues (Figures 1A, 1B; Table S1). In contrast, of the exons that exhibit nSR100-dependent skipping in human and mouse cells, only five overlapped (Figure 1C), one of which is neural-enriched (Figure 1A and 1B, black circles), suggesting that many of these changes may be the consequence of indirect effects. RT-PCR assays validated 100% (40/40 events in 20 genes) of the analyzed nSR100-regulated AS events detected by RNA-Seq profiling (Figures 1D, 1E, S1A, S1B; data not shown).
Consistent with previous observations (Calarco et al., 2009; Ellis et al., 2012), the set of 157 conserved nSR100-regulated exons are enriched in genes that function in cytoskeleton remodeling. However, we also observe enrichment in genes associated with “GTPase activity”, “synaptic membrane”, “neuron projection”, “establishment of cell polarity”, and “cell junctions”. Additional nSR100-regulated exons are predicted to impact genes that function in transcriptional control of nervous system development, including the myocyte enhancer factor 2 (Mef2) family of transcription activators (Figures 1D, 1E), which are critical for the formation of cortical neuronal layers and synaptic plasticity (Akhtar et al., 2012; Li et al., 2008).
Identification of cis-regulatory motifs associated with nSR100-dependent exons
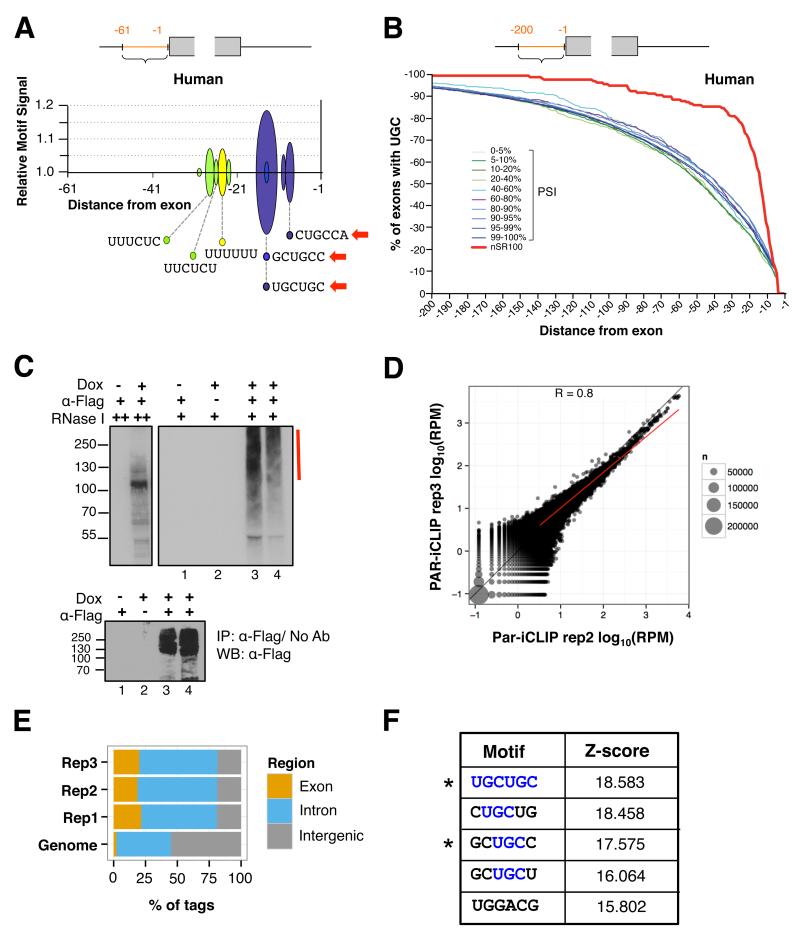
PEAKS software (Bellora et al., 2007) was used to identify motifs with significant positional biases relative to the splice sites of the set of 157 conserved nSR100-dependent exons. While significant enrichment of motifs in downstream intronic sequence was not observed, two types of enriched motifs were detected in the upstream introns, with similar sequence and positional biases in human and mouse (Figures 2A, S2A S2B). One of these corresponds to C/U-rich hexamers that resemble binding sites of PTBP1/PTBP2, and the other consists of motifs containing one or more UGC triplets.
Figure 2. UGC-containing motifs are enriched adjacent to target exons and are directly bound by nSR100 in vivo.
(A) PEAKS analysis plot showing sequences and distances of peaks of significantly enriched hexamers in intron sequences upstream of the 3′ splice sites of conserved, nSR100-regulated human exons. Red arrows, hexamers with UGC triplets.
(B) Cumulative distribution plots indicating the position of the first UGC triplet within 200 nucleotides upstream of conserved, nSR100-regulated (red) and control alternative or constitutive human exons with different PSI ranges (blue, purple and green lines).
(C) Top panel, protein gel autoradiographs of nSR100-RNA 32P-labeled complexes after high (++) and low (+) RNase I digestion in 293T cells induced to express Flag-nSR100. Red bar, region of the gel excised for library generation; Bottom panel, western blot confirming immunoprecipitation of Flag-nSR100 protein.
(D) Scatterplot showing correlation between mappings of reads in 100 base pair genomic bins using data from two biological replicates of nSR100 293T PAR-iCLIP experiments.
(E) Bar plots showing the distributions of nSR100 293T PAR-iCLIP tags in exon, intron, and intergenic sequences from three independent PAR-iCLIP experiments. “Genome” indicates the distribution of a subset of randomly chosen genomic positions.
(F) Top five-most enriched hexamers and their corresponding Z-scores identified using PAR-iCLIP analyses. Asterisks, motifs that were also identified using PEAKS. See also Figure S2.
We compared the cumulative distributions of the first occurrence of a UGC triplet in mouse and human introns upstream of the nSR100-dependent exons, versus sets of control alternative exons that are not regulated by nSR100 but that have comparable ranges of PSI levels (Figures 2B, S2C). This reveals that 3′ splice sites of nSR100-regulated exons are significantly more often associated with proximal UGC triplets than are control exons (p < 0.0001, all comparisons, Mann-Whitney test), but these motifs are not enriched within nSR100-regulated exons relative to control exons (data not shown). Consistent with these findings, UGC motifs were implicated in the regulation of AS events affected in the nSR100-mutant Bronx waltzer mouse strain (Nakano et al., 2012). However, based on the analysis of a much smaller set of regulated exons, it was concluded that these motifs are enriched at a greater distance from 3′ splice sites and their positional relationship with distal C/U-rich motifs was not detected. Moreover, whether nSR100 functions via binding to the UGC motifs in vivo was not determined.
nSR100 binds intronic UGC motifs adjacent to the 3′ splice site of target exons in vivo
To investigate whether nSR100 binds to the UGC, C/U-rich, and/or other motifs, we used PAR-iCLIP, which employs incorporation of the photoreactive nucleoside analog 4-thiouridine in nascent RNA to increase crosslinking efficiency to protein, followed by the iCLIP protocol (Hafner et al., 2010; Huppertz et al., 2014). Three independent PAR-iCLIP experiments were performed using 293T cells expressing Flag-nSR100 under dox-inducible control, in conjunction with immunoprecipitation with anti-Flag antibody. Immunoprecipitated complexes were specifically detected following nSR100 induction and were susceptible to RNase I, indicating successful in vivo crosslinking of nSR100 to RNA (Figures 2C, S2D). Pairwise comparisons of the mapped tag sequences revealed that the biological replicates correlate well (mean R=0.8, Figures 2D, S2E). Analysis of the genomic distributions of the PAR-iCLIP tags further revealed that these preferentially map to exonic and intronic sequence relative to intergenic sequence, after normalizing for length differences between genomic regions (Figure 2E).
Clusters of 293T cell PAR-iCLIP tags were analyzed for significantly enriched hexamer sequences, using randomized cluster sequences as a background. Notably, the top four most enriched hexamers contain one or more UGC triplets (Figures 2F, S2F), and two of the hexamers matched those detected by the PEAKS analysis (compare Figure 2F with 2A). We generated a merged in vivo “binding map” (Licatalosi et al., 2008) displaying the mean, normalized distributions of nSR100 293T cell PAR-iCLIP crosslink sites within 400-nucleotide windows surrounding the 157 conserved target exons, and a set of ~400 control exons that are not nSR100 targets but that have a comparable PSI distribution. We observed a strong enrichment for nSR100 binding to intronic sequences proximal to the 3′ splice sites of target exons relative to control exons, with a pronounced peak approximately -15 nucleotides from the 3′ splice site (Figures 3A, S3A). Furthermore, PAR-iCLIP analysis using an N2A cell line expressing Flag-nSR100 under dox-inducible control also revealed that nSR100 preferentially crosslinks to intronic UGC containing motifs concentrated approximately -14 nucleotides from the 3′ splice site of the conserved nSR100 regulated exons (Figures S3B-D and data not shown). Importantly, these binding peaks coincide precisely with the preferred location of UGC motifs identified using PEAKS analysis (compare with Figure S2B).
Figure 3. nSR100 binds directly upstream of suboptimal 3′ splice sites to promote neural exon inclusion.
(A) nSR100 RNA binding map showing the mean, normalized density of crosslinked sites in 400 nucleotide windows encompassing nSR100-regulated exons (blue) and control non-regulated exons (gray) in 293T cells; ss, splice site.
(B) Top panel, genome browser view of the raw density of nSR100 293T PAR-iCLIP tags surrounding the UGC motif (orange box) upstream of the REST neural exon. Bottom panel, RT-PCR assay monitoring inclusion levels of the REST neural exon upon transfection of wild-type (W) or mutant (M) minigene reporters into 293T cells with and without dox-induced nSR100 expression.
(C) Box plots comparing the 3′ and 5′ splice site strengths of conserved nSR100 target exons (blue) and control, non-regulated exons (white). Asterisks represent significant differences; p values, Wilcoxon rank sum test.
(D) Plots comparing polypyrimidine tract lengths and distances between 3′ splice sites and inferred branch points of nSR100 regulated exons (red), and of PSI-matched control exons (gray) in human; p values, Kolmogorov-Smirnov test.
(E) RT-PCR assays monitoring the effects of mutating UGC motif(s) on inclusion levels of the DAAM1 and MEF2D neural exons. Dox-inducible nSR100-expressing 293T cells were transfected with wild-type (W) minigene reporters (lanes 1-2). 293T cells (not dox-inducible) were transfected with wild-type or mutant (M, M1, M2) reporters, and control (lanes 3, 5, 7) or PTBP1 and PTBP2 siRNAs (lanes 4, 6, 8). PPT, polypyrimidine tract. Asterisk represents a product from usage of an alternative splice site. See also Figure S3.
UGC motifs are critical for nSR100-dependent neural-specific splicing
To confirm whether UGC motifs bound by nSR100 in vivo are important for nSR100-dependent AS, we asked if mutation of these motifs disrupts the inclusion of nSR100 target exons. The PAR-iCLIP data reveals an accumulation of reads surrounding a UGC motif upstream of the neural exon of REST (Figure 3B), which we previously defined as a functionally important nSR100 target (Raj et al., 2011). REST minigene reporters were constructed with the neural exon and 300 nucleotides of flanking intronic sequence, with and without a G to A point mutation in the UGC motif, and were transfected into the dox-inducible, nSR100-expressing 293T cell line. In the absence of dox induction, the neural exon is skipped, whereas it is efficiently included in “wild-type” (W) reporter transcripts when nSR100 expression is induced (Figure 3B). In contrast, in the mutant (M) reporter, nSR100-mediated exon inclusion is severely abrogated. Similar results were obtained when introducing G to A point mutations into UGC motifs in additional minigene reporters containing nSR100-regulated exons from the MEF2C and MEF2D genes (Figure S3E).
The combined analyses of cis-motifs associated with a network of conserved nSR100-dependent AS, PAR-iCLIP-Seq data, and mutant minigene reporters, demonstrate that binding of nSR100 to UGC motifs proximal to 3′ splice sites is required for the regulation of an extensive program of neural-specific AS.
Distinct features of core splicing signals associated with nSR100-dependent AS
Next, we examined whether properties of the core splicing signals associated with nSR100 target exons, including the 5′ splice site, 3′ splice site, polypyrimidine tract region, and branch site, are significantly different from the corresponding signals associated with control alternative exons that have matching PSI levels. Using maximum entropy scoring of splice site strength (Yeo and Burge, 2004), we observe that the 5′ splice sites of nSR100-regulated exons are moderately stronger than those of the control alternative exons (Figure 3C; median 5′ splice site scores: 8.8 [nSR100 exons] vs 8.0 [control exons], p = 6.6e-7, Wilcoxon rank sum test) and comparable in strength to those of constitutive exons (Figure S3F). In contrast, the 3′ splice sites of nSR100-regulated exons are significantly weaker than those of the control alternative exons (Figure 3C; median 3′ splice site scores: 5.3 [nSR100 exons] vs. 7.6 [control exons], p = 5.91e-14, Wilcoxon rank sum test). Similar differences between 3′ and 5′ splice site strengths are also observed when comparing sets of nSR100-regulated and control exons over a range of different PSI levels (Figure S3F).
The strength of a 3′ splice site is influenced by the distance between the branch site and the acceptor AG dinucleotide (Chiara et al., 1997; Reed, 1989). Relative to PSI-matched control exons, nSR100-regulated exons are associated with predicted branch sites that are more distal from 3′ AG acceptor sites, and are therefore expected to have weakened splicing activity (Figure 3D). The use of distal branch sites is further influenced by the quality of the polypyrimidine tract; increased length and pyrimidine content of the polypyrimidine tract, and its proximity to the AG dinucleotide, all contribute to increased 3′ splice site strength (Chiara et al., 1997; Merendino et al., 1999; Reed, 1989). Although nSR100-regulated exons are associated with longer polypyrimidine tracts (Figure 3D), there is a lower density of pyrimidine nucleotides immediately upstream of the AG acceptor site relative to the control exons (Figures 3D, S3G), in part due to the insertion of UGC motifs. Collectively, this unique arrangement of cis-features is expected to result in skipping of the nSR100 regulated exons when nSR100 is absent (i.e. in non-neural cells).
To directly test whether nSR100-binding UGC motifs weaken 3′ site recognition, we constructed minigene reporters containing an nSR100-dependent human DAAM1 neural exon and 300 nucleotides of flanking intronic sequence, with and without G to C point mutations in two neighboring UGC motifs located between the polypyrimidine tract and 3′ acceptor site. Since the murine Daam1 nSR100-regulated exon is repressed by Ptbp1 and Ptbp2 (Calarco et al., 2009), we also tested the effect of these point mutations following knockdown of these factors. When the “wild-type” (W) reporter was transfected into the dox-inducible nSR100-expressing 293T cell line, we observed nSR100-dependent exon inclusion (Figure 3E, lanes 1-2). In the presence of PTBP1/2 but absence of nSR100 expression, the neural exon is efficiently skipped, whereas it shows modest inclusion when PTBP1/2 are depleted (Figure 3E, lanes 3-4). In contrast, in the mutant (M) reporter, there is a pronounced switch to inclusion of the neural exon in the absence of nSR100, which is further enhanced upon PTBP1/2 depletion (Figure 3E, lanes 5-6). Similarly, we observed increased exon inclusion when either deleting or introducing a G to C mutation in the UGC motif adjacent to the MEF2D exon (Figure 3E, bottom lanes 5-8). These results confirm that UGC motifs provide a dual function of weakening 3′ site recognition leading to exon skipping of neural exons in non-neural cells, while forming intronic splicing enhancers (ISEs) that mediate efficient nSR100-dependent exon inclusion in neural cells.
nSR100-interacting co-factors that function in neural exon inclusion
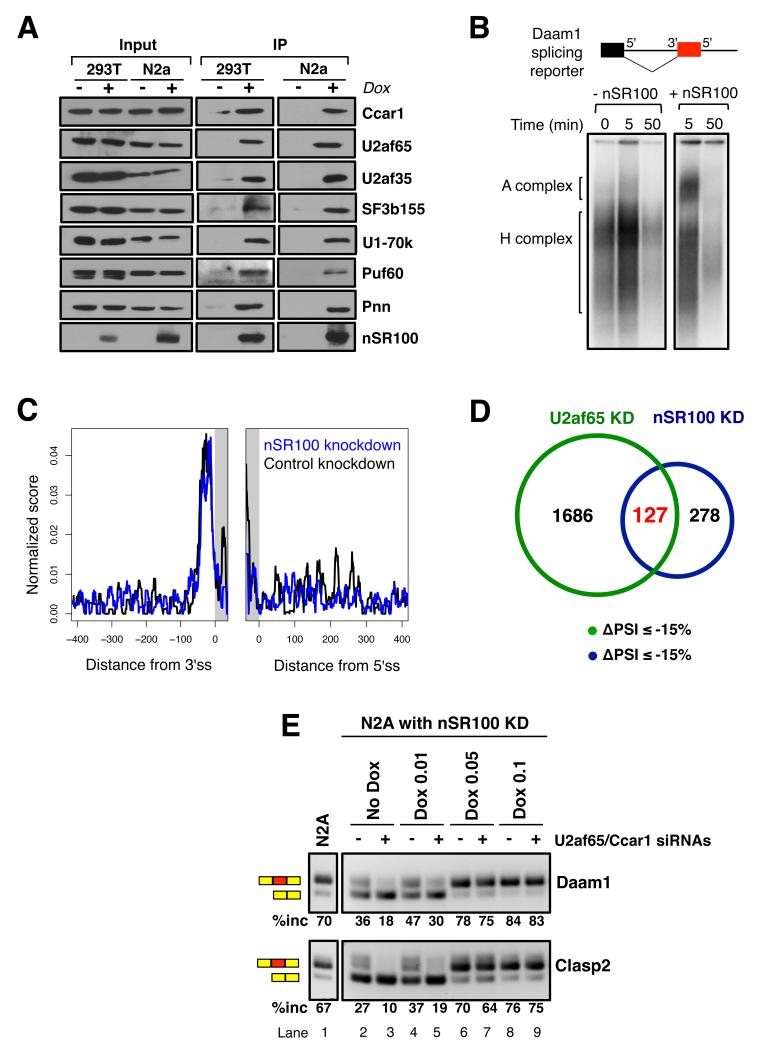
Immunoaffinity purification coupled to mass spectrometry (AP-MS) was next used to identify proteins that may mediate nSR100 regulatory activity. Complexes associated with Flag-tagged nSR100 expressed under dox-inducible control in 293T and N2A cells were immunoprecipitated with anti-Flag antibody in the presence of RNase and DNase, and then subjected to ion-trap MS. We scored 84 and 117 proteins in 293T and N2A cells, respectively, as candidate nSR100 partner proteins (Table S2), with 24 proteins in common between these datasets (p<0.0001, Fisher’s exact test). Gene Ontology analysis revealed that nSR100 interacting proteins are significantly enriched in “mRNA splicing” (FDR q-value = 1.72e-11). Co-immunoprecipitation western blot analysis validated all (7/7) tested Flag-nSR100 interaction partners in 293T and N2A cells (Figure 4A). Moreover these interactions were also validated in human neural Weri-Rb1 cells using an antibody specific for endogenous nSR100 protein (Figure S4A).
Figure 4. nSR100 interaction partners are enriched in early-acting splicing complex proteins.
(A) Co-immunoprecipitation western blot assays validating AP-MS-detected interactions between nSR100 and binding partners in 293T and N2A cells, with and without dox-induced Flag-nSR100 expression. Flag-nSR100 complexes were immunoprecipitated with anti-Flag antibody and blots were probed with antibodies specific for the interaction partners, as indicated.
(B) Native agarose gel electrophoresis of pre-spliceosomal complexes. A radiolabeled Daam1 reporter was incubated with Weri-Rb1 splicing extracts in the presence of ATP at 30 °C for the indicated lengths of time, and the complexes were run out on a 2% agarose gel.
(C) U2af65 RNA binding map showing the mean, normalized density of crosslinked sites in 400 nucleotide windows encompassing nSR100-regulated exons in N2A cells expressing nSR100-targeting (blue), or control (black) shRNAs; ss, splice site.
(D) Venn diagram representing the overlap between AS events promoted by U2af65 and nSR100. Exons displaying increased skipping upon U2af65 or nSR100 knockdowns were compared (ΔPSI ≤ -15%).
(E) A stable N2A cell line expressing nSR100-targeting shRNAs and an shRNA-resistant, dox-inducible nSR100 cDNA was transfected with non-targeting control siRNA (lanes 2, 4, 6, 8) or U2af65 and Ccar1 siRNAs (lanes 3, 5, 7, 9). nSR100 expression was induced by titrating dox concentration from 0 μg/ml (lanes 2-3) to 0.1 μg/ml (lanes 8-9). RT-PCR assays were used to monitor changes in inclusion levels of nSR100-regulated exons upon altering U2af65, Ccar1 and nSR100 levels. Lane 1 shows splicing levels in a parental N2A cell line for comparison. See also Figure S4 and Tables S2 and S3.
Among the detected nSR100 interaction partners are the two subunits of the U2 snRNP auxiliary splicing factor, U2AF65/U2AF2 and U2AF35/U2AF1, which bind directly to the polypyrimidine tract and 3′ splice site AG dinucleotide, respectively (Merendino et al., 1999; Wu et al., 1999; Zamore et al., 1992; Zorio and Blumenthal, 1999), and promote U2 snRNP binding to the pre-mRNA branch site during spliceosome assembly (Ruskin et al., 1988). Spliceosomal proteins detected in nSR100 complexes include multiple U2 snRNP (SF3) subunits, U1 snRNP-specific 70K protein (U1-70K), the splicing co-activator complex subunits SRm160/SRRM1 and SRm300/SRRM2 – which function in bridging interactions between U1 and U2 snRNPs across introns, and CCAR1, which interacts with U2AF65 (Hegele et al., 2012).
Since the interaction partners are enriched in early spliceosomal components, we used native gel electrophoresis (Das and Reed, 1999) to investigate whether nSR100 enhances formation of the U2 snRNP-containing pre-spliceosomal ‘A’ complex (Figure 4B). Complex assembly was assayed using an in vitro transcribed, radiolabeled, two-exon splicing reporter containing the Daam1 neural exon, and splicing extracts from neural Weri-Rb1 cells. In the presence of limiting amounts of splicing extract, complex formation was inefficient. In contrast, addition of purified recombinant nSR100 promoted formation of a complex that displays hallmark characteristics of A complexes, including the requirement for ATP, and an accumulation after incubation (at 30 °C) for 5 min, but disappearance after prolonged incubation periods (Figure 4B and data not shown). Taken together with results from analyzing cis-elements associated with nSR100 regulation and interacting factors, the results suggest that nSR100 promotes neural exon inclusion at suboptimal 3′ splice sites by promoting the formation of prespliceosomal complexes.
Since nSR100 interacts with both U2AF subunits, we investigated whether its binding to intronic enhancers may promote exon inclusion by recruiting U2AF65 to weak polypyrimidine tracts. In U2af65 PAR-iCLIP experiments, the average crosslinking density of U2af65 adjacent to nSR100-regulated targets did not show an appreciable difference upon nSR100 depletion in N2A cells compared to the control knockdown (Figure 4C). Furthermore, using U2AF65 iCLIP data from HeLa cells (Zarnack et al., 2013), in which nSR100 is not expressed and nSR100-regulated exons are skipped, we detected widespread crosslinking of U2AF65 to the polypyrimidine tracts upstream of nSR100 target exons (Figure S4B). Therefore, binding of U2AF65 to polypyrimidine tracts upstream of nSR100 target exons occurs in the absence of nSR100, yet is not sufficient to promote exon inclusion.
We next investigated whether U2af65 participates in the inclusion of nSR100-target exons. Since U2af65 interacts with Ccar1 (Figure S4C), and the role of Ccar1 in regulated AS is unclear, we also investigated the contribution of this factor to nSR100-dependent AS. We used siRNA pools to efficiently knockdown U2af65 (Figure S4D, ~75% depletion) and Ccar1 (Figure S4E, >90% depletion) individually or together in N2A cells, then assayed for effects on the inclusion of nSR100-dependent exons by RT-PCR. All (11/11) analyzed exons displayed increased skipping upon knockdown of U2af65 and/or Ccar1 (Figure S4F), with U2af65 depletion having a greater effect on exon inclusion in general. Thus, at least some nSR100-regulated exons are dependent on U2af65 and Ccar1 for inclusion.
To further assess the dependency of nSR100 target exons on U2af65, we performed an RNA-Seq analysis of AS changes following U2af65 knockdown in N2A cells. As expected for a factor that promotes splicing, knockdown of U2af65 resulted in approximately five times more exons displaying increased skipping than increased inclusion (|ΔPSI| ≥15%). Of the exons that undergo skipping upon nSR100 depletion in N2A cells, 31% also display skipping upon U2af65 knockdown (Figure 4D; p < 0.001, Chi-Square test; Table S3), thus indicating that nSR100-regulated neural exons have variable dependencies on U2af65 for inclusion.
We next determined whether increased expression of nSR100 promotes inclusion of exons that are skipped when U2af65 and Ccar1 are knocked down. N2A cell lines were generated that express nSR100-targeting shRNAs in which we additionally transfected inducible Flag-nSR100 cDNA vectors that are resistant to the shRNAs. Knockdown of nSR100 resulted in increased skipping of exons, and this was further exacerbated when U2af65/Ccar1 were co-depleted (Figures 4E, S4G; compare lane 1 with lanes 2 and 3). While low levels of dox-induced nSR100 expression did not rescue exon inclusion, higher levels of nSR100 expression were sufficient to restore inclusion of target exons to levels seen in control cells (Figures 4E, S4G; compare lanes 4-9 with lane 1). Therefore, while U2af65 interacts with nSR100 and promotes the inclusion of many nSR100-regulated target exons, nSR100 can activate efficient neural exon inclusion under conditions where U2af65 is rate limiting (see Discussion).
Widespread dominance of nSR100 over PTBP1-repressed neural exon splicing
Previously, we showed that Ptbp1, and to some extent Ptbp2, represses the inclusion of several analyzed neural exons that are positively regulated by nSR100 (Calarco et al., 2009). These findings, taken together with the observations in Figures 2A and S2A showing that intronic sequences upstream of nSR100 target exons are enriched for C/U-rich motifs (which resemble Ptbp1 binding sites), suggested that nSR100 may act more widely during neurogenesis to overcome the negative activity of Ptbp1.
To investigate this possibility, we used RNA-Seq following knockdown of Ptbp1 in N2A cells to identify neural exons that are differentially regulated by nSR100 and Ptbp1 (Figure 5A, ~85% depletion). Consistent with reports showing that Ptbp1 primarily functions to repress exon splicing (Boutz et al., 2007; Han et al., 2014; Llorian et al., 2010), knockdown of Ptbp1 resulted in approximately twice as many exons showing increased inclusion compared to skipping (Figure S5A; |ΔPSI| ≥15%). Furthermore, we observe significant overlap between the exons that are negatively regulated by Ptbp1 and positively regulated by nSR100; 130 (~30%) of nSR100-dependent exons are also repressed by Ptbp1 (p < 0.0001, Chi-square test), of which ~90% are neural-enriched (Figures 5B, 5C, S5B; Table S4). In contrast, only three exons are repressed by both proteins.
Figure 5. Opposing regulation of neural exons by PTBP1 and nSR100.
(A) N2A cells were transfected with control or Ptbp1 siRNAs, and RT-PCR assays and western blots were used to confirm Ptbp1 knockdown. Tubulin and gapdh detection was used to control for loading.
(B) Venn diagrams showing the overlap between AS events repressed by Ptbp1 and promoted by nSR100 (left), or repressed by Ptbp1 and nSR100 (right) in N2A cells. Exons displaying increased inclusion upon Ptbp1 depletion (ΔPSI ≥ 15%) were compared with those that change upon nSR100 knockdown.
(C) RT-PCR assays monitoring changes in inclusion levels of target exons upon knockdown of Ptbp1 or nSR100 in N2A cells.
(D) Western blots monitoring nSR100 and Ptbp1 protein levels in mouse cortex tissues during development (E – embryonic, P – postnatal). Tubulin detection was used as a loading control.
(E) Line graphs showing the relative levels of gene expression of nSR100 (blue), Ptbp1 (orange), and the median PSI levels (red) of alternative exons subjected to opposing regulation by Ptbp1 and nSR100 during in vitro differentiation of embryonic stem cells (ESC) to cortical glutamatergic neurons. NESC, neuroepithelial stem cells; Radial NP, radial neural progenitors; DIV, days in vitro. See also Figure S5 and Table S4.
To establish when during development nSR100 might act in opposition to Ptbp1 to promote its program of neural-specific AS, we compared the levels of the two proteins in mouse cortex tissues at different developmental stages (Figure 5D). As shown previously (Zheng et al., 2012), Ptbp1 protein is most abundant at embryonic stage day 12 (E12) when the cortex is populated by mostly undifferentiated neural progenitor cells, and then rapidly decreases after the onset of neurogenesis (~E12.5). In contrast, nSR100 protein levels are very low at E12, but readily detected at E15 and at subsequent time points (to postnatal day 3, P3). Analysis of RNA-Seq data generated from in vitro differentiation of mouse embryonic stem cells (ESCs) to cortical glutamatergic neuronal cells (Hubbard et al., 2013) also revealed dramatic changes in the expression patterns of nSR100 and Ptbp1, with nSR100 showing a marked increase from almost no expression in ESCs to its highest level at approximately day 7 of neuronal differentiation, with a subsequent decline in expression as neurons mature (Figure 5E). In contrast, Ptbp1 expression is highest in ESCs and then steadily decreases to almost no expression in mature neurons. Both genes show maximal co-expression in neural precursor cells and immature neurons. Moreover, PSI levels of neural exons regulated in the opposite manner by Ptbp1 and nSR100, as detected in the RNA-Seq analysis described above, showed changes that are consistent with the mRNA expression dynamics and antagonistic activities of these proteins during differentiation (Figures 5E, S5C).
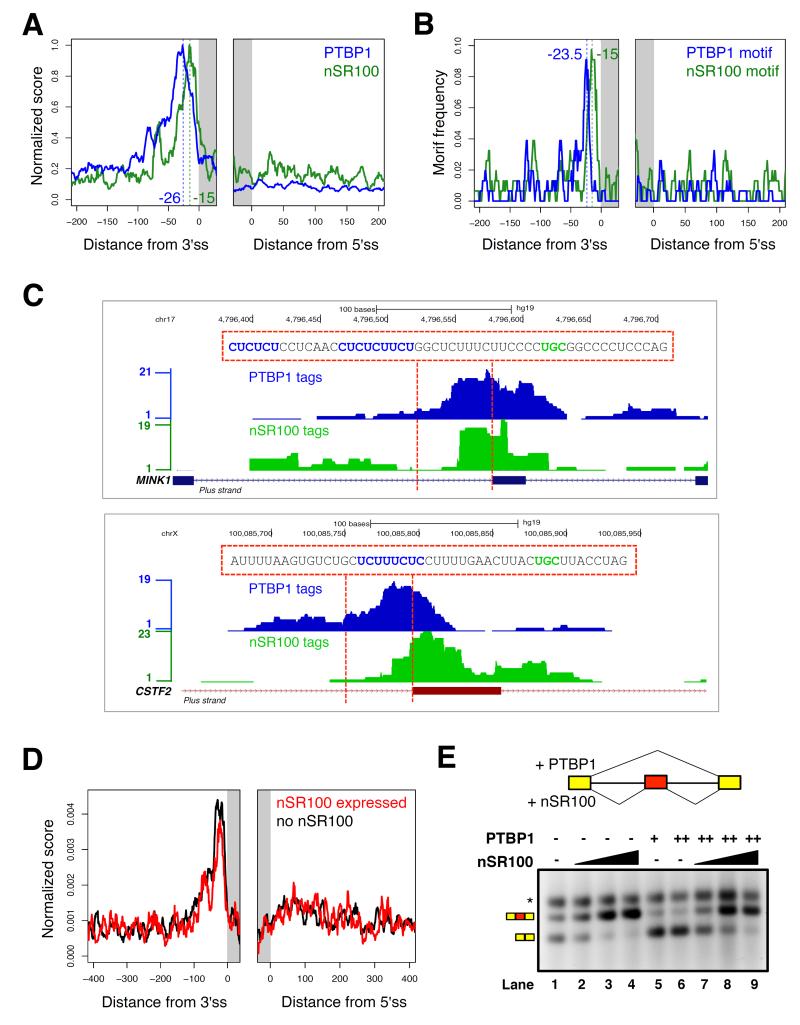
To investigate the mechanism by which nSR100 positively regulates PTBP1-repressed exons, we performed a PAR-iCLIP analysis to globally map sites of PTBP1 binding in 293T cells (Figure S6A). PTBP1 preferentially cross-linked to intronic sequences enriched in C/U-rich motifs upstream of nSR100 target exons (Figure S6B). When directly comparing the in vivo binding profiles of PTBP1 and nSR100 in 293T cells, these factors have crosslink peaks -26 and -15 nucleotides upstream of 3′ splice sites, respectively (Figures 6A, 6C). Moreover, plotting the distributions of PAR-iCLIP PTBP1 and nSR100 motifs within 200 nucleotide windows overlapping nSR100-regulated exons captured similar profiles (Figures 6B, 6C), which are also remarkably similar to the results of the PEAKS analysis (Figures 2A, S2B). In contrast, for exons that are repressed by PTBP1 but not activated by nSR100, PTBP1 forms crosslink peaks -10 nucleotides from the 3′ splice site (Figure S6C). Therefore, PTBP1 binding to intronic sequences upstream of nSR100-dependent exons is, on average, shifted by approximately 15 nucleotides upstream of the 3′ splice site. This unique motif arrangement likely evolved to accommodate the aforementioned dual, antagonistic regulation of neural-specific exons by nSR100 and PTBP1. It is possible that binding of nSR100 to UGC motifs -15 nucleotides from 3′ splice sites results in physical displacement of proximal PTBP1 binding. On the other hand, nSR100 may overcome the negative activity of PTBP1 without affecting PTBP1 binding to pre-mRNA. To distinguish between these possibilities, we analyzed the PTBP1 binding profile in vivo, with and without expression of nSR100. Induction of nSR100 in 293T cells did not appreciably affect the crosslinking density or positional binding preference of PTBP1 (Figures 6D, S6D, S6E), indicating that nSR100 can overcome the repressive effects of PTBP1 without generally interfering with its binding.
Figure 6. nSR100 directly overcomes PTBP1-mediated repression to promote exon inclusion.
(A) Merged RNA binding map showing the mean, normalized density of PTBP1 (blue) and nSR100 (green) crosslinked sites in 200 nucleotide windows encompassing conserved nSR100-regulated exons in 293T cells; ss, splice site.
(B) Distribution of PTBP1 and nSR100 binding motifs in 200 nucleotide windows surrounding conserved nSR100 target exons in human.
(C) Representative genome browser tracks showing the raw density of PTBP1 (blue) and nSR100 (green) PAR-iCLIP tags flanking the MINK1 (top) and CSTF2 (bottom) target exons. Genomic sequences between the dotted lines are displayed with binding sites for PTBP1 (blue) and nSR100 (green) highlighted.
(D) RNA binding maps of the mean, normalized density of PTBP1 crosslinked sites surrounding conserved nSR100-regulated exons in control uninduced (black) and dox-induced nSR100-expressing (red) 293T cells.
(E) In vitro splicing assay monitoring AS of the Daam1 neural exon in the presence and absence of purified nSR100 and/or PTBP1 proteins in Weri-Rb1 splicing extracts. The splicing reporter consists of constitutively spliced 5′ and 3′ MINX exons and the Daam1 neural exon flanked by its native intron sequences. Amount of proteins used: Lane 1: no protein; lanes 2, 3 and 4: 50, 100 and 200 ng nSR100, respectively; lanes 5 and 6: 100 and 200 ng PTBP1, respectively; lanes 7, 8 and 9: 200 ng PTBP1 and 50, 100 or 200 ng nSR100, respectively. Asterisk, non-specific band. See also Figure S6.
Finally, to investigate whether nSR100 directly promotes neural exon inclusion in the presence of PTBP1, we performed an in vitro splicing assay using a pre-mRNA substrate containing the nSR100-dependent Daam1 neural exon, and recombinant PTBP1 and nSR100 proteins (Figure 6E, S6F). Incubation of the pre-mRNA in Weri-Rb1 splicing extracts resulted in high inclusion of the neural exon, which was further stimulated by addition of nSR100 protein (Figure 6E). In contrast, addition of increasing amounts of PTBP1 resulted in efficient skipping of the exon. However, simultaneous addition of nSR100 and PTBP1 resulted in enhanced inclusion of the exon.
Collectively, our findings demonstrate that the levels of nSR100 and Ptbp1 are tightly and dynamically controlled during nervous system development, such that nSR100 can directly counteract Ptbp1 activity to ensure the activation of a network of neural exons with extensive roles in the formation of the vertebrate nervous system.
DISCUSSION
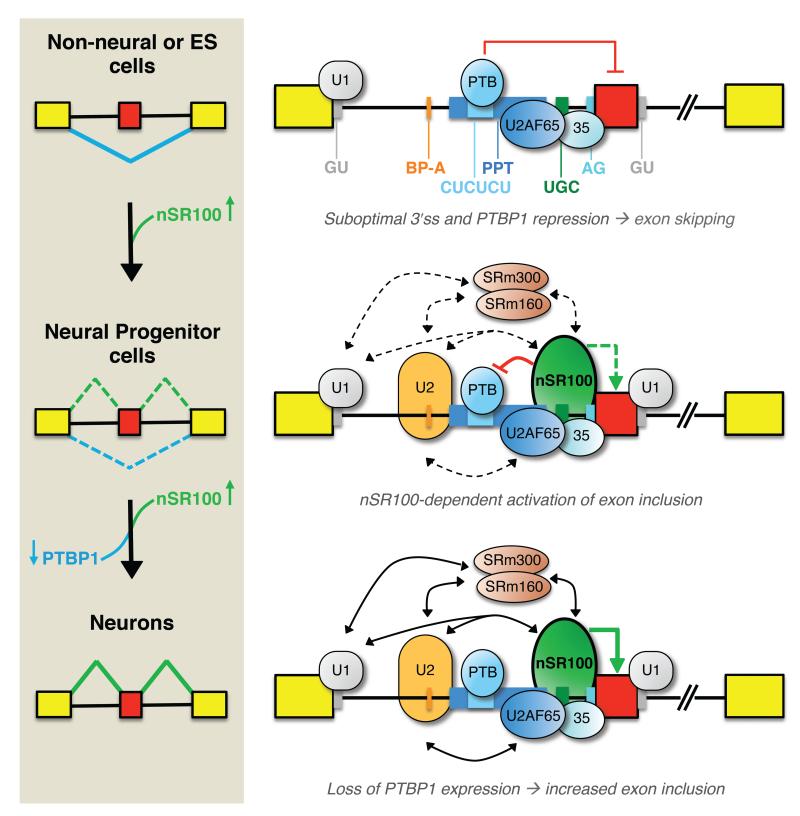
Although AS is particularly widespread in the nervous system, the mechanisms underlying the regulation of neural-specific exon networks are not well understood. We define a novel regulatory mechanism by which nSR100/SRRM4 activates the inclusion of a large program of conserved brain-enriched exons during neurogenesis. A key feature of this mechanism is the unique arrangement of intronic cis-elements upstream of target exons that control 3′ splice site recognition, including predicted branch sites that are more distal to acceptor sites, a lower density of pyrimidine residues upstream of the acceptor sites, and the insertion of UGC-containing ISEs. Remarkably, the guanosine nucleotide in these ISEs is sufficient to weaken 3′ splice site recognition and cause exon skipping of at least some target exons when nSR100 is absent. Combined with the additional elements negatively controlling 3′ splice site selection (see below), the dual function of the ISE in promoting efficient exon inclusion in neural cells and exon skipping in non-neural cells provides an effective mechanism for ensuring strong cell-type specificity in the differential regulation of the nSR100 neural exon network.
The mechanism by which nSR100 regulates neural exon inclusion bears interesting similarities with a previously proposed “coactivator model” for enhancer-dependent splicing. In this model, a complex of the SR-related proteins SRm160 and SRm300, which we detect as nSR100 interacting proteins, promotes splicing by forming cross-intron and cross-exon interactions involving U1 snRNP bound to the 5′ splice site, U2 snRNP bound to the branch site, and SR family proteins bound to exonic splicing enhancer (ESE) sequences (Eldridge et al., 1999). Moreover, SRm160/300 and other SR proteins have been shown to promote ESE-dependent splicing through interactions that are independent of those required for binding of U2AF65 to the polypyrimidine tract (Kan and Green, 1999; Li and Blencowe, 1999; Sabine Guth, 1999).
Similarly, while nSR100 associates with both U2AF subunits, its knockdown does not appear to alter in vivo crosslinking of U2AF65 to polypyrimidine tracts adjacent to target exons, and crosslinking of U2AF65 to these sites is detected in cells that do not express nSR100. Furthermore, induced expression of nSR100 in non-neural cells can promote efficient neural exon inclusion under conditions where U2AF65 is rate limiting for splicing of the same exons. It is therefore possible that nSR100, through its interactions with SRm160/300, U1-70K, and U2 snRNP components, also promotes efficient splicing in a manner that acts in conjunction with - but is largely independent of - interactions required for binding of U2AF to the polypyrimidine tract and 3′ AG acceptor site (Figure 7). However, it is noteworthy that purified recombinant nSR100 does not bind stably to transcripts in vitro (data not shown), indicating that one or more associated co-factors are important for its sequence-specific binding to RNA in vivo. Regardless of its specific mechanism(s) of action, a unique feature of nSR100 is that it acts in a strong positive manner to promote exon inclusion via binding to upstream ISEs, whereas all other tissue-regulated AS factors characterized to date repress exon inclusion when binding to upstream intronic elements.
Figure 7. Mechanistic model for nSR100-dependent regulation of neural exon alternative splicing.
Alternative exons in the nSR100-regulated network are associated with a unique arrangement of cis-elements that weaken 3′ splice sites, and act in conjunction with negative regulation mediated by PTBP1 to cause skipping of target exons in non-neural cells. When nSR100 is expressed in differentiating neural precursors and mature neurons it binds to intronic enhancers proximal to 3′ splice sites, and also interacts with multiple early-acting spliceosomal components, to promote exon inclusion. These interactions are sufficient to potently outcompete PTBP1-mediated repression. As neurons develop, PTBP1 is no longer expressed, enabling maximal nSR100-dependent neural exon inclusion.
nSR100 also provides strong dominant-positive activity over Ptbp1 during neurogenesis through its multifaceted cis- and trans-acting interactions. While nSR100 promotes the inclusion of an exon required for the neuronal expression of the Ptbp1 paralog, Ptbp2 (Calarco et al., 2009), which is thought to contribute to neural exon inclusion (Makeyev et al., 2007; Zheng et al., 2012), Ptbp2 expression is not sufficient for the activation of nSR100 target exons. Rather, nSR100 binding effectively overcomes PTBP1-mediated repression without displacing it from its adjacent, upstream binding sites (Figure 7). Not only does this bipartite system of enhancer and repressor afford tight control over neural AS, it also enables the rapid transition between states of exon repression and activation, as there is no requirement for displacement or turnover of PTBP1. This feature of nSR100 is also apparent from its pronounced and dynamic expression changes during neuronal differentiation. Notably, nSR100 and its target exons display striking increases in expression and inclusion, respectively, at early stages of differentiation when PTBP1 is still expressed. As neurons mature, nSR100 expression shows a partial decline while the inclusion levels of its target exons are maintained. At this stage, the level of nSR100 expression is presumably sufficient for full target exon inclusion levels because PTBP1 is no longer expressed (see Introduction).
In conclusion, the identification and characterization of multifaceted interactions involving an expanded network of conserved nSR100 target exons have provided insight into the mechanism by which an extensive program of AS is activated during neurogenesis. The identification of this expanded network of exons, and of its associated global regulatory elements, further provide a valuable resource for future investigations into the molecular basis by which coordinated AS changes contribute to the formation and function of the vertebrate nervous system.
EXPERIMENTAL PROCEDURES
Cell lines
Generation of N2A cell lines expressing nSR100/SRRM4- or GFP-targeting shRNAs, and the 293T cell line with inducible Flag-nSR100 expression have been previously described (Calarco et al., 2009; Raj et al., 2011). To establish inducible Flag-nSR100 N2A cell lines, the full-length mouse nSR100 ORF with an N-terminal 3xFlag tag was cloned into a PiggyBac vector (gift from Dr. Andras Nagy, Lunenfeld-Tanenbaum Research Institute). N2A cells were transfected with nSR100-PiggyBac cDNA vector and the transposase plasmid, pBase (gift from Dr. Andras Nagy). Stable clonal populations were then derived. For one set of experiments, the clonal N2A Flag-nSR100 line was additionally transduced with lentiviruses expressing nSR100-targeting shRNA (Addgene Plasmid #35174). nSR100 expression was induced using 2 μg/ml dox for 24 h unless otherwise stated.
siRNA transfections
N2A cell lines were transfected with 10 nM of U2af65, Ccar1 or Ptbp1 ON-TARGETplus siRNA pools (Thermo Scientific-Dharmacon) using RNAiMax (Life Technologies), as recommended by the manufacturer. 293T cells were transfected with 25 nM each of PTBP1 and PTBP2 siRNAs (Sigma-Aldrich) using RNAiMax. A non-targeting siRNA pool was used as control. N2A and 293T cells were harvested 48 h and 72 h, respectively, post-transfection.
RNA extraction and RT-PCR assays
Total RNA was extracted from cells using TRI Reagent (Sigma-Aldrich) and RT-PCR assays were performed using the OneStep RT-PCR kit (Qiagen) as per the manufacturer’s instructions. Reaction products were separated on a 1%-2.5% agarose gels, or on 6% denaturing polyacrylamide gels.
RNA-Seq Analyses
RNA-Seq libraries were generated from Poly(A)+ mRNA and sequenced using the Illumina Hi-Seq 2000 or 2500 machine to generate ~100 million x 100 bases, paired end reads. Reads were mapped to the mouse (mm9) or human (hg19) genomes. Details of the data analysis pipeline are provided in the Supplemental Experimental Procedures.
PAR-iCLIP experiments
PAR-iCLIP experiments were performed as described previously (Huppertz et al., 2014) with anti-Flag, anti-U2af65 and anti-PTBP1 antibodies. Further details and computational analyses are described in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Gonatopoulos-Pournatzis, S. Gueroussov, H. Han, M. Quesnel-Vallières, A. Lapuk, and J. Calarco for comments on the manuscript, A. Sherker for experimental assistance, and J. Valcárcel for helpful discussions. We also gratefully acknowledge D. Torti and D. Leung in the Donnelly Sequencing Centre for sequencing samples. This work was supported by grants from the Canadian Institutes of Health Research (B.J.B. and A.-C.G.), a grant from the ORF (B.J.B. and A.-C.G.), and a Natural Sciences and Engineering Research Council of Canada (NSERC) Polyani Award (B.J.B). E.E. was supported by grants BIO2011-23920, Consolider RNAREG (CSD2009-00080), a visiting scholar grant “Salvador Madariaga” from the Spanish Government, and by the Sandra Ibarra Foundation for Cancer; B.R. was supported by NSERC Alexander Graham Bell and University of Toronto Open fellowships; M.I and U.B were supported by Human Frontiers Science Program Organization fellowships; and U.B. was also part supported by an EMBO fellowship. A.-C.G holds the Canada Chair in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics. B.J.B. holds the Banbury Chair in Medical Research at the University of Toronto.
Footnotes
Accession Numbers The Gene Expression Omnibus accession number for the RNA-Seq and PAR-iCLIP datasets is GSE57278. The AP-MS datasets are deposited in the MassIVE repository and have been assigned the accession number MSV000078609.
REFERENCES
- Akhtar MW, Kim M-S, Adachi M, Morris MJ, Qi X, Richardson JA, Bassel-Duby R, Olson EN, Kavalali ET, Monteggia LM. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS ONE. 2012;7:e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Änkö M-L. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin. Cell Dev. Biol. 2014;32:11–21. doi: 10.1016/j.semcdb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Çolak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Bellora N, Farré D, Mar Albà M. PEAKS: identification of regulatory motifs by their position in DNA sequences. Bioinformatics. 2007;23:243–244. doi: 10.1093/bioinformatics/btl568. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin C-H, Chawla G, Ostrow K, Shiue L, Ares M, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O'Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of Vertebrate Nervous System Alternative Splicing and Development by an SR-Related Protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Chiara MD, Palandjian L, Feld Kramer R, Reed R. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. Embo J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. Rna. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge AGA, Li YY, Sharp PAP, Blencowe BJB. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl. Acad. Sci. USA. 1999;96:6125–6130. doi: 10.1073/pnas.96.11.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JD, Barrios-Rodiles M, Çolak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM, et al. Tissue-Specific Alternative Splicing Remodels Protein-Protein Interaction Networks. Mol. Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp A-C, Munschauer M, et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Stoilov P, Linares AJ, Zhou Y, Fu X-D, Black DL. De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function. PLoS Comput. Biol. 2014;10:e1003442–e1003442. doi: 10.1371/journal.pcbi.1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U. Dynamic Protein-Protein Interaction Wiring of the Human Spliceosome. Mol. Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- Hubbard KS, Gut IM, Lyman ME, McNutt PM. Longitudinal RNA sequencing of the deep transcriptome during neurogenesis of cortical glutamatergic neurons from murine ESCs. F1000Res. 2013;2:35. doi: 10.12688/f1000research.2-35.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 2012;24:323–332. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Kan JL, Green MR. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang Y-J, Nakanishi N, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zheng S, Han A, Lin C-H, Stoilov P, Fu X-D, Black DL. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Blencowe BJ. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J. Biol. Chem. 1999;274:35074–35079. doi: 10.1074/jbc.274.49.35074. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian MM, Schwartz SS, Clark TAT, Hollander DD, Tan L-YL, Spellman RR, Gordon AA, Schweitzer ACA, la Grange, de PP, Ast GG, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 Promotes Neuronal Differentiation by Triggering Brain-Specific Alternative Pre-mRNA Splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino LL, Guth SS, Bilbao DD, Martínez CC, Valcárcel JJ. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Jahan I, Bonde G, Sun X, Hildebrand MS, Engelhardt JF, Smith RJ, Cornell RA, Fritzsch B, Bánfi B. A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS Genet. 2012;8:e1002966. doi: 10.1371/journal.pgen.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, O'Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ. Cross-Regulation between an Alternative Splicing Activator and a Transcription Repressor Controls Neurogenesis. Mol. Cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3′splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Guth Sabine, M. C, G. RK, V. J. Evidence for Substrate-Specific Requirement of the Splicing Factor U2AF35 and for Its Function after Polypyrimidine Tract Recognition by U2AF65. Mol. Cell. Biol. 1999;19:8263. doi: 10.1128/mcb.19.12.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW. Crossregulation and Functional Redundancy between the Splicing Regulator PTB and Its Paralogs nPTB and ROD1. Mol. Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon Y-S, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide Analysis of PTB-RNA Interactions Reveals a Strategy Used by the General Splicing Repressor to Modulate Exon Inclusion or Skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comp. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct Competition between hnRNP C and U2AF65 Protects the Transcriptome from the Exonization of Alu Elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O'Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012;15:381–3S381. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DAD, Blumenthal TT. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.