Abstract
The anaphase-promoting complex (APC/C) is a multimeric RING E3 ubiquitin ligase that controls chromosome segregation and mitotic exit. Its regulation by coactivator subunits, phosphorylation, the mitotic checkpoint complex, and interphase inhibitor Emi1 ensures the correct order and timing of distinct cell cycle transitions. Here, we used cryo-electron microscopy to determine atomic structures of APC/C-coactivator complexes with either Emi1 or a UbcH10-ubiquitin conjugate. These structures define the architecture of all APC/C subunits, the position of the catalytic module, and explain how Emi1 mediates inhibition of the two E2s UbcH10 and Ube2S. Definition of Cdh1 interactions with the APC/C indicates how they are antagonized by Cdh1 phosphorylation. The structure of the APC/C with UbcH10-ubiquitin reveals insights into the initiating ubiquitination reaction. Our results provide a quantitative framework for the design of experiments to further investigate APC/C functions in vivo.
The activities of the diverse proteins that orchestrate the sequential biochemical and morphological changes intrinsic to the cell division cycle are controlled through the integration of protein phosphorylation, proteolysis and changes in gene expression. Two cullin-RING E3 ubiquitin ligases, the APC/C and SCF, catalyze the ubiquitination of multiple cell cycle proteins to regulate their proteasome-mediated proteolysis. By regulating the ordered degradation of substrates such as cyclins, securin, mitotic kinases, and microtubule motors and assembly factors, the APC/C controls sister chromatid segregation, cytokinesis and the initiation of chromosome duplication 1-3.
The APC/C is a large assembly comprising 19 subunits 4. Its activity depends on the association of one of two coactivator subunits (either Cdc20 or Cdh1) that specify substrate recognition and stimulate the catalytic activity of APC/C – E2 complexes 4-7. Coactivators engage the APC/C through a conserved N-terminal C box motif and a C-terminal Ile-Arg (IR) tail 2,3. Cyclin-dependent kinase (CDK) phosphorylates the APC/C promoting Cdc20 association 8,9. In contrast, Cdh1 is negatively regulated by phosphorylation 8,10,11.
APC/C activity is repressed by the spindle assembly checkpoint (SAC) to ensure accurate sister chromosome segregation 3. The SAC effector, the mitotic checkpoint complex (MCC), inhibits mitotic APC/CCdc20 whereas Emi1 (early mitotic inhibitor1) inhibits APC/CCdh1 during interphase 12. Similar to the MCC, Emi1 blocks D box recognition by APC/C-coactivator complexes. However, Emi1 also antagonizes the two E2s, UbcH10 and Ube2S, that pair with the APC/C and which are responsible for catalyzing ubiquitin chain initiation and elongation, respectively 13-17.
A quantitative mechanistic understanding of the APC/C requires structural information to atomic resolution. We previously described the human APC/CCdh1-substrate complex (APC/CCdh1.Hsl1) from a cryo-EM reconstruction allowing definition of the overall architecture of the multi-subunit assembly 4. Still, the atomic structures of several large and most small APC/C subunits, and the regulatory N-terminal domain (NTD) of Cdh1 were unknown, which together with the flexibility of the catalytic module comprising the RING subunit Apc11 and the C-terminal domain of Apc2 (Apc2CTD) limited our understanding of the complex.
Here, we used recombinant APC/CCdh1 in complex with Emi1 (APC/CCdh1.Emi1) to determine an atomic model of the APC/C to 3.6 Å resolution (Extended Data Figs 1 and 2 and Extended Data Table 1). This structure, together with an APC/CCdh1 – UbcH10 complex, reveals details of the initiating ubiquitination reaction and how Emi1 inhibits UbcH10 and Ube2S 13,14.
Atomic structure of APC/CCdh1.Emi1
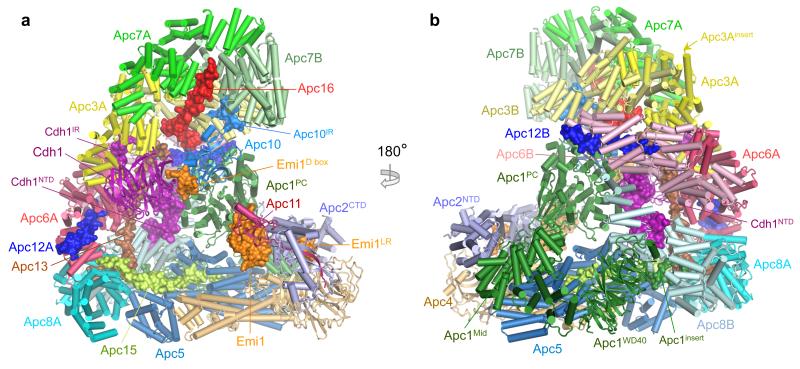
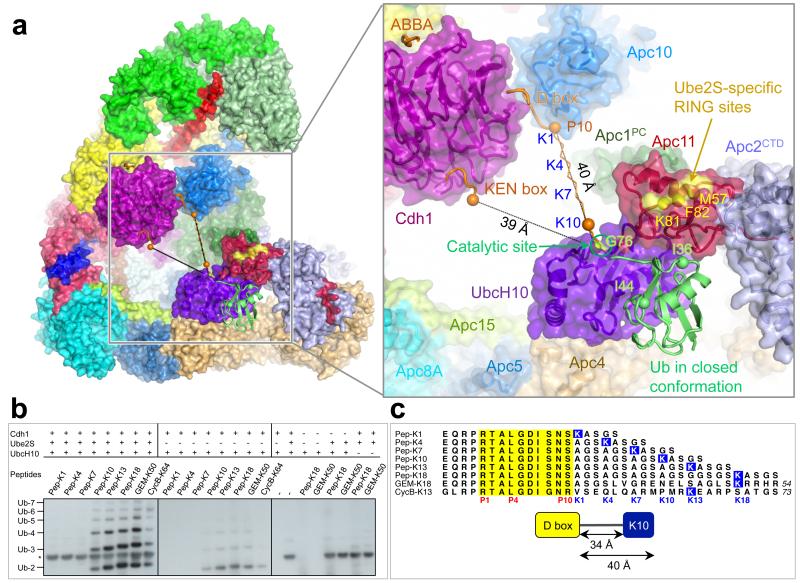
Figs 1a, b show two views of the atomic model of APC/CCdh1.Emi1 (Supplementary Video 1). A local resolution map indicates that most of the static regions of the complex are at ~3.2 Å resolution (Extended Data Fig. 1). Specific regions of the molecule, including the catalytic module and the Cdh1WD40 domain are more mobile and visualized at lower resolution (Extended Data Fig. 1).
Figure 1. EM reconstructions of the APC/CCdh1.Emi1 complex.
(a,b) Two views of the atomic structure of APC/CCdh1.Emi1. Large subunits are shown in cartoon, whereas the four small subunits, Emi1, Cdh1NTD, Cdh1IR and Apc10IR are shown as surface representations. Emi1 interacts with the substrate-recognition and catalytic modules.
Similar to APC/CCdh1.Hsl1 (Ref. 4), 60% of molecules are the coactivator-bound APC/CCdh1.Emi1 complex, with the remainder in the apo state. APC/CCdh1.Emi1 adopts the active conformation with the platform sub-domain, incorporating Apc2CTD-Apc11, in the upward position (Fig. 1a). However, in contrast with APC/CCdh1.Hsl1 where Apc2CTD-Apc11 is flexible 4, Emi1 stabilizes Apc2CTD-Apc11 by bridging it to Apc1 (Fig. 1a).
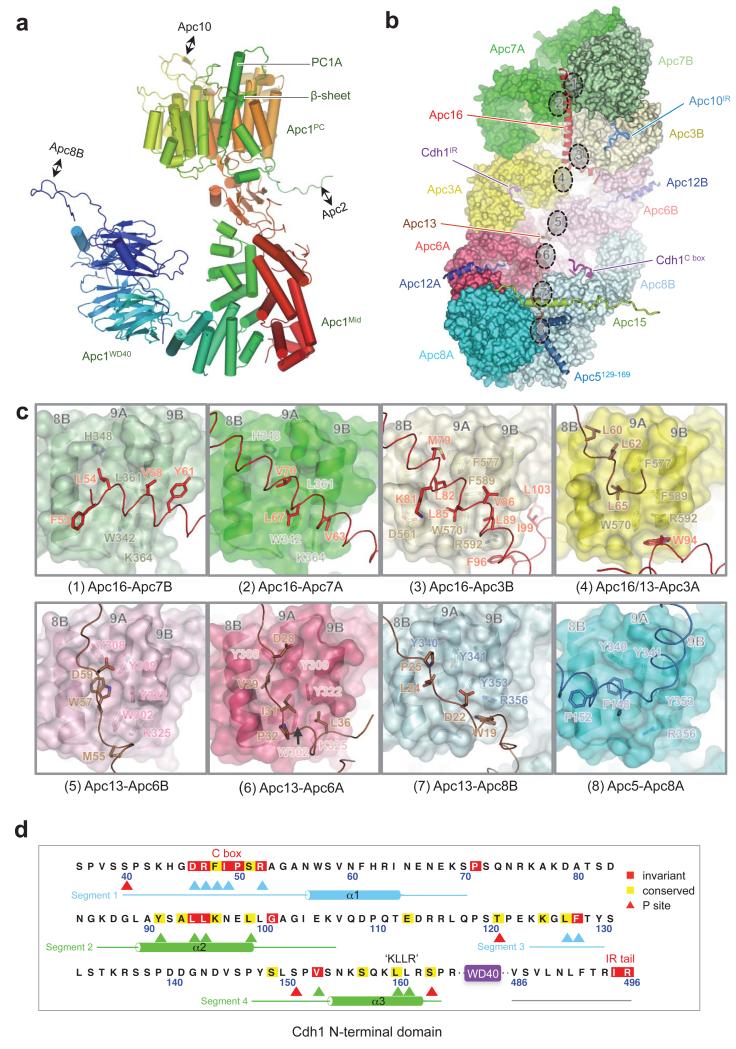
With the APC/CCdh1.Emi1 EM map at 3.6 Å resolution we built a complete atomic model of the complex. Existing crystal structures, complemented by newly determined structures of Apc4 and the N-terminal domain of Apc5 (unpublished data), together with homology models (Extended Data Tables 2 & 3), were used to guide building of most large APC/C subunits. For Apc1, fitting to the N-terminal Apc1WD40 domain and the densities N- and C-terminal to its central PC domain (Apc1PC) were performed ab initio. These two regions coalesce to form Apc1Mid that connects Apc1WD40 with Apc1PC and comprises an α-solenoid capped by a β-sandwich (Extended Data Fig. 3a). Apc13 and Apc16 of the TPR lobe interact with structurally homologous symmetry-related sites on the four TPR homo-dimers (Extended Data Figs 3b, c). The refined APC/CCdh1.Emi1 model has excellent stereochemistry and is in good agreement with the EM density map (Extended Data Fig. 2 and Extended Data Table 1).
Cdh1NTD binds to Apc1 and Apc8
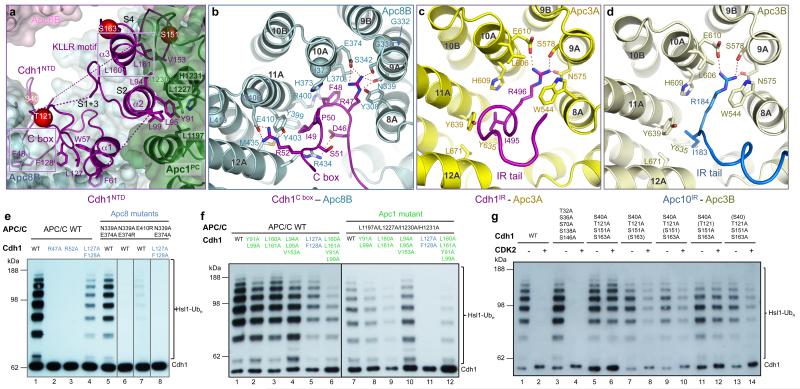
In the APC/CCdh1.Emi1 EM map, the Cdh1NTD-assigned region shows clearly resolved side chain features (Extended Data Fig. 1g). Four distinct segments of Cdh1NTD are organized into two mini-domains (Fig. 2a). Segments 1 and 3 interact with Apc8B, whereas segments 2 and 4 interact with Apc1PC. Segment 1 incorporates the seven-residue C box (residues 46-52) (Extended Data Fig. 3d), which through extensive contacts with Apc8 is a key determinate of Cdh1NTD – APC/C interactions. The C box adopts a hairpin conformation that inserts into the TPR superhelix of Apc8 (Figs 2a, b). Asp46, Arg47 and Arg52 of the C box form a network of electrostatic interactions, whereas Phe48 and Ile49 mediate non-polar contacts. Strikingly, Arg47 and Ile49 bind a site on Apc8 that is structurally homologous to the IR tail-binding site of Apc3, a paralog of Apc8 (Figs 2b-d). In both TPR proteins, three invariant residues (Asn339, Ser342 and Glu374 of Apc8) coordinate the Arg guanidinium group, with the Arg’s aliphatic moiety sandwiched between a conserved aromatic residue and an invariant Leu (Figs 2b-d). The aliphatic side chain of the C box-Ile49 is flanked by two tyrosines (both conserved in Apc3) and a Met (Leu in Apc3). Segment 3’s main role is to stabilize the conformation of segment 1 by contributing Leu127 and Phe128 to a cluster of hydrophobic residues at the Apc8 – Cdh1NTD interface (Fig. 2a). Mutation of both residues severely abrogates Cdh1 activity (Fig. 2e [lanes 1 and 4]).
Figure 2. The C box of Cdh1 and IR tails of Cdh1 and Apc10 interact with structurally related sites on Apc8 and Apc3.
(a) Cdh1NTD binds to Apc8 and Apc1. Segments 1 and 3 of Cdh1NTD including the C box interact with Apc8, whereas segments 2 and 4 interact with Apc1PC. Segments 1-4 are labelled as S1-4. Phosphorylation sites S40, S151 and S163 as red spheres. (b) Details of the C box-interactions. The R47 and I49 interaction sites are structurally related to the IR tail-binding sites for Cdh1 and Apc10 in Apc3 shown in (c, d). S. cerevisiae CDC23 (Apc8) temperature-sensitive mutations 23 map to the C box-binding site (Cα of mutant residues shown as spheres in (b)). (e) Mutation of C box-residues Arg47 and Arg52 eliminates APC/CCdh1 activity, as do mutation of Apc8 residues that interact with either Arg47 or Arg52. (f) At the Cdh1NTD – Apc1PC interface multiple residues cooperate to mediate APC/C – Cdh1 interactions. (g) S40, S151 and S163 mediate the negative regulation of Cdh1 by CDK phosphorylation.
Segment 2 forms a leucine zipper-like α-helical interface with two outer α-helices of Apc1PC, an interaction mediated by three conserved leucines (Fig. 2a). The α-helix of segment 4 aligns antiparallel to segment 2 (Fig. 2a). It includes the KLLR sequence (Extended Data Fig. 3d), structurally similar to the KILR motif of Cdc20NTD that interacts with Mad2 as a β-strand 18, and is implicated in mediating APC/C – Cdc20 interactions 19. Leu160 and Leu161 of the KLLR sequence, together with Val153 of segment 4, contribute to the hydrophobic interface of segment 2 and Apc1PC (Figs 2a, f).
The C-terminal ten residues of Cdh1 form the IR tail. This is well ordered and adopts a mainly α-helical conformation that inserts into the TPR superhelix of Apc3A (Fig. 2c and Extended Data Fig. 3b). The universally invariant C-terminal Arg496 contacts residues of Apc3A analogous to how Arg47 of the C box contacts Apc8 (Figs 2b, c). The adjacent Ile of the IR tail packs into a hydrophobic pocket on Apc3. Through its C-terminal IR residues, Apc10 interacts with a similar conformation to the symmetry-related IR tail-binding site on the Apc3 homo-dimer (Apc3B) (Fig. 2d).
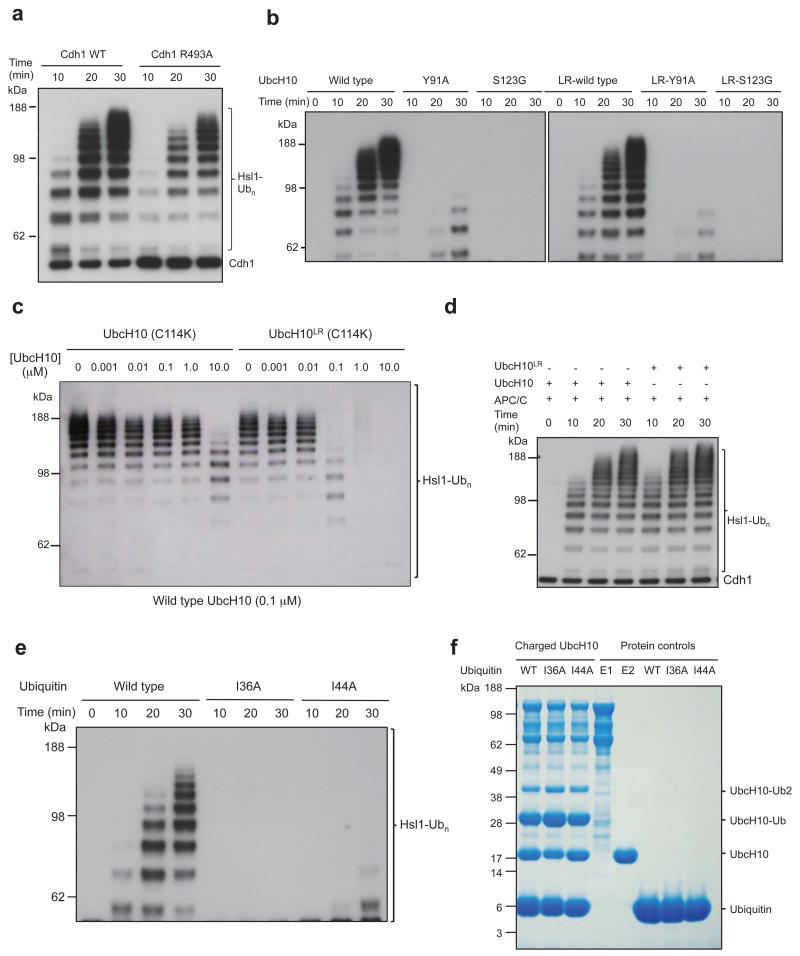
Both C box-Arg47 and its coordinating Asn339 residue (Fig. 2b) are required for coactivator binding to the APC/C 20-22, and the biochemically defined interaction of Cdc20NTD. Disruption of Apc3 residues Asn575 and Leu606 impaired coactivator binding 21. Furthermore, temperature-sensitive cell cycle mutations of S. cerevisiae CDC23 (Apc8) (Ref. 23) can be rationalized due to their disruption of the C box-receptor (Fig. 2b). In support of these previous data, mutating Asn339 and Glu374 of Apc8 that engage Arg47 abolished the capacity of Cdh1 to activate the APC/C (Fig. 2e). By contrast, mutation of the structurally related-Arg493 of the IR tail only reduced APC/CCdh1 activity (Extended Data Fig. 4a). Arg52 of the C box forms a salt bridge with Glu410 of Apc8 (Fig. 2b), and disrupting either residue eliminates APC/C activity (Fig. 2e).
To explore the basis for the negative regulation of coactivators by CDK phosphorylation 8,10,11, we first identified which of the nine potential CDK phosphorylation sites in Cdh1NTD account for this control. Four of these sites (S40, T121, S151, S163) (Extended Data Fig. 3d) are both necessary and sufficient to mediate CDK-regulation of Cdh1 (Fig. 2g, lanes 1-6), in agreement with 24. Although S40, S151 and S163 (but not T121) phosphorylation each contributes individually to a degree of Cdh1 inactivation (Fig. 2g, lanes 7-14), simultaneous phosphorylation of all sites was required to inactivate Cdh1 completely (Fig. 2g, lanes 1-4). Phosphorylation of these sites would destabilize Cdh1NTD – APC/C interactions through electrostatic repulsion and steric clashes (Fig. 2a). Ser40 is immediately N-terminal to the C box, whereas the side chains of Ser151 and Ser163 flank the KLLR sequence of segment 4 and contact Apc1PC (Fig. 2a).
Emi1 inhibits UbcH10 and Ube2S binding
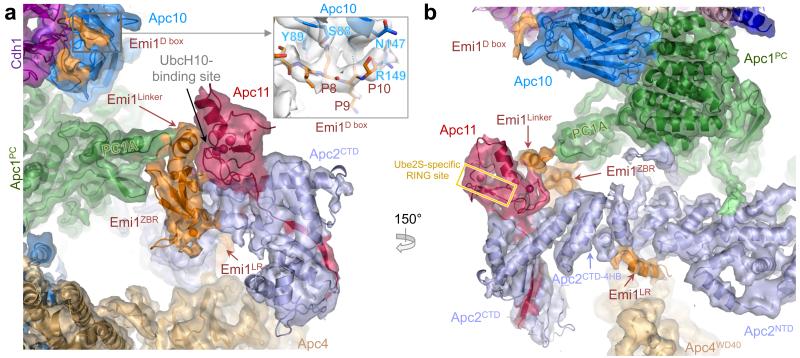
Four functional elements of Emi1 mediate APC/CCdh1 inhibition 13,14,25. A D box motif that occludes substrate recognition is connected through a linker to a zinc-binding region (ZBR) (Emi1ZBR) that interferes with UbcH10-dependent APC/C activity 13,14,25. A C-terminal LRRL sequence (LR tail: Emi1LR), identical to the LRRL motif required for Ube2S association with the APC/C 4,7,14, antagonizes Ube2S 13,14. In the APC/CCdh1.Emi1 structure we identified EM density corresponding to all four elements (Figs 3a, b). This agrees with a previous EM study at lower resolution that had assigned Emi1 density bridging the substrate recognition module and platform 13. In our structure, the Emi1-D box is engaged by the D box co-receptor of Cdh1 and Apc10 (Figs 1a and 3a). We identified EM density matching the In-Between-RING (IBR) architecture of Emi1ZBR (Ref. 13) associated with Apc2CTD-Apc11 (Figs 3a, b). The domain connects these subunits to an insertion within Apc1PC. In the EM map, part of the Emi1Linker forms an α-helix that packs against the Emi1ZBR β-sheet and docks onto the UbcH10-binding site of Apc11RING (Figs 3a, b). This blocks UbcH10 association with Apc11RING.
Figure 3. Interactions of Emi1 with Apc2CTD-Apc11RING and D box receptor of Cdh1 and Apc10.
(a) and (b) views of the EM density and underlying secondary structure. Emi1 elements are shown: D box (Emi1D-box); Emi1Linker and Emi1ZBR interconnects Apc1PC with Apc2CTD and Apc11RING. Both Emi1Linker and Emi1ZBR block the UbcH10-binding site on Apc11, but not the Ube2S-specific site 26. The C-terminal LRRL tail (Emi1LR) of Emi1 binds to Apc2CTD.
We assigned Emi1LR as a helical density feature to a site on Apc2CTD at the interface of its four-helix bundle and α/β sub-domain, adjacent to Apc4WD40 (Fig. 3b & Extended Data Fig. 2f), and in agreement with biochemical studies 14,26. Emi1ZBR does not contribute to inhibiting Ube2S-catalyzed ubiquitination reactions 13,14, and our structure shows that Ube2S-specific sites on Apc11RING (Ref. 26) are not blocked by Emi1ZBR (Fig. 3b).
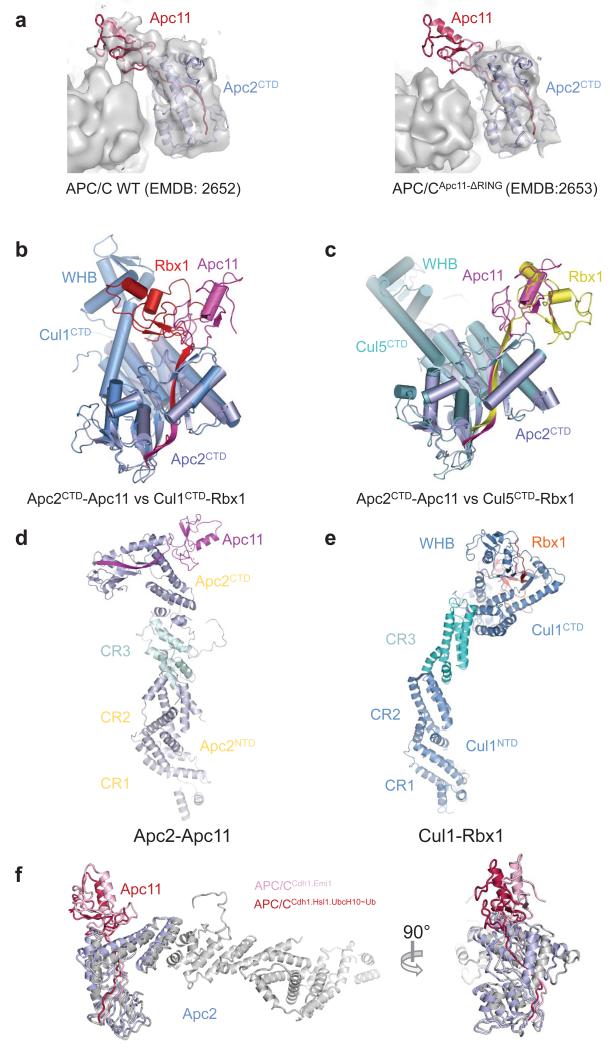
The Apc11RING-assigned density of APC/CCdh1.Emi1 was a perfect match to the crystal structure of Apc11RING (Ref. 26) (Figs 3a, b and Extended Data Figs 5a). Assignment of EM density to Apc2CTD-Apc11 indicated disorder of the Apc2CTD WHB sub-domain. The juxtaposition of Apc11RING and Apc2CTD is similar to the swung out conformation of Rbx1RING in activated Cul5-Rbx1 (Ref. 27) (Extended Data Figs 5b-e).
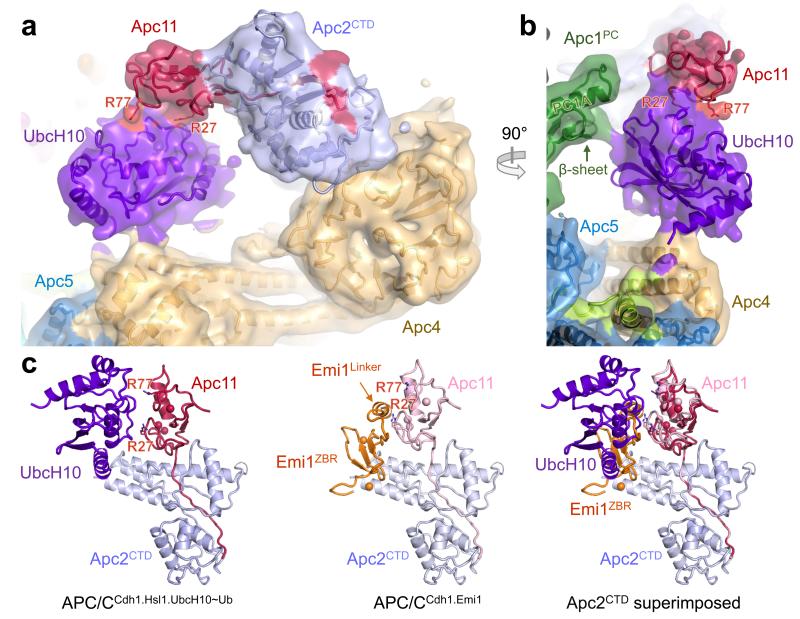
Apc11RING positions UbcH10-ubiquitin
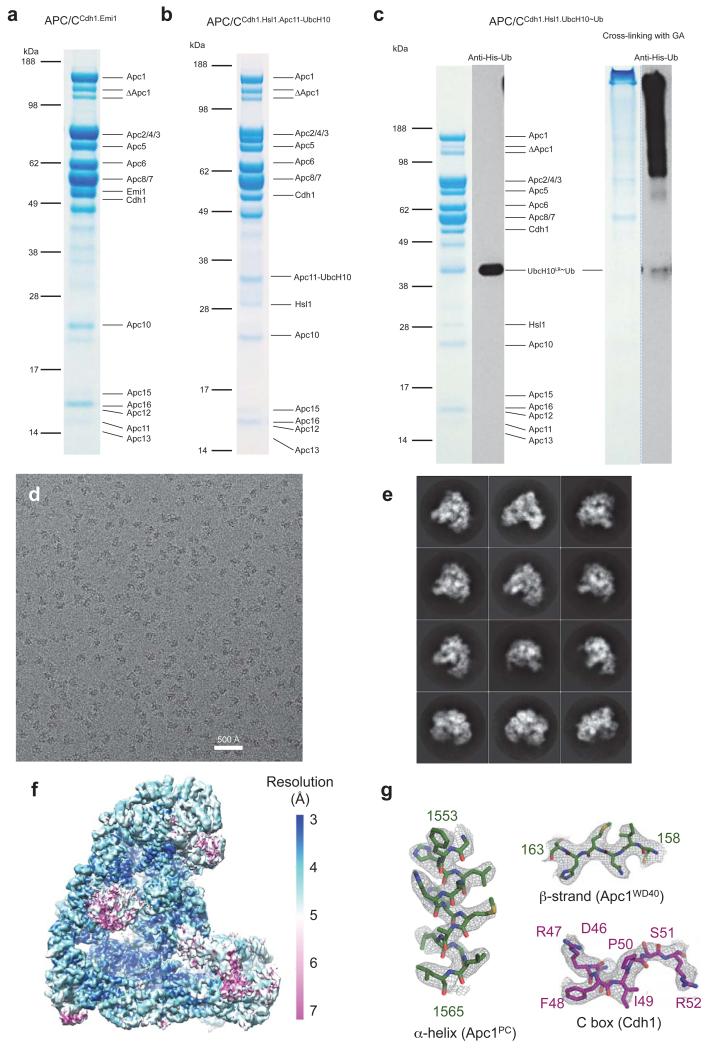
Apc11RING stimulates ubiquitination reactions catalyzed by the initiating E2s – UbcH10 and UbcH5, and the elongating E2 – Ube2S 26. Activation of UbcH10 involves a canonical interaction between its UBC domain and Apc11RING (Ref. 26), whereas for Ube2S, Apc11RING is repurposed to position the acceptor ubiquitin for modification by the Ube2S-ubiquitin conjugate 26. To understand substrate ubiquitination catalyzed by the APC/C – UbcH10 pair, we determined a 3D reconstruction of an APC/CCdh1-substrate complex (APC/CCdh1.Hsl1) with a UbcH10-ubiquitin conjugate. Because of the low affinity of APC/C – UbcH10-ubiquitin interactions 4, to facilitate generation of APC/CCdh1.Hsl1.UbcH10-Ub complexes suitable for cryo-EM, we fused the LR tail of Ube2S to the C-terminus of UbcH10 (UbcH10LR) (Extended Data Fig. 1c). This approach is similar to that used to target the heterologous E2 Ube2G2 to the APC/C 28. The UbcH10LR fusion protein exhibited a 100-fold slower dissociation rate and a 10-fold higher affinity for the APC/C (Extended Data Table 3). Competition and mutagenesis analyses indicated that the UBC domain of UbcH10LR binds the physiological site on the APC/C (Extended Data Figs 4b, c). Ubiquitination assays showed that UbcH10LR and UbcH10 had indistinguishable activity (Extended Data Fig. 4d).
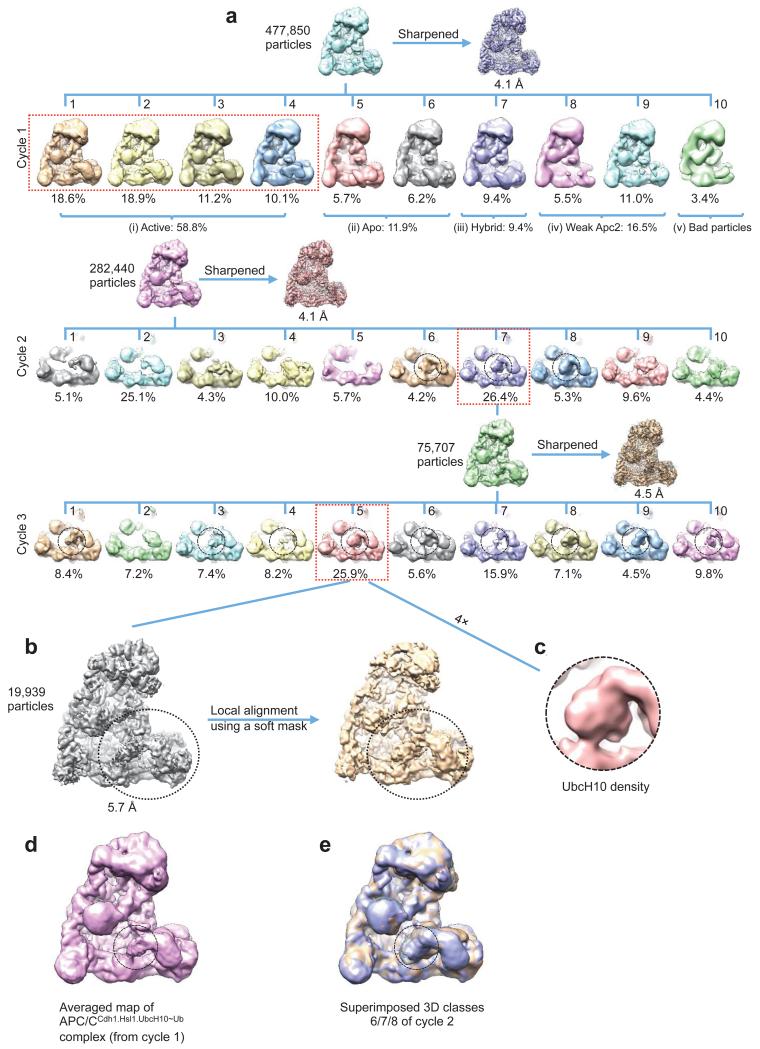
The APC/CCdh1.Hsl1.UbcH10-Ub complex was determined to 4.1 Å resolution (Extended Data Fig. 2c & Extended Table 1b). Similar to APC/CCdh1.Emi1, 3D classification revealed both ternary (coactivator-bound) and apo states (Extended Data Fig. 6a – cycle 1). Visible EM density for the LR tail in the ternary state indicated stoichiometric association of UbcH10-Ub (Extended Data Fig. 2f). However, density corresponding to UbcH10, connected to Apc2CTD-Apc11RING, was weak.
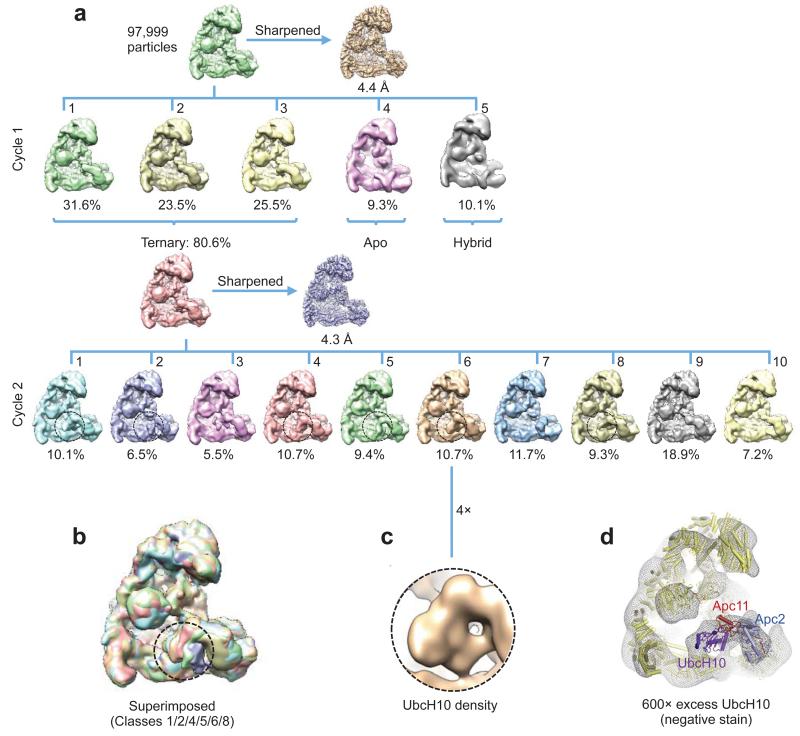
Extensive 3D classification of APC/CCdh1.Hsl1.UbcH10-Ub revealed structural variability of the UbcH10 density consistent with low occupancy of the UBC domain (~36%), coupled with minor conformational variability of the Apc2CTD-Apc11-UbcH10 catalytic module (Extended Data Fig. 6a – cycle 2). Subjecting a major 3D class for further classification generated a 3D class with clearly resolved UbcH10 density (Extended Data Fig. 6a – cycle 3 – class 5 and Extended Data Figs 6b, c). This structural class was suitable for docking UbcH10 coordinates (Figs 4a, b) and is representative of other APC/C-UbcH10 conformational states (Extended Data Figs 6d, e). Relative to APC/CCdh1.Emi1, Apc2CTD-Apc11RING tilts slightly at the Apc2NTD-Apc2CTD interface (Extended Data Fig. 5f), and Apc11RING tilts relative to Apc2CTD (Fig. 4c). UbcH10 and Apc11RING interact through a canonical E2-RING mechanism 26, thus docking of UbcH10 – Apc11RING was guided by crystal structures of RING-UbcH5-ubiquitin complexes 29,30 (Figs 4a, b). Importantly, this showed that density for ubiquitin was not visible in the EM map, indicating that the ubiquitin moiety is flexible (Figs 4a, b and Fig. 5a). Consistent with this conclusion, a reconstruction of an APC/CCdh1.Hsl1.Apc11-UbcH10 fusion complex (Extended Data Figs 1b & 2d & Extended Table 1b), and a negative stain structure of APC/CCdh1.Hsl1 with a huge excess of UbcH10, showed that the shape of the UbcH10-assigned density resembled the UbcH10-Ub density in the APC/CCdh1.Hsl1.UbcH10-Ub map (Extended Data Figs 6c & 7).
Figure 4. Structure of the APC/CCdh1.Hsl1.UbcH10-Ub complex reveals the location of UbcH10.
(a) and (b) views of the EM density at the catalytic centre with UbcH10-ubiquitin. No density for the ubiquitin moiety was recovered. R27 and R77 of Apc11, required for UbcH10 interactions 26 are indicated. (c) The UbcH10- and Emi1-binding sites of Apc11RING overlap. The Apc11RING orientation differs slightly in the two complexes.
Figure 5. The relative position of the catalytic and substrate-recognition modules defines the target lysine.
(a) View of APC/CCdh1.Hsl1.UbcH10-Ub with close-up view of the catalytic and substrate-recognition modules. A UbcH10-ubiquitin conjugate is modelled in the closed conformation 29. The Ube2S-specific site of Apc11RING (for distal Ub positioning) 26 is indicated. KEN box and ABBA motif modelled on 39. The distance from the D and KEN boxes to the catalytic centre of UbcH10 (measured as Cα-CO distance: P10 of D box and ‘N’ of KEN box to Gly76 of ubiquitin) is indicated. A ten-residue linker is modelled connected to P10 of the D box with Lys at K10. Spheres denote Cα-atoms of D box-P10 and linker-K10. (b) The reactivity of defined peptides with progressively longer spacers separating the D box and accepter lysine residue (sequences shown in (c)). Only peptides with a lysine ten or more residues C-terminal to the D box are substrates. *: Ube2S-specific band. (c) Peptide sequences used in ubiquitination assays. D box in yellow, target lysines in blue.
Since UbcH10 and Apc11RING interact through a canonical E2-RING interface (Figs 4a, b), the APC/C likely stimulates the intrinsic catalytic activity of UbcH10-ubiquitin 31 by promoting a closed conformation that primes the UbcH10-ubiquitin thioester bond 15,29,30,32-36. In agreement with this, mutation of Ile36 that contacts Apc11RING (Refs 29,30,34) and Ile44 that contacts UbcH10 (Refs 15,29,30,32,34) (Fig. 5a) ablates APC/C-catalyzed ubiquitination (Extended Data Figs 4e, f). Nevertheless, the absence of EM density for the ubiquitin moiety of the UbcH10LR-ubiquitin conjugate indicates that APC/C does not stabilize a static closed UbcH10-ubiquitin conformation 29,30,34. Modelling shows that only Apc11RING would interact with ubiquitin in the closed UbcH10-ubiquitin conformation (Fig. 5a), whereas in crystal structures of RING–UbcH5-ubiquitin conjugates where ubiquitin is static, either a second RING subunit 29,30 or a non-RING element 34 also contact ubiquitin to enhance catalytic efficiency. The APC/C is reminiscent of other single domain RING and U box E3s that bias the E2-ubiquitin conformation from multiple extended states to the closed state 33,37. An interesting possibility is that substrate initiation motifs that promote lysine ubiquitination 38 may induce a closed UbcH10-ubiquitin conformation.
Defining target lysine ubiquitination
The APC/CCdh1.Hsl1.UbcH10-Ub model shows that the UbcH10 catalytic Cys is some 40 Å from the C-terminus of a D box at the D box receptor 39 (Fig. 5a). An extended linker of ten residues would place a lysine C-terminal to the D box at the UbcH10-Ub catalytic site (Fig. 5a). Consistent with this, peptides with a lysine at positions K10, K13 and K18 relative to the D box were ubiquitinated, but not peptides with lysines at K1, K4 and K7 (Figs 5b, c). Peptides modelled on known APC/C substrates: geminin (Lys50, equivalent to K18, [Ref. 38]) and cyclin B (Lys64, equivalent to K13) were also ubiquitinated.
Conclusions
The atomic structure of the APC/C provides a quantitative framework for understanding APC/C mechanisms and the basis for future studies to investigate mechanisms of control by phosphorylation and the MCC. We speculate that in the APC/CCdc20 complex inhibited by MCC, the second C box-receptor of the Apc8 homo-dimer may bind the Cdc20 subunit of the MCC 3,40. TAME is an Ile-Arg dipeptide APC/C inhibitor 41. Its proposed mechanism is to inhibit coactivator association by competing for the IR tail-binding site on Apc3. The homology of the C box-receptor of Apc8 and the IR tail-receptor of Apc3 suggests that TAME may also disrupt C box – APC/C interactions, a notion consistent with the more important role of the C box for APC/C activity and cell viability 20.
Methods
Cloning, expression and purification of recombinant human APC/C
The genes for recombinant human APC/CCdh1.Emi1 were cloned into a modified MultiBac system as described previously 42. Two pU2 plasmids were prepared: pU2-1 harbouring genes for Apc1 and Apc11, and pU2-2 containing genes for Apc5, Apc8, Apc10, Apc13 and Apc15. Two pF2 plasmids were prepared: pF2-1 harbouring genes for Apc3, Apc6, Apc7, Apc12 and Apc16, and pF2-2 containing genes for Apc2, Apc4, Cdh1 and Emi1. A C-terminal tev cleavable double Strep-II tag was added to Apc4 for purification of the complex. Two bacmids were prepared from these four plasmids, resulting in two viruses harbouring all 16 genes with each gene flanked by its own promoter and terminator. The APC/CCdh1.Emi1 ternary complex was co-expressed from these two viruses in High 5 insect cells. The complex was purified by a combination of Strep-Tactin (Qiagen), anion exchange chromatography Mono Q and Superose 6 size-exclusion chromatography (GE Healthcare) similar to 42.
APC/CCdh1.Hsl1.Apc11-UbcH10 was cloned, expressed and purified as described previously 4, except that Apc11 was replaced with Apc11 fused to UbcH10(C114K) at its C-terminus by a 30-residue linker of GSA repeats. A similar version of APC/CCdh1.Hsl1.Apc11-UbcH10 fusion complex was also prepared using wild type UbcH10 instead of UbcH10(C114K) for activity studies.
UbcH10LR was constructed by fusion of residues 154-222 of Ube2S to the C-terminus of UbcH10(C114K) in the pET28 plasmid. An N-terminal His6-tev site was added to UbcH10LR. In the purified ubiquitin and UbcH10LR proteins the His6 tag was retained on ubiquitin, but cleaved from UbcH10LR. The UbcH10LR-Ub conjugate was generated as previously described 4 with minor modifications. UbcH10LR (200 μM) was incubated with His6-tagged ubiquitin (400 μM) at 25 °C for 16 h in a buffer containing 5 μM UBA1, 3 mM ATP, 5 mM MgCl2, 0.8 mM TCEP, 150 mM NaCl, 50 mM Tris.HCl (pH 10.0). Additional 2 mM ATP and 2 μM UBA1 were added to the reaction and incubated at 25 °C for another 8 h. The UbcH10LR-Ub conjugate was separated from unconjugated UbcH10LR by Ni-NTA and separated from free ubiquitin by S75 size-exclusion chromatography (GE Healthcare). APC/CCdh1.Hsl1 was purified as described previously 4. The purified APC/CCdh1.Hsl1 was concentrated to 3 μM. APC/CCdh1.Hsl1.UbcH10-Ub was reconstituted by incubating APC/CCdh1.Hsl1 with UbcH10LR-Ub conjugate in a 1:3 molar ratio on ice for 30 min in buffer of 20 mM HEPES (pH 8.0), 150 mM NaCl, 1 mM DTT. Just prior to applying to EM grids, the sample was cross-linked with glutaraldehyde (GA) by adding 0.03% GA, and incubated on ice for 40 min. The reaction was quenched by adding 40 mM Tris.HCl (pH 8.0). The quenched sample was incubated on ice for 30 min before further purification with size-exclusion Superose 6. A similar UbcH10LR protein was made by using wild type UbcH10 for use in activity tests. Surface plasmon resonance experiments for UbcH10LR and APC/C were performed as described previously 4. The Y91A and S123G mutants of UbcH10 and UbcH10LR were prepared as for wild type UbcH10.
For ubiquitination reactions, APC/C and its variants with mutant subunits (Apc1 and Apc8) were cloned, expressed and purified as for apo APC/C 42 by replacing the target subunit with its mutant. These include APC/C with Apc1L1197A/L1227A/I1230A/H1231A, APC/C with Apc8N339A/E374A, APC/C with Apc8N339A/E374R, APC/C with Apc8E410R.
Human Cdh1 was cloned into pU1 with an N-terminal His6 followed by an HA tag. Based on this plasmid, variants of Cdh1 mutations were made. These were Cdh1L160A/L161A, Cdh1Y91A/L99A, Cdh1L127A/F128A, Cdh1L94A/L95A/V153A, Cdh1L160A/L161AY91A/L99A, Cdh1 L160A/L161A/L127A/F128A, Cdh1Y91A/L99A/L127A/F128A, Cdh1L160A/L161A/Y91A/L99A/L127A/F128A, Cdh1R47A, Cdh1R52A, Cdh1R493A, Cdh1T121A/S151A/S163A, Cdh1S40A/S151A/S163A, Cdh1S40A/T121A/S163A, Cdh1S40A/T121A/S151A, Cdh1S40A/T121A/S151A/S163A, Cdh1T32A/S36A/S70A/S138A/S146A. All Cdh1 and its mutations were expressed in High 5 insect cells. To purify Cdh1, the cells were lysed in a buffer of 50 mM Tris.HCl (pH 7.3), 500 mM NaCl, 20 mM imidazole (Buffer A), with a protease inhibitor cocktail. After loading, the Ni-NTA column was washed with 10 column volumes of buffer A, followed by a gradient wash to 200 mM imidazole in buffer A and further washing with 10 column volumes of 200 mM imidazole in buffer A. The protein was eluted in buffer B of 300 mM imidazole (pH 7.3), 500 mM NaCl. Cdh1 was finally purified using S-200 size exclusion chromatography in 20 mM HEPES (pH 7.0), 200 mM NaCl, 1 mM DTT.
Human ubiquitin cDNA was cloned into a pET28 plasmid with an N-terminal His6 tag and TEV cleavage site. Based on this plasmid, ubiquitin I36A and ubiquitin I44A mutants were prepared. Wild type and mutant ubiquitin were expressed in B834rare2 strain and purified by Ni-NTA and S75 size-exclusion chromatography (GE Healthcare).
APC/C Ubiquitination assays
Human APC/C and its different mutant derivatives, UBA1, UbcH10 and its mutants, Ube2S, ubiquitin and its mutants, and yeast Hsl1667-872 were buffer exchanged into 20 mM HEPES (pH 8.0), 150 mM NaCl, 1 mM DTT. Human Cdh1 and its mutants were in 20 mM HEPES (pH 7.0), 200 mM NaCl, 1 mM DTT. Ubiquitination assays were performed as described 4. For the UbcH10 competition assays, UbcH10 C114K and UbcH10 C114K-LR at the following concentrations (0.001, 0.01, 0.1, 1, 10 μM) were used to compete with UbcH10 (0.1 μM). Cdh1 was at 20 nM. To test the UbcH10-RING domain interface, the following UbcH10 and UbcH10LR mutants were prepared and tested (Y91A and S123G). Single time point assays are at 10 mins.
Cdh1 Phosphorylation assays
CDK2-dependent phosphorylation of Cdh1 and its mutants was performed in buffer containing 50 mM HEPES (pH 8.0), 10 mM MgCl2 2mM ATP, 10 mM NaF, 10 mM glycerol phosphate, and 150 mM NaCl. The reactions were incubated at 23 °C for 30 min with 500 nM of Cdh1 and 50 nM of active phosphorylated CDK2-cyclin A3. The phosphorylation was terminated using the CDK1/2 inhibitor (Calbiochem). Non-CDK2 treated Cdh1 samples were set up in parallel with phosphorylating reactions by adding buffer instead of CDK2-cyclin A3. The activity assays were carried out as described for ubiquitination assays.
Peptide assays
Peptides for D box-lysine distance analysis were synthesized by Designer BioScience (Cambridge, UK). The peptides were modified by N-terminal biotinylation. Peptides used are Pep-1K: (EQRPRTALGDISNSKASGS), Pep-4K: (EQRPRTALGDISNSAGSKASGS), Pep-7K: (EQRPRTALGDISNSAGSAGSKASGS), Pep-10K (EQRPRTALGDISNSAGSAGSAGSKASGS), Pep-13K: (EQRPRTALGDISNSAGSAGSAGSAGSKASGS), Pep-18K: (EQRPRTALGDISNSAGSAGSAGSAGSAGSGSKASGS), GEM-K50: (EQRPRTALGDISNSASGSLVGRENELSAGLSKRRHR), and CycB-K64: (GLRPRTALGDIGNRVSEQLQARMPMRKEARPSATGS). The peptides were dissolved at a concentration of 6.25 mM in 100% DMSO. The ubiquitination assays were performed as described above except that 125 μM of peptide instead of Hsl1 was used as substrate. Previous work showed that at 100 μM D box-only peptides bind the D box receptor of APC/CCdh1 (Refs 43,44). The reactions were separated in 4-12% NuPAGE Bis-Tris gels (Life Technology) by 60 v for 2 h, followed by 150 v for 30 min for better resolution of peptides and ubiquitination products. After blotting, the membranes were blocked in 10% nonfat dry milk in PBS overnight at 4 °C. After washing the blot four times with PBS-2% Tween 20 buffer, ubiquitinated peptide products were visualized by Western blotting with PBS-buffered Streptavidin-HRP (Sigma) with 1% BSA.
Electron microscopy
Freshly purified recombinant human APC/C complexes (APC/CCdh1.Emi1, APC/CCdh1.Hsl1.UbcH10-Ub, and APC/CCdh1.Hsl1.Apc11-UbcH10) were first visualized by negative stain electron microscopy to check homogeneity. Complexes were then diluted to ~200 μg/ml and aliquots of 2 μl were applied to Quantifoil R2/2 grids coated with a second layer of home-made thin carbon film (estimated to be ~50 Å thick), treated with a 9:1 argon:oxygen plasma for 20 s before use. The grids were incubated for 30 s at 4 °C and 100% humidity and then blotted for 6 s and plunged into liquid ethane using an FEI Vitrobot III. The grids were loaded into an FEI Tecnai Polara electron microscope at an accelerating voltage of 300 kV. Images were taken manually using low-dose mode at a nominal magnification of × 78,000 (yielding a pixel size of 1.36 Å pixel−1) for APC/CCdh1.Emi1 and APC/CCdh1.Hsl1.Apc11-UbcH10, and × 59,000 (yielding a pixel size of 1.77 Å pixel−1) for APC/CCdh1.Hsl1.UbcH10-Ub. Images were recorded using a FEI Falcon II direct electron detector with a defocus range of ~2.5 to 4 μm. Each image was exposed for 2 s at dose rate of 27 e−/Å2/s at a nominal magnification of × 78,000 and 16 e−/Å2/s at a nominal magnification of × 59,000. 34 movie frames were recorded for each image as previously described 45.
To prepare a negative stain reconstruction of APC/CCdh1.Hsl1-UbcH10 (with excess wild type UbcH10), we incubated freshly purified APC/CCdh1.Hsl (0.04 μM) with a 1500-fold excess of UbcH10 (60 μM, 200-fold in excess of Kd) for 1 h on ice. Images were collected on an FEI Spirit electron microscope at an accelerating voltage of 120 kV at −1.5 μm defocus. The map was reconstructed using RELION without CTF correction.
Image processing
All movie frames were first aligned by motioncorr program 46 before subsequent processing. Contrast transfer function parameters were calculated with CTFFING3 (Ref. 47). Particles in 264×264 pixels (or 200×200 pixels for data collected at × 59,000 magnification) were initially selected automatically by e2boxer.py in EMAN2 (Ref. 48) from a small portion of the same data set (typically ~300 micrographs). After 2D classification using RELION 49, 10 images in different views were selected from the 2D averages and used as reference for automatic particle picking for the whole data set by RELION 1.3 (Ref. 50). The following procedures were performed in RELION to exclude bad particles from the final reconstruction. First, particles in each micrograph were displayed and manually screened to delete wrongly picked ice contamination. Secondly, extracted particles were sorted by similarity to reference images and particles with low Z scores were deleted (roughly ~10% particles were deleted in this step). Thirdly, 2D classification was performed and particles in bad classes (with poor structural features) were removed (roughly ~10-20% particles were deleted in this step). The resultant particles were then used for 3D classifications. Conformational heterogeneity and remaining bad particles were excluded from the final reconstruction in this step. A summary of particles used for final refinements is listed in Extended Data Table 1c.
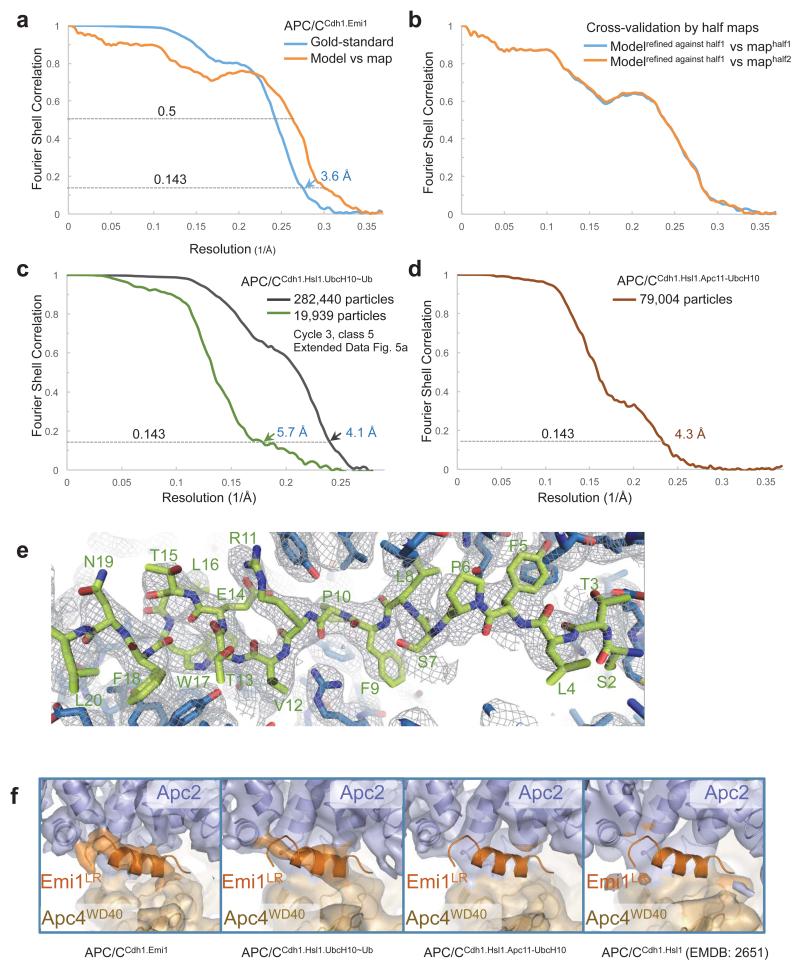
Beam-induced particle motion was corrected using statistical movie processing in RELION as previous described 45. For APC/C, a running average of 9 movie frames for frames within the first 32 e− dose and a standard deviation of one pixel for the translational alignment were used. This step improved the resolution from 4.2 Å to 3.9 Å. Following this, particle polishing in RELION 1.3 were used to further improve the resolution 51. This program fits all movie frames as linear tracks, considering movements of neighboring particles to improve signal for smaller particles. Furthermore, each frame is weighted by B-factor (estimated by each frame reconstruction) to treat radiation damage problem. Particle polishing was then used to improve the resolution from 3.9 Å to 3.6 Å. All resolution estimations were from gold-standard FSC calculations to avoid over-fitting and reported resolutions are based on the FSC = 0.143 criterion 52,53. Final FSC curves were calculated using a soft spherical mask (with a 5-pixel fall-off) of the two independent reconstructions. To visualize high-resolution details, all density maps were corrected for modulation transfer function (MTF) of the detector and sharpened by applying negative B-factors, estimated using automated procedures 52.
Motion correction was performed after 3D classification for APC/CCdh1.Emi1 which showed ~ 60% ternary complex and ~ 40% apo state APC/C. For APC/CCdh1.Hsl1.UbcH10-Ub and APC/CCdh1.Hsl1.Apc11-UbcH10, all particles were refined against one model and motion corrected as described above. The polished particles were used for 3D classification as shown in Extended Data Figure 6 and 7. The first cycle of classification was done by global search and sampling angular interval of 7.5° to sort out large conformational changes and cycle 2-4 were executed by local search within 15° and a smaller sampling angular interval of 3.7°. 3D classifications were run for 25 iterations with class number set at 10 and regularization parameter T set at 4.
To further improve the density map of flexible parts (RING domain and UbcH10) in the complexes, a soft mask was used for local alignment during refinement. A soft mask was generated according to combined coordinates of Apc1/Apc2/Apc4/Apc5/Apc11/UbcH10 (approximately half of the mass) and a restrained angular search of 3° was used. Local resolution was estimated using ResMap 54.
Model building of APC/C
Modelling of the structures of APC/C was performed in COOT 55. Initially, available crystal structures and homology models as described in our previous work 4 were fitted into the cryo-EM map in Chimera. These structures include (1) crystal structures of human Apc10 (residues 2-162, PDB code: 1JHJ) 56, (2) N-terminal seven helices of Apc7 (residues 21-166, PDB code: 3FFL) 57, (3) S. pombe Cut9-Hcn26 complex (Apc6-Apc12 homologue, PDB code: 1XPI) 58, (4) N-terminal dimerization domains of E. cuniculi Cdc27 (Apc3 homologue, PDB code: 3KAE) 59 (5) S. pombe Cut23 (Apc8 homologue, PDB code: 3ZN3) 60, (6) Cdh1-D-KEN (PDB code: 4BH6) 39 (7) Apc5 TPR domain and C-terminal TPR helices of Apc3, Apc8 and Apc7 (model using the crystal structure of S. pombe Cut9 as template), (8) crystal structures of Apc4 and Apc5NTD (residues 1-160) (unpublished data), (9) NMR structure of Emi1 ZBR domain 13 (PDB:2M6N) and Apc11 RING domain 26 (PDB code: 4R2Y). For Apc2-Apc11N, the structure was modelled using a crystal structure of Cul1 (PDB code: 1U6G) 61 as a template. For Apc1 PC repeats, the structure was predicted with I-Tassar 62 using the PC domain structure of Rpn2 (PDB code: 4ADY) 63 as a reference. All fitted structures were rebuilt according to the cryo-EM map. The majority of Apc1, small subunits (Apc13, Apc15, Apc16), Ile-Arg (IR) C-terminal tails of Apc10 and Cdh1, N-terminal domain of Cdh1 (Cdh1NTD) and some loops not seen in crystal structures and homology were built ab initio. Details given in Extended Data Table 2.
The model was refined by REFMAC v.5.8 (Ref. 64). A REFMAC weight of 0.04 was defined by cross-validation using half reconstructions 65. The resolution limit for refinement was set at 3.5 Å. All high-resolution crystal structures or NMR structures were used for secondary structure restraints. A summary of model refinement is listed in Extended Data Table 1a.
Rigid-body fitting was performed to UbcH10 by ‘fit in map’ program in Chimera 66. Crystal structure of Rnf4 - UbcH5a - ubiquitin heterotrimeric complex (PDB: 4AP4) 29 was used to model ubiquitin not seen directly from the reconstruction due to flexibility. A summary of model building is listed in Extended Data Table 2.
All regions of the APC/C EM density map were assigned to APC/C subunits. However, the intrinsically disordered regions of some APC/C subunits were not visible in the EM map (defined in Extended Data Table 2). Altogether, 78% APC/C residues were fitted to the EM map. Inter-subunit contact areas were calculated using CCP4 (Ref. 67).
Map visualization
Figures and movie were generated using Pymol and Chimera 66.
Structural conservation was calculated using CONSURF 68 based on a multiple sequence alignment of H. sapiens, M. musculus, X. laevis, C. elegans, D. melanogaster, A. thaliana, S. cerevisiae, S. pombe, APC/C subunit sequences.
Protein similarity searches were performed using DALI 69, and protein alignment figures created using ALSCRIPT 70.
Extended Data
Extended Data Figure 1. Preparations and EM images of APC/C complexes.
(a) Coomassie blue stained SDS gel of APC/CCdh1.Emi1. (b) Coomassie blue stained SDS gel of APC/CCdh1.Hsl1.Apc11-UbcH10. (c) Coomassie blue stained SDS gel and Western blot analysis (anti-His antibody - only ubiquitin in the complex contains His tag) of APC/CCdh1.Hsl1.UbcH10-Ub without or with cross-linking by glutaraldehyde (GA). (d) A typical cryo-EM micrograph of APC/CCdh1.Emi1. Representative of 3328 micrographs. (e) Gallery of 2D averages of APC/CCdh1.Emi1 showing different views. Representative of 100 2D averages. (f) Local resolution map of APC/CCdh1.Emi1 showing resolution range. (g) Details of EM density for segments of α-helix and β-strand of Apc1 and the C box of Cdh1.
Extended Data Figure 2. Resolution estimation and example of de novo model building.
(a) Gold standard Fourier Shell Correlation (FSC) curve and FSC curve between cryo-EM map and final atomic model of the APC/CCdh1.Emi1. (b) Cross-validation of model refinement by half maps. Shown are FSC curves between the atomic model and the half map (maphalf1) it was refined against and FSC curve between the atomic model and the other half map (maphalf2) that was not used during refinement. (c) Gold standard FSC curve of APC/CCdh1.Hsl1.UbcH10-Ub. (d) Gold standard FSC curves of human APC/CCdh1.Hsl1.Apc11-UbcH10. (e) de novo model building of Apc15 N-terminal loop. The surrounding Apc5 residues are also shown. (f) EM density for the LR tail common to Emi1 and Ube2S is only observed in APC/C complexes with subunits incorporating the LR tail. (i) APC/CCdh1.Emi1. (ii) An APC/C complex with an LR tail-bearing subunit (UbcH10LR of APC/CCdh1.Hsl1.UbcH10-Ub). (iii) No LR tail density in APC/CCdh1.Hsl1.Apc11-UbcH10 fusion and (iv) APC/CCdh1.Hsl1 (Ref. 4).
Extended Data Figure 3. Apc1 structure and the TPR lobe interacts with multiple subunits.
(a) Cartoon of Apc1 colour-ramped from blue to red for N- to C-terminus. Insertions that interact with Apc2, Apc8 and Apc10 are labelled. Apc1Mid adopts a novel architecture. (b) TPR lobe with TPR subunits shown as surface representations, with the small TPR-accessory subunits (Apc12, Apc13, Apc15 and Apc16), a segment of Apc5, an insert of Apc1, IR tails of Cdh1 and Apc10, and Cdh1 C box that interact with the TPR lobe are shown as cartoons. The N-termini of Apc12, Apc13 and Apc15 are buried. The eight structurally homologous and symmetry-equivalent sites on the TPR lobe that bind Apc13, Apc16 and Apc5 are indicated and shown in detail in (c). View is similar to Fig 1a. (c) The eight TPR subunits interact with Apc13, Apc16 and Apc5 mainly through contacts to four conserved aromatic residues present on most TPR subunits (Y308, Y309, W302, Y322 of Apc6 [panels 5 and 6]). (d) Sequence of the ordered region of Cdh1NTD bound to Apc1 and Apc8 (ordered regions shown as lines and α-helices). Critical Apc1 and Apc8 contact residues are indicated with green and blue arrows. Phosphorylation sites indicated with red arrows.
Extended Data Figure 4. APC/C ubiquitination assays.
(a) Mutation of Arg493 of the IR tail reduces APC/CCdh1 activity. (b) Mutations at the RING domain interface of UbcH10 and UbcH10LR disrupt ubiquitination activity. (c) Ubiquitination assay shows that both UbcH10(C114K) and UbcH10LR(C114K) compete with wild type UbcH10. UbcH10LR(C114K) is a more potent inhibitor. (d) The APC/C-UbcH10-mediated substrate ubiquitination activities of UbcH10 and UbcH10LR are indistinguishable. (e) The ubiquitin (I36A) and ubiquitin (I44A) mutants were defective for APC/C-UbcH10-mediated substrate ubiquitination. (f) UbcH10 charging by the ubiquitin (I36A) and ubiquitin (I44A) mutants was unchanged relative to wild type ubiquitin.
Extended Data Figure 5. The position of Apc11RING in the APC/C is more similar to Rbx1RING of activated cullin-Rbx1 structures.
(a) Identification of Apc11 in apo APC/C. Left panel: EM density map for apo APC/C with the coordinates of Apc2CTD-Apc11 fitted (from APC/CCdh1.Emi1 structure). Right panel. EM density for APC/CApc11-ΔRING. The difference density corresponds to Apc11RING. EM density maps from 4. (b) Superimposed Apc2CTD onto Cul1CTD (PDB: 1LDK) 61. (c) Superimposed Apc2CTD onto Cul5CTD (PDB: 3DQV) 27. In the inactive conformation of Cul1-Rbx1, Rbx1RING packs against WHB. In APC/CCdh1.Emi1 the location of Apc11RING remains in contact with Apc2CTD but has rotated ~180° relative to inactive CRL structures being similar to the swung out conformation of Rbx1RING of neddylated and activated Cul5-Rbx1 27. (d and e) The relative orientation of Apc2NTD and Apc2CTD is also dramatically different from Cul1 (Ref. 61). This is due to a 70° rotation within cullin repeat 3 (between helices A-B and C-D-E), and a ~20° rotation around the 4HB - cullin repeat 3 interface. Similar less pronounced structural variations are observed within the CRL family. (d) Apc2-Apc11 (this study). (e) Cul1-Rbx1 (PDB: 1LDK) 61. (f) The position of the Apc2CTD-Apc11 module differs slightly about the Apc2NTD-Apc2CTD interface between APC/CCdh1.Emi1 and APC/CCdh1.Hsl1.UbcH10-Ub.
Extended Data Figure 6. Three dimensional classification of APC/CCdh1.Hsl1.UbcH10-Ub.
(a) The 3D classification (Cycle 1) started with 477,850 motion corrected particles, which were divided into ten classes. The resultant classes were grouped into five categories: (i) 58.8% in the active ternary state with coactivator and substrate (Hsl1); (ii) 11.9% in the apo inactive state; (iii) 9.4% in a class with Cdh1 bound but with the catalytic module in the apo inactive conformation (hybrid state); (iv) 16.5% with weak Apc2 density and (v) 3.4% were a poor reconstruction due to bad particles. Examination of the hybrid state (iii) showed that density for Apc1WD40 was absent, explaining the lack of Cdh1-induced conformational change of the catalytic module. Particles in the ternary state (reconstructed to an overall resolution of 4.1 Å) were subjected to further 3D classification (Cycle 2). A major class (class 7, 26.4% of particles) showed improved density for UbcH10 (circled) and Cycle 3 classification was performed on particles in this class. The major class of Cycle 3 (class 5, 25.9% particles) showed further improved UbcH10 density. Further 3D classification of this class did not improve the UbcH10 density. (b) Particles in the best class (Cycle 3, class 5, 19,939 particles) were refined in RELION and resulted in a map at 5.7 Å resolution (Extended Data Fig. 2c). The UbcH10 density was improved by local alignment using a soft mask (indicated by circles) as described in Methods. (c) Enlarged view of UbcH10 density. (d) Enlarged view of the averaged APC/CCdh1.Hsl1.UbcH10-Ub reconstruction from cycle 1 of the 3D classification (59% of particles). UbcH10 density is circled. (e) Superimposition of classes 6, 7 and 8 of cycle 2 of the 3D classification (from (a)), showing the structural variability of the three 3D classes that indicate UbcH10 density. UbcH10 density is circled.
Extended Data Figure 7. Three dimensional classification of APC/CCdh1.Hsl1.Apc11-UbcH10.
(a) The 3D classification started with 97,999 motion corrected particles, which were divided into five classes. The resultant classes were grouped into three categories: (i) 80.6% in the active ternary state with coactivator and substrate (Hsl1); (ii) 9.3% in the apo state and (iii) 10.1% in a hybrid state. Particles in the active ternary state (reconstructed to an overall resolution of 4.3 Å) were subjected to Cycle 2 classification with ten classes. UbcH10 density in the resultant classes is indicated with circles. (b) Classes with UbcH10 density in Cycle 2 classification are superimposed, showing variability of UbcH10. (c) Enlarged view of UbcH10 density. (d) A negative stain EM reconstruction of an APC/CCdh1.Hsl1 complex at ~25 Å resolution with a 1500-fold excess of UbcH10. The molecular surface is shown as a mesh representation and the coordinates of the APC/CCdh1.Hsl1.UbcH10-Ub were docked into the EM reconstruction. The UbcH10 coordinates fit new EM density proximal to Apc11.
Extended Data Table 1. Statistics of structure determination.
| a. Statistics of APC/CCdh1.Emi1 structure determination | |
|---|---|
| Data collection | |
| EM: | FEI Polara, 300keV |
| Detector: | FEI Falcon II |
| Pixel size (Å): | 1.36 |
| Defocus range (μm): | 1.5-4 |
| Reconstruction | |
| Software: | RELION 1.3 |
| Accuracy of rotations (degrees): | 1.073 |
| Accuracy of translations (pixels): | 0.691 |
| Final Resolution (Å): | 3.6 |
| Refinement | |
| Software: | RefMac 5.8 |
| Refmac weight: | 0.04 |
| Resolution limits (Å): | 3.5 |
| Residue number | 8408 |
| Average Fourier shell correlation | 0.8315 |
| R factor: | 0.222 |
| Rms BondLength: | 0.0135 |
| Rms BondAngle: | 1.8098 |
| Validation | |
| Ramachandran plot: | |
| Preferred: | 7669 (92.88%) |
| Allowed: | 352 (4.26%) |
| Outliers: | 236 (2.86%) |
| b. Statistics of all cryo-EM data | ||||
|---|---|---|---|---|
| Samples | Micrographs | Particles (after movie correction) | Resolution (Å) | |
| APC/CCdh1.Emi1 | 3328 | In total:363,774 | - | |
| Ternary | 202,084 (55.6%) | 3.6 | ||
| Apo | 161,690 (44.4%) | 4.3 | ||
| *Others (Hybrid, Weak Apc2 and Bad particles) |
*52,178 particles deleted before movie correction |
- | ||
| APC/CCdh1.Hsl1-UbcH10RL~Ub | 4524 | In total: 477,850 | - | |
| Ternary | 282,440 (59.1%) | 4.1 | ||
| Apo | 56,865 (11.9%) | - | ||
| Others (Hybrid, Weak Apc2 and Bad particles) |
138,545 (29.0%) | - | ||
| APC/CCdh1.Hsl1-UbcH10Apc11-UbcH10 | 869 | In total: 97,999 | - | |
| Ternary | 79,004 (80.6%) | 4.3 | ||
| Apo | 9,114 (9.3%) | - | ||
| Others (Hybrid, Weak Apc2 and Bad particles) |
9,801 (10.1%) | - | ||
Extended Data Table 2. Summary of model building for APC/C subunits.
| Subunit | Length | Domain/Region 1 | Domain/Region 2 | Domain/Region 3 | Disordered regions | Phosphorylation sites |
|---|---|---|---|---|---|---|
| Apc1 | 1,944 | WD40 domain (1-612) De novo |
Mid-N (613-986) Mid-C (1617-1944) De novo |
PC domain (1013-1616) Homology. PDB: 4ADY Inserts: De novo |
1-10,59-70,135-145,193-205,227- 235,296-401,415-423,514-581,671- 756,815-838,883-923,987-1012,1334- 1346,1438-1451,1712-1726,1930-1944 |
202,286,291,341,343,355, 362,364,373,377,537,571,681, 688 |
| Apc2 | 822 | NTD (1-432) Homology. PDB: 1LDK |
CTD(433-822) Homology. PDB: 1LDK |
- | 1-14, 30-52,66-69,103-106, 192-195, 221-231,305-319,444-450,459-475 731-822 (WHB) |
218,314,532,534 |
| Apc3 | 824 | TPR dimer interface TPR motifs 1-7 (1-535) Homology. PDB: 3KAE |
TPR superhelix TPR motifs 8-14 (536-824) Homology. PDB: 2XPI |
- | 171-450,767-824 (A) 171-450,782-824 (B) |
205,209,220,244,264,291, 302,304,313,334,336,364, 366,369,383,384,386,426, 430,435,438,446,447 |
| Apc4 | 808 | WD40 domain/4HB X ray: unpublished Inserts: De novo |
- | - | 429-438,458-469,758-808 | 469,777 |
| Apc5 | 755 | NTD (1-169) X ray: unpublished |
TPR superhelix TPR motifs 1-13 (206-755) Homology. PDB: 2XPI Inserts: De novo |
- | 1-26,170-205 | 15,232,195 |
| Apc6 | 620 | TPR dimer interface TPR motifs 1-7 (1-261) Homology. PDB: 2XPI |
TPR superhelix TPR motifs 8-14 (262-620) Homology. PDB: 3HYM |
- | 96-126,528-620 (A) 96-123,534-620 (B) |
112,490,559,560,580,581, 585,586,599 |
| Apc7 | 599 | TPR dimer interface TPR motifs 1-3 (21-166) X ray. PDB: 3FFL |
TPR dimer interface TPR motifs 4-7 (167-359) Homology. PDB: 2XPI |
TPR superhelix TPR motifs 8-14 (360-599) Homology. PDB: 2XPI |
1-35,111-131,553-599 (A) 1-35,111-131,541-599 (B) |
51,57,64,119,120,125 |
| Apc8 | 597 | TPR dimer interface TPR motifs 1-7 (1-287) Homology. PDB: 3ZN3 |
TPR superhelix TPR motifs 8-14 (288-597) Homology. PDB: 2XPI |
- | 1-25,501-508,558-597 (A) 1-25,501-508,558-597 (B) |
267,556,576,578,582,590 |
| Apc10 | 185 | Doc homology (2-162) X-ray. PDB: 1JHJ |
IR tail (163-185) De novo |
- | - | - |
| Apc11 | 84 | β-strand (1-18) Homology. PDB: 1LDK |
RING domain (21-84) X-ray. PDB: 4R2Y |
- | - | - |
| Apc12 | 85 | N-term (1-25) X-ray. PDB: 3HYM |
- | - | 26-85 | - |
| Apc13 | 74 | De novo | - | - | 40,47,68-79 | |
| Apc15 | 121 | De novo | - | - | 57-121 | - |
| Apc16 | 110 | De novo | - | - | 1-51 | |
| Cdh1 | 496 | NTD (1-163) De novo |
WD40 domain (169-470) Homology: 4BHY |
IR tail (483-496) De novo |
1-41,68-87,109-124,133-145,164- 168,471-482 |
- |
| Emi1 | 447 | D box (319-335) X ray/De novo |
ZBR (356-372) NMR. PDB: 2M6N |
Linker (356-372), De novo LR tail (431-447), De novo |
1-318,336-355,373-430 | - |
| UbcH10 | 179 | UBC domain (30-179) X-ray. PDB: 1I7K |
- | - | 1-29 | - |
| Ubiquitin | 76 | X-ray. PDB: 4ADY | - | - | - | - |
Extended Data Table 3.
| a. Inter-subunit contact surface area Å2. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subunit | Apc1 | Apc2 | Apc3 | Apc4 | Apc5 | Apc6 | Apc7 | Apc8 | Apc10 | Apc11 | Apc12 | Apc13 | Apc15 | Apc16 | Cdh1 | Emi1 |
| Apc1 | - | |||||||||||||||
| Apc2 | 2520 | - | ||||||||||||||
| Apc3 | 360 | 0 | 5746 | |||||||||||||
| Apc4 | 0 | 714 | 0 | - | ||||||||||||
| Apc5 | 8744 | 0 | 0 | 6226 | - | |||||||||||
| Apc6 | 1056 | 0 | 4590 | 0 | 0 | 5244 | ||||||||||
| Apc7 | 0 | 0 | 5354 | 0 | 0 | 0 | 5026 | |||||||||
| Apc8 | 4290 | 0 | 0 | 0 | 3722 | 3772 | 0 | 4558 | ||||||||
| Apc10 | 2616 | 0 | 2510 | 0 | 0 | 0 | 0 | 0 | - | |||||||
| Apc11 | 0 | 3402 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - | ||||||
| Apc11RING | 0 | 666 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Apc12 | 246 | 0 | 0 | 0 | 0 | 5970 | 0 | 0 | 0 | 0 | - | |||||
| Apc13 | 0 | 0 | 836 | 0 | 0 | 2068 | 0 | 3240 | 0 | 0 | 0 | - | ||||
| Apc15 | 0 | 0 | 0 | 224 | 3446 | 340 | 0 | 1082 | 0 | 0 | 0 | 0 | - | |||
| Apc16 | 40 | 0 | 2514 | 0 | 0 | 222 | 1502 | 0 | 0 | 0 | 0 | 0 | 0 | - | ||
| Cdh1 | 1960 | 0 | 1906 | 0 | 0 | 712 | 0 | 3022 | 0 | 0 | 0 | 0 | 0 | 0 | - | |
| Cdh1NTD | 1960 | 0 | 0 | 0 | 0 | 500 | 0 | 3022 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cdh1WD40 | 0 | 0 | 82 | 0 | 0 | 212 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cdh1IR | 0 | 0 | 1824 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Emi1 | 732 | 762 | 0 | 0 | 0 | 0 | 0 | 0 | 704 | 1300 | 0 | 0 | 0 | 0 | 754 | - |
| Total | 24524 | 8064 | 25722 | 7164 | 34010 | 24686 | 11882 | 26708 | 5830 | 4702 | 6216 | 6144 | 5092 | 4278 | 8354 | 4252 |
| b. DALI Results | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subunit/domain | Protein match | RCSB PDB | Z | RMSD Å | Aligned N | Res N | ID % | Subunit/domain | Protein match | RCSB PDB | Z | RMSD Å | Aligned N | Res N | ID % |
| Apc1WD40 | Sc Cdh1 | 4BH6-E | 15.9 | 4.0 | 259 | 303 | 8 | Apc6 | Sp Apc6 | 2XPI-D | 36.9 | 2.2 | 481 | 520 | 37 |
| Apc1PC | Sc Rpn2 | 4ADY-B | 27.8 | 3.2 | 406 | 802 | 15 | Apc7 | Sp Apc6 | 2XPI-D | 24.9 | 7.8 | 464 | 518 | 15 |
| Apc2NTD | Hs Cul3 | 4HXI-B | 13.8 | 7.4 | 301 | 346 | 7 | Apc8 | Sp Apc6 | 2XPI-D | 25.5 | 7.2 | 443 | 518 | 19 |
| Apc2CTD | Hs Cullin1 | 4F52-A | 23.5 | 2.2 | 237 | 258 | 18 | Apc3A | Apc3B | - | 1.6 | 482 | 497 | 100 | |
| Apc5TPR | Sp Apc6 | 2XPI-A | Apc6A | Apc6B | - | 4.0 | 486 | 503 | 100 | ||||||
| Apc3 | Sp Apc6 | 2XPI-A | 29.6 | 4.9 | 456 | 518 | 19 | Apc7A | Apc7B | - | 2.6 | 483 | 483 | 100 | |
| Apc8A | Apc8B | - | 3.7 | 389 | 490 | 100 | |||||||||
| c. Surface Plasma Resonance Data | ||||||
|---|---|---|---|---|---|---|
| Ligand | Analyte | koff (s−1) | kon (M−1 s−1) | Keqd (nM) | Kkin d (nM) | Reference |
| UbcH10 | Ternary | 0.42 ± 0.02 | 1.78 ± 0.2 × 106 | 215 | 236 ± 29 | 4 |
| UbcH10LR | Ternary (fast) | 4.0 ± 0.2 × 10−2 | 1.70 ± 0.1 × 106 | 31 | 24 ± 2 | This study |
| (slow) | 4.6 ± 0.2 × 10−3 | 1.70 ± 0.1 × 105 | 23 ± 5 | |||
Kkind is the ratio of the observed microscopic rate constants.
Calculated using CCP4. (b) Pair-wise structural comparison (DALI) of APC/C subunits. (c) Surface Plasma Resonance data. Table comparing APC/CCdh1.Hsl1 (ternary APC/C), dissociation constants for UbcH10 (Ref. 4), and UbcH10LR (this work). Keqd: Equilibrium dissociation constant; Kkind: kinetic dissociation constant = koff/kon. Standard errors of the fit are listed.
Supplementary Material
Supplementary Video 1. Video showing APC/CCdh1.Emi1 and APC/CCdh1.Hsl1.UbcH10-Ub reconstructions.
Acknowledgments
This work was funded by a Cancer Research UK grant to DB. We thank W.J. Chazin and members of the Barford group for helpful discussions, and X. Bai and S. Scheres for their help with RELION; C. Savva and S. Chen for EM facilities; P. Emsley for help with COOT; G. Murshudov for help with REFMAC; G. McMullan for his assistance in movie data capture; J. Grimmett and T. Darling for computing and A. Boland for advice with COOT and PyMol.
Footnotes
Author information. EM maps are deposited with the EM-DB with accession codes: 2924 (APC/CCdh1.Emi1), 2925 (APC/CCdh1.Hsl1.UbcH10-Ub), 2926 (APC/CCdh1.Hsl1.Apc11-UbcH10). APC/CCdh1.Emi1 coords have accession code 4ui9.
The authors declare no competing financial interests.
References
- 1.Meyer HJ, Rape M. Processive ubiquitin chain formation by the anaphase-promoting complex. Seminars in cell & developmental biology. 2011;22:544–550. doi: 10.1016/j.semcdb.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 3.Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. The Journal of cell biology. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimata Y, Baxter JE, Fry AM, Yamano H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Molecular cell. 2008;32:576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Van Voorhis VA, Morgan DO. Activation of the APC/C ubiquitin ligase by enhanced E2 efficiency. Current biology: CB. 2014;24:1556–1562. doi: 10.1016/j.cub.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin chain elongation requires e3-dependent tracking of the emerging conjugate. Molecular cell. 2014;56:232–245. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Molecular biology of the cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudner AD, Murray AW. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. The Journal of cell biology. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Current biology: CB. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 11.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 12.Reimann JD, et al. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 13.Frye JJ, et al. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nature structural & molecular biology. 2013;20:827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Kirschner MW. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nature cell biology. 2013;15:797–806. doi: 10.1038/ncb2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit e2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson A, et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu T, et al. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. The EMBO journal. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. The Journal of cell biology. 2012;199:27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornton BR, et al. An architectural map of the anaphase-promoting complex. Genes & development. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Molecular cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nature cell biology. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikorski RS, Michaud WA, Hieter P. p62cdc23 of Saccharomyces cerevisiae: a nuclear tetratricopeptide repeat protein with two mutable domains. Mol Cell Biol. 1993;13:1212–1221. doi: 10.1128/mcb.13.2.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukas C, et al. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- 25.Miller JJ, et al. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes & development. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown NG, et al. Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nature structural & molecular biology. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Molecular cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruneda JN, et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Molecular cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nature structural & molecular biology. 2013;20:982–986. doi: 10.1038/nsmb.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott DC, et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soss SE, Klevit RE, Chazin WJ. Activation of UbcH5c~Ub is the result of a shift in interdomain motions of the conjugate bound to U-box E3 ligase E4B. Biochemistry. 2013;52:2991–2999. doi: 10.1021/bi3015949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williamson A, et al. Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Molecular cell. 2011;42:744–757. doi: 10.1016/j.molcel.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He J, et al. Insights into Degron Recognition by APC/C Coactivators from the Structure of an Acm1-Cdh1 Complex. Molecular cell. 2013;50:649–660. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2014 doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng X, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, et al. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) The Biochemical journal. 2013;449:365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 43.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Molecular cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 44.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 48.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. Journal of structural biology. 2014 doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife. 2014;3:e03665. doi: 10.7554/eLife.03665. doi:10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of molecular biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 53.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Wendt KS, et al. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat Struct Biol. 2001;8:784–788. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- 57.Han D, et al. Crystal structure of the N-terminal domain of anaphase-promoting complex subunit 7. The Journal of biological chemistry. 2009;284:15137–15146. doi: 10.1074/jbc.M804887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. The EMBO journal. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, et al. Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture. Journal of molecular biology. 2010;397:1316–1328. doi: 10.1016/j.jmb.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, et al. The four canonical tpr subunits of human APC/C form related homo-dimeric structures and stack in parallel to form a TPR suprahelix. Journal of molecular biology. 2013;425:4236–4248. doi: 10.1016/j.jmb.2013.04.004. doi:10.1016/j.jmb.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 62.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He J, et al. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure. 2012;20:513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 64.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Z, et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. Journal of structural biology. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 68.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic acids research. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. Protein engineering. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 71.Hegemann B, et al. Systematic phosphorylation analysis of human mitotic protein complexes. Science signaling. 2011;4:rs12. doi: 10.1126/scisignal.2001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kraft C, et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. The EMBO journal. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steen JA, et al. Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: a quantitative proteomic analysis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6069–6074. doi: 10.1073/pnas.0709807104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. Video showing APC/CCdh1.Emi1 and APC/CCdh1.Hsl1.UbcH10-Ub reconstructions.