Abstract
Background
Increasing evidence has demonstrated that long non-coding RNAs (lncRNAs) play essential roles in the occurrence and development of human cancers, including gastric cancer (GC). However, the functional and clinical significance of lncRNAs are still poorly understood.
Methods
In this study, the expression of LncRNA HNF1A antisense RNA 1 (HNF1A-AS1) was first examined by lncRNAs microarray analysis in 6 GC tissues, and was then further verified by real-time quantitative reverse transcription PCR (qRT-PCR) both in 3 GC cell lines and 161 cases of GC tissues. We also evaluated the association between HNF1A-AS1 expression and clinicopathological features of patients with GC.
Results
LncRNAs microarray analysis results exhibited that HNF1A-AS1 was downregulated in GCs tissues (mean fold change 2.06, p < 0.05), which was further confirmed by qRT-PCR. The results from qRT-PCR showed that the expression of HNF1A-AS1 was not only downregulated in three GC cell lines (AGS, BGC-823, and MKN-45) relative to that in a normal gastric mucosal epithelial cell line (GES-1), but also decreased in GC tissues relative to that in paired adjacent non-neoplastic tissues (low expression, 94 of 161; low expression rate, 58.38 %). Furthermore, low HNF1A-AS1 expression was associated with tumor size/diameter (p = 0.005, multivariate analysis), levels of serum carcinoembryonic antigen (CEA), and carbohydrate antigen 19-9 (CA19-9), and RRM1 expression in tissue samples (p = 0.028, p = 0.009, and p = 0.006, respectively).
Conclusions
Taken together, our data indicate that lncRNA HNF1A-AS1 may be a regulator of GC, and thus, it may have potential as a novel biomarker and treatment target for this type of cancer.
Keywords: Gastric cancer, lncRNA, HNF1A-AS1, Biomarker
Background
Gastric cancer (GC) is one of the most common cancers and can occur in any section of the human stomach. These tumors are frequently malignant and have the ability to invade perineural tissue [1]. Data from the latest global annual survey have shown that there are almost 1 million new cases and 740,000 deaths from this cancer per year worldwide. The incidence of GC in China is higher than that in most countries in the world, with more than 400,000 new cases each year and approximately 300,000 deaths. Studies have shown that if this illness can be diagnosed at an early stage, good prognosis can be achieved through complete surgical resection of the primary tumor tissue [2]. Unfortunately, patients with GC show few symptoms in the early stages of disease and diagnosis is usually difficult. The 5-year survival rate of GC at advanced stages remains poor and is stubborn at approximately 30 % [3]. Cancer biomarkers allow cancer to be diagnosed and can serve as therapeutic targets, as well as can be used to explore the pathogenesis of disease; therefore, identification of cancer biomarkers would be of great value.
Long non-coding RNAs (lncRNAs) are non-coding RNAs greater than 200 nucleotides [4]. These RNAs do not encode proteins and are the main components of the human transcriptome [5]. Emerging evidence indicates that lncRNAs are important regulatory molecules whose main functions are related to modulation of gene expression networks [6]. Many of these networks are closely associated with cancer development and progression [6–9]. Data from different groups have determined that changes in expression of lncRNAs are related to the pathogenesis of diverse human cancers. Recent findings from Wang et al., Song et al., and Shao et al. have demonstrated that changes in the expression profiles of lncRNAs are linked to gastric cancer carcinogenesis and progression and that lncRNAs could be used as early biomarkers or as treatment targets for GC [10–12].
In order to identify the abnormal expression of tumor-related LncRNAs and those LncRNAs with potential values as biomarkers, in this study, the lncRNAs expression profile microarray analysis was performed in six GC tissue and their paired adjacent non-neoplastic gastric tissues. LncRNA HNF1A antisense RNA 1 (HNF1A-AS1) was one of the LncRNAs that were identified to be downregulated in GC tissues by the microarray analysis. HNF1A-AS1, a single-exon gene at band 12q24.31 of chromosome 12, is a bidirectional lncRNA of 2455 nucleotides [13]. The transcriptional start site of this lncRNA is very close to that of hepatic nuclear factor 1 alpha gene (HNF1A; HNF1A-AS1’s start site is approximately 5 kb downstream of HNF1A) [13]. To our knowledge, the expression of HNF1A-AS1 in GC tissues remains unreported. Therefore, in this work, we determined the expression levels of HNF1A-AS1 in 3 GC cell lines and in 161 cases of GC tissues and their paired adjacent non-neoplastic gastric tissues. We also investigated the potential association between the expression of HNF1A-AS1 and clinicopathological features of patients with GC. Our data illustrate the potential of this lncRNA as a novel biomarker and a treatment target for GC.
Methods
Tissue samples and clinical data collection
One hundred and sixty-one fresh paired tissue samples (GC tissue and non-neoplastic tissues collected 5 cm from the cancer’s edge) were obtained from Fuzhou General Hospital, Fujian, China, in 2014 and 2015. All of the samples were obtained immediately after surgical operation and preserved in RNAlater (Qiagen, Germany) at −80 °C until use. Serum samples used for detection of the serum tumor biomarkers and paraffin-embedded tissue samples used for detection of the immunohistochemical markers were also collected from the same patients. Every tissue sample was histopathologically diagnosed and confirmed by at least two pathologists. The standards for tumor-node-metastasis (TNM) stage and histological grade were in accordance with the guidelines of the International Union Against Cancer (UICC; 5th Ed) and the National Comprehensive Cancer Network’s (NCCN) Clinical Practice Guidelines in Oncology (V.1.2011), respectively. None of the patients received treatment prior to resection.
Ethics statement
Informed consent was obtained from all individual participants included in the study, and this study was approved by the Ethics Committee of Fuzhou General Hospital.
LncRNAs microarray assay
The lncRNAs microarrays used in this study were the Human LncRNAs Microarray V3.0 (Arraystar Inc., MD, USA), which was composed of lncRNAs and mRNAs from the human genome. lncRNAs microarray assay and the data analysis were performed by KANGCHEN Bio-tech (Shanghai, China) based on the instructions of the manufacturer.
Cell culture
AGS, BGC-823, and MKN-45 GC cell lines and normal human gastric epithelial cell line (GES-1) were obtained from the American Type Culture Collection, Manassas, VA, Shanghai Institute of Biochemistry and Cell Biology, the Chinese Academy of Sciences in Shanghai, China, Japanese Cancer Research Bank and Beijing Cancer Institute (Beijing, China), respectively. The cells were grown in F12, RP1640, or DMEM medium (Invitrogen, Grand Island, NY, USA), all of which contained 10 % fetal bovine serum, in an incubator at 37 °C with 5 % CO2.
Total RNA extraction and qRT-PCR
Total RNAs were isolated from both tissues and cultured cells by using TRIzol reagent (Invitrogen). Reverse transcription was then performed by using a Reverse Transcription kit (Promega) in accordance with the instructions of the manufacturer. Real-time quantitative reverse transcription PCR (qRT-PCR) was carried out by using the SYBR Green Mix kit (Promega, Madison, WI, USA) in a Step One Real-time PCR System (ABI, Grand Island, NY, USA). The primers for 18s and HNF1A-AS1 qPCR were as follows: (forward) 5′-TCAAGAAATGGTGGCTAT-3′ and (reverse) 5′-GCTCTGAGACTGGCTGAA-3′ for HNF1A-AS1; (forward) 5′-AGAAACGGCTACCACATCCA-3′ and (reverse) 5′-CACCAGACTTGCCCTCCA-3′ for 18s. The conditions for PCR were as follows: stage 1, 95 °C for 10 min; stage 2, 40 cycles at 95 °C for 15 s, 57 °C for 1 min; stage 3 (dissociation stage), 95 °C for 15 s, 60 °C for 15 s, 95 °C for 15 seconds. The recorded cycle threshold (Ct) values included both HNF1A-AS1 and 18 s; the latter served as a control. The expression level of HNF1A-AS1 was obtained by using the ΔCt method. Higher ΔCt values indicated lower HNF1A-AS1 expression. The results were obtained from three independent experiments and expressed as the mean ± standard deviation (SD).
Measurement of AFP, CA-125, CEA, and CA19-9 concentrations in the serum of patients with GC
The concentrations of alpha-fetoprotein (AFP), carcinoembryonic antigen (CEA), cancer antigen 125 (CA-125), and carbohydrate antigen 19-9 (CA19-9) were detected in the serum of all of 161 cases of this group patients by using the Quantitative Kit for Tumor Marker (Protein Chip-Chemiluminescence) (HealthDigit, Huzhou, China) with the HD-2001A ChipReader System (HealthDigit). In this study, the normal reference values for healthy individuals were <20.0 ng/ml, <5.0 ng/ml, <35.0 U/ml and <35.0 U/ml for AFP, CEA, CA-125, and CA19-9, respectively.
IHC
We used immunohistochemistry (IHC) in paraffin-embedded sections to determine the expression of vascular endothelial growth factor (VEGF), human epidermal growth factor receptor 2 (C-erbB-2 or HER2), thymidylate synthase (TS), breast cancer 1 (BRCA1), excision repair cross-complementation group 1 (ERCC1), Ki67 antigen (Ki67), and ribonucleotide reductase subunit M1 (RRM1), synaptophysin (Syn), neuronal cell adhesion molecules (CD56), and chromogranin A (CgA); 150 cases of the paraffin-embedded tissue samples of this group patients were available for the IHC assay. IHC was performed by pathologists in accordance with the instructions of the manufacturers. The antibodies and other reagents for IHC used in this work were purchased from Maixin (Fujian, China) and ZSGB-BIO (Beijing, China). With the exception of Ki67, the intensity of staining was graded from 0 to 4, with 0 being no staining; 1, less than 25 % of cells positive; 2, 25 to 50 % of cells positive; 3, 51 to 75 % of cells positive; and 4, >76 % cells positive. For Ki67, the grades were directly judged as the percent positive for Ki67 expression. The slides were graded by at least two pathologists.
Statistical analysis
Statistical Program for Social Sciences (SPSS) 16.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, LaJolla, CA, USA) were used for all statistical analyses. One-way analysis of variance (ANOVA), the rank-sum test, and the Student’s t test were main statistical methods used in this work, the p < 0.05 was regarded as statistically significant.
Results
LncRNAs microarray analysis exhibits that HNF1A-AS1 expression is downregulated in GCs tissues
As shown in Fig. 1, the results of lncRNAs expression profile microarray analysis gotten from six GC tissues and their paired adjacent non-neoplastic gastric tissues exhibited that HNF1A-AS1 expression was downregulated in all of the six GC tissues (mean fold change 2.06, p < 0.05), which indicated that this lncRNA may have the potential to be one of the tumor-related LncRNAs. Thus, we chose it for further analysis.
Fig. 1.
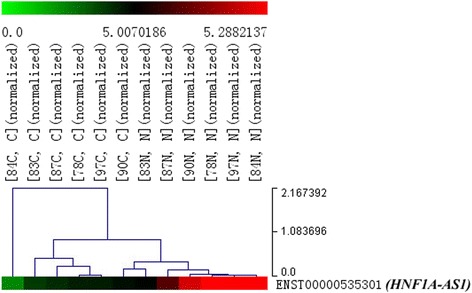
HNF1A-AS1 was downexpressed in six GC tissues identified by lncRNAs microarray analysis. 84C, 83C, 87C, 78C, 97C, and 90C indicated six GC tissues (84, 83, 87, 78, 97, and 90 were the six cases of tissue number, C cancer); 83N, 87N, 90N, 78N, 97N, and 84N indicated the six corresponding paired adjacent non-neoplastic gastric tissues (N non-neoplastic gastric tissues). Cluster analysis based on the microarray results exhibited that HNF1A-AS1 expression in GC tissues was downregulated compared with that of paired non-neoplastic gastric tissues (mean fold change 2.06, p < 0.05)
qRT-PCR verification further confirms that HNF1A-AS1 expression is downregulated in GC cell lines and tissues
We first investigated the expression of lncRNA HNF1A-AS1 in three GC cell lines: AGS, BGC-823, and MKN-45. By using qRT-PCR, we confirmed that the expression of HNF1A-AS1 was significantly decreased in all three GC cell lines relative to that in normal gastric epithelial cells (GES-1 cells; AGS, p < 0.0001; BGC-823, p < 0.05; MKN-45, p < 0.05; Fig. 2). We also examined HNF1A-AS1 expression in GC tissue samples and paired adjacent non-neoplastic tissue samples. Our results showed that HNF1A-AS1 was downregulated in 58.38 % (94/161) of GC tissues relative to that in the non-neoplastic tissues (p < 0.01, Fig. 3). Taken together, these data indicate that HNF1A-AS1 is downregulated in human GC.
Fig. 2.
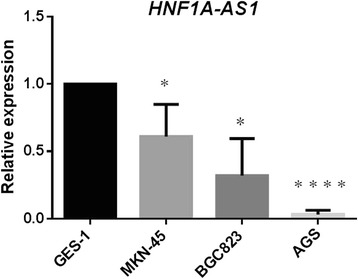
Decreased expression of HNF1A-AS1 in GC cells relative to that in normal gastric epithelial cells. qRT-PCR was used to compare the expression of HNF1A-AS1 in BGC-823, MKN-45, and AGS GC cells to that of non-neoplastic GES-1 cells. Data shown are from three independent experiments and expressed as the mean ± SD. Asterisks indicate values with a statistically significant difference versus the value in GES-1 cells. *p < 0.05, ****p < 0.0001. GC gastric cancer, qRT-PCR quantitative real-time PCR, SD standard deviation
Fig. 3.

Expression of HNF1A-AS1 in neoplastic and non-neoplastic gastric tissues. The expression of HNF1A-AS1 in GC tissues was compared to that in adjacent non-neoplastic tissues (n = 161, **p < 0.01). T GC tissues, N non-neoplastic tissues. Data shown are from three independent experiments and expressed as the mean ± SD (∆Ct value). GC gastric cancer, SD standard deviation
Association of HNF1A-AS1 expression with clinicopathological features of patients with GC
To assess the clinical significance of HNF1A-AS1, we analyzed the relationship between its expression and clinicopathological features of patients with GC. As shown in Tables 1 and 2, the univariate analysis exhibited that the expression of HNF1A-AS1 was associated with tumor size/diameter (p = 0.002), invasion depth (T stages, p = 0.032), lymphatic metastasis (N stages, p = 0.006), venous invasion (p = 0.001), and perineural invasion (PNI, p = 0.014). However, after controlling affect factors including age, sex, tumor location, tumor size/diameter, differentiation, invasion (T stages), lymphatic metastasis (N stages), venous invasion, and PNI, the multivariate analysis further confirmed that tumor size persisted to be significantly correlated to HNF1A-AS1 expression (p = 0.005), which meant that patients with tumor size ≥5 cm had lower levels of HNF1A-AS1. These data suggest that HNF1A-AS1 may play roles during cancer occurrence and progression and may be a new biomarker of gastric cancer.
Table 1.
Association of HNF1A-AS1 expression (ΔCt) with clinicopathological features of patients
| Characteristics | No. of case (%) | Mean ± SD | p value |
|---|---|---|---|
| Age (years) | 0.119 | ||
| ≥60 | 82(50.9) | 16.17 ± 2.90 | |
| <60 | 79(49.1) | 15.47 ± 2.71 | |
| Gender | 0.688 | ||
| Male | 123(76.4) | 15.78 ± 2.88 | |
| Female | 38(23.6) | 15.99 ± 2.66 | |
| Tumor location | 0.560 | ||
| Sinuses ventriculi | 57(35.4) | 16.06 ± 3.13 | |
| Cardia | 33(20.5) | 15.87 ± 3.04 | |
| Corpora ventriculi | 45(28.0) | 15.92 ± 2.50 | |
| Others | 26(16.1) | 15.11 ± 2.36 | |
| Diameter(cm) | 0.002 | ||
| ≥5 | 73(45.3) | 16.59 ± 2.8 | |
| <5 | 88(54.7) | 15.2 ± 2.70 | |
| Differentiation | 0.396 | ||
| Well | 2(1.2) | 16.15 ± 0.71 | |
| Moderate | 34(21.1) | 15.24 ± 2.66 | |
| Poor | 125(77.7) | 15.98 ± 2.87 | |
| Invasion | 0.032 | ||
| T1 | 13(8.1) | 16.13 ± 1.95 | |
| T2 | 14(8.7) | 14.09 ± 2.24 | |
| T3 | 16(9.9) | 14.85 ± 3.71 | |
| T4 | 118(73.3) | 16.13 ± 2.76 | |
| Lymphatic metastasis | 0.006 | ||
| N0 | 38(23.6) | 14.74 ± 2.77 | |
| N1-3 | 123(76.4) | 16.16 ± 2.76 | |
| Venous invasion | 0.001 | ||
| Absent | 86(53.4) | 15.14 ± 2.58 | |
| Present | 75(46.6) | 16.61 ± 2.90 | |
| Perineural invasion (PNI) | 0.014 | ||
| Absent | 89(55.3) | 15.34 ± 2.77 | |
| Present | 72(44.7) | 16.43 ± 2.79 | |
| CEA | 0.028 | ||
| Normal | 130(80.7) | 15.59 ± 2.74 | |
| High | 31(19.3) | 16.83 ± 2.97 | |
| CA19-9 | 0.009 | ||
| Normal | 140(87.0) | 15.60 ± 2.70 | |
| High | 21(13.0) | 17.32 ± 3.22 | |
| AFP | 0.936 | ||
| Normal | 150(93.2) | 15.82 ± 2.80 | |
| High | 11(6.8) | 15.89 ± 3.25 | |
| CA-125 | 0.494 | ||
| Normal | 154(95.7) | 15.79 ± 2.84 | |
| High | 7(4.3) | 16.55 ± 2.33 |
Italicized values are significant at p < 0.05
Table 2.
Comparison of the p values obtained from univariate and multivariate analyses of the association of HNF1A-AS1 expression (ΔCt) with clinicopathological features of patients
| Characteristics | Univariate | Multivariate |
|---|---|---|
| p value | p value | |
| Age (years) | 0.119 | 0.197 |
| Gender | 0.688 | 0.174 |
| Diameter (cm) | 0.002 | 0.005 |
| Differentiation | 0.396 | 0.603 |
| Invasion | 0.032 | 0.580 |
| Lymphatic metastasis | 0.006 | 0.235 |
| Venous invasion | 0.001 | 0.057 |
| Perineural invasion (PNI) | 0.014 | 0.192 |
| Tumor location | 0.560 | 0.342 |
Italicized values are significant at p < 0.05
Association of HNF1A-AS1 expression with serum or immunohistochemical markers of GC
HNF1A-AS1 expression in GC tissues was associated with two serum markers of digestive tract tumors, CEA (p = 0.028) and CA19-9 (p = 0.009), but there was no significant association with other digestive tract cancer serum marker AFP or with the epithelial tumor marker CA125 (Table 1). The expression of HNF1A-AS1 in tumor samples was also significantly correlated with one immunohistochemical markers of cancer, RRM1 (p = 0.006), but not with VEGF, C-erbB-2, TS, BRCA1, ERCC1, Ki67, CD56, Syn, or CgA (Table 3).
Table 3.
Association of HNF1A-AS1 expression (ΔCt) with immunohisochemical features of GC tissue
| Characteristics | No. of case (%) | Mean ± SD | p value |
|---|---|---|---|
| VEGF | 0.809 | ||
| 0 | 15 (10.0) | 15.27 ± 3.69 | |
| 1 | 22 (14.7) | 15.57 ± 2.92 | |
| 2 | 33 (22.0) | 15.83 ± 2.72 | |
| 3 | 59 (39.3) | 15.94 ± 2.54 | |
| 4 | 21 (14.0) | 16.39 ± 3.32 | |
| C-erbB-2 | 0.312 | ||
| 0 | 129 (86.0) | 15.70 ± 2.84 | |
| 1 | 5 (3.3) | 16.37 ± 2.96 | |
| 2 | 4 (2.7) | 15.77 ± 1.01 | |
| 3 | 12 (8.0) | 17.30 ± 3.21 | |
| TS | 0.543 | ||
| 0 | 22 (14.7) | 16.41 ± 2.45 | |
| 1 | 53 (35.3) | 15.99 ± 2.97 | |
| 2 | 52 (34.7) | 15.33 ± 2.73 | |
| 3 | 21 (14.0) | 16.25 ± 3.34 | |
| 4 | 2 (1.3) | 15.92 ± 1.82 | |
| BRCA1 | 0.893 | ||
| 0 | 10 (6.7) | 16.59 ± 1.83 | |
| 1 | 31 (20.7) | 16.07 ± 2.91 | |
| 2 | 73 (48.6) | 15.78 ± 2.57 | |
| 3 | 34 (22.7) | 15.61 ± 3.57 | |
| 4 | 2 (1.3) | 15.77 ± 5.01 | |
| ERCC1 | 0.585 | ||
| 0 | 19 (12.7) | 15.81 ± 2.39 | |
| 1 | 22 (14.7) | 16.51 ± 3.61 | |
| 2 | 44 (29.3) | 15.96 ± 2.85 | |
| 3 | 41 (27.3) | 15.85 ± 3.05 | |
| 4 | 24 (16.0) | 15.11 ± 2.02 | |
| RRM1 | 0.006 | ||
| 0 | 81 (54.0) | 15.79 ± 2.41 | |
| 1 | 10 (6.7) | 13.27 ± 4.11 | |
| 2 | 17 (11.3) | 15.51 ± 3.08 | |
| 3 | 35 (23.3) | 16.98 ± 2.93 | |
| 4 | 7 (4.7) | 15.48 ± 2.67 | |
| Ki67 | 0.328 | ||
| 1 | 14 (9.3) | 16.54 ± 1.67 | |
| 2 | 35 (23.3) | 16.44 ± 2.79 | |
| 3 | 57 (38.0) | 15.48 ± 2.97 | |
| 4 | 44 (29.4) | 15.66 ± 3.02 | |
| Syn | 0.463 | ||
| 0 | 128 (85.3) | 15.99 ± 2.95 | |
| 1 | 16 (10.7) | 15.15 ± 2.17 | |
| 2 | 3 (2.0) | 13.98 ± 0.87 | |
| 3 | 3 (2.0) | 15.59 ± 3.06 | |
| CD56 | 0.364 | ||
| 0 | 137 (91.4) | 15.94 ± 2.86 | |
| 1 | 9 (6.0) | 15.66 ± 3.09 | |
| 2 | 2 (1.3) | 12.72 ± 0.84 | |
| 3 | 2 (1.3) | 14.22 ± 0.97 | |
| CgA | 0.980 | ||
| 0 | 141 (94.0) | 15.87 ± 2.91 | |
| 1 | 7 (4.7) | 15.69 ± 1.83 | |
| 2 | 2 (1.3) | 15.63 ± 3.26 |
Italicized values are significant at p < 0.05
Discussion
Several lines of evidence have demonstrated that changes in expression of lncRNAs play important roles in carcinogenesis and cancer development [14, 15] and that altered expression of lncRNA is known to be significantly associated with several clinicopathological features of human tumors, such as lymph node metastasis, TNM stage, distant metastasis, and the grade of tissue differentiation. For example, decreased expression of gastric cancer-associated transcript 1 (GACAT1) is significantly associated with metastasis, invasion, and tissue differentiation in patients with GC [16]. In addition, elevated expression of homeobox (HOX) transcript antisense RNA (HOTAIR), a long intergenic non-coding RNA, is associated with the grade of endometrial carcinoma (EC), metastasis, depth of myometrial invasion, and poorer overall survival [17]. Finally, low expression of HMlincRNA717 in GC is associated with distal metastasis and invasion of the venous and nervous systems [12]. All of these findings are evidence of the important roles of lncRNAs in carcinogenesis and of the potential value in their clinical application.
In this study, we determined that the expression level of HNF1A-AS1 in three gastric cancer cell lines was significantly decreased relative to that in normal gastric epithelial cells. More importantly, it was downregulated in 58.38 % of gastric cancer tissue samples relative to that in adjacent non-neoplastic tissues. The findings from Yang et al. show that this lncRNA was upregulated in esophageal adenocarcinoma (EAC) [13]. Carcinogenesis and cancer progression is a complex pathological process that is mediated by gene expression networks. Cancer occurrence in different organs or tissues may be regulated by differentially expressed levels of the same genes or the same lncRNAs [18, 19]. This may explain why HNF1A-AS1 exhibits divergent expression profiles in different types of cancer cells. Further investigation may be necessary to reveal the precise molecular mechanism for these differences.
It is well known that cancer invasion and metastasis frequently occurs via three common pathways: lymph node metastasis, travel through the vasculature, and local seeding and growth. PNI is an additional means of invasion that is usually correlated with local recurrence. Because local recurrence is closely associated with poor prognosis, identifying patients at high risk of PNI has great clinical significance.
The prognosis and survival of patients with GC may also be affected by the size of the tumor, which is related to its time of growth. Smaller tumors exhibit less infringement into the stomach than do larger tumors. Moreover, increased depth of invasion by larger tumors increases the likelihood that the lymphatic system will be breached. Increasing lymph node metastasis allows the cancer to grow more easily and results in poor prognosis.
Both CEA and CA19-9 are well-known tumor markers that are usually upregulated in serum or tissue samples from patients with early or primary malignant tumors of the digestive tract. RRM1 is one of the three known human ribonucleotide reductase with the functions of regulating the cancer cell proliferation via mediating DNA synthesis and repair, and detection of the expression of RRM1 in GC tissues by IHC has been identified to have prognostic significance in patients with GC [20].
Conclusions
In summary, we analyzed the association of the expression level of HNF1A-AS1 with the clinicopathological features of patients with GC. Our results showed that the expression of HNF1A-AS1 was significantly downregulated in both GC tissues and cell lines. Moreover, decreased expression was associated with tumor size/diameter and levels of serum CEA, CA19-9, and tissue RRM1. These findings indicate that HNF1A-AS1 may be involved in suppression of gastric cancer occurrence and development and that it has the potential to serve as a novel treatment target and biomarker for the evaluation of disease severity in patients with GC. Because the sample size of this study was relatively small, further verification may be necessary to confirm HNF1A-AS1’s true clinical value.
Acknowledgements
This work was supported by the Innovation team Foundation of Fuzhou General Hospital for Lie Wang (2014CXTD04); the Postdoctoral Foundation of Fuzhou General Hospital (43853) for Yuan Dang; the National Natural Science Foundation of China (Grant No. 81372788), the Medical Scientific Research Key Foundation of Nanjing Military Command (No. 11Z032), and the Natural Science Foundation of Fujian Province (No. 2014J01427) for Qiaojia Huang; and The Medical Scientific Innovation Foundation of Nanjing Military Command (No. 2013-MS122) for Yinghao Yu.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YD performed the experiments and data analysis. FL guided some experiments. XO, KW, and YL participated in samples collected. YY guided the formalin-fixed paraffin-embedded tissue collected and immunohistochemical assay. LW and YW guided major fresh samples collected. LW also contributed to the major funding support. QH designed the study. QH and YD wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yuan Dang, Email: dangyuan830307@hotmail.com.
Fenghua Lan, Email: fhlan@mail.fjmu.edu.cn.
Xiaojuan Ouyang, Email: 798951352@qq.com.
Kai Wang, Email: posuly@sina.com.
Youdong Lin, Email: 2444734512@qq.com.
Yinghao Yu, Email: yuyinghao0808@126.com.
Lie Wang, Email: fzptwk@xmu.edu.cn.
Yu Wang, Email: flyfishwang@hotmail.com.
Qiaojia Huang, Email: huangqj100@126.com.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Yamashita K, Sakuramoto S, Nemoto M, Shibata T, Mieno H, Katada N, et al. Trend in gastric cancer: 35 years of surgical experience in Japan. World J Gastroenterol. 2011;17:3390–3397. doi: 10.3748/wjg.v17.i29.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang Y, Wang YC, Huang QJ. Microarray and next-generation sequencing to analyse gastric cancer. Asian Pac J Cancer Prev. 2014;15:8033–8039. [PubMed] [Google Scholar]
- 4.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Xuan Z, Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci. 2013;14:18790–18808. doi: 10.3390/ijms140918790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Feng X, Jia R, Liu G, Zhang M, Fan D, et al. Microarray expression profile analysis of long non-coding RNAs of advanced stage human gastric cardia adenocarcinoma. Mol Genet Genomics. 2014;289:291–302. doi: 10.1007/s00438-013-0810-4. [DOI] [PubMed] [Google Scholar]
- 11.Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. doi: 10.1186/1479-5876-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao Y, Chen H, Jiang X, Chen S, Li P, Ye M, et al. Low expression of lncRNA-HMlincRNA717 in human gastric cancer and its clinical significances. Tumour Biol. 2014;35:9591–9595. doi: 10.1007/s13277-014-2243-z. [DOI] [PubMed] [Google Scholar]
- 13.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Inter J Biochem Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 15.Tang JY, Lee JC, Chang YT, Hou MF, Huang HW, Liaw CC, et al. Long noncoding RNAs-related diseases, cancers, and drugs. Scientific World Journal. 2013;943539. [DOI] [PMC free article] [PubMed]
- 16.Sun W, Wu Y, Yu X, Liu X, Song H, Xia T, et al. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013;34:2697–2701. doi: 10.1007/s13277-013-0821-0. [DOI] [PubMed] [Google Scholar]
- 17.He X, Bao W, Li X, Chen Z, Che Q, Wang H, et al. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med. 2014;33:325–332. doi: 10.3892/ijmm.2013.1570. [DOI] [PubMed] [Google Scholar]
- 18.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma P, et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14(1):165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Liu X, Zhou J, Huang Y, Zhang S, Shen J, et al. Ribonucleotide reductase large subunit M1 predicts poor survival due to modulation of proliferative and invasive ability of gastric cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070191. [DOI] [PMC free article] [PubMed] [Google Scholar]


