Abstract
Background
Genetic variants have been associated with the risk of coronary heart disease (CHD). We tested whether a composite of these variants could identify the risk of both incident as well as recurrent CHD events and distinguish individuals who derived greater clinical benefit from statin therapy.
Methods
A community-based cohort and four randomized controlled trials of both primary (JUPITER and ASCOT) and secondary (CARE and PROVE IT-TIMI 22) prevention with statin therapy totaling 48,421 individuals and 3,477 events were included in these analyses. We examined the association of a genetic risk score based on 27 genetic variants with incident or recurrent CHD, adjusting for established clinical predictors. We then investigated the relative and absolute risk reductions in CHD events with statin therapy stratified by genetic risk. Data from studies were combined using meta-analysis.
Findings
When individuals were divided into low (quintile 1), intermediate (quintiles 2-4), and high (quintile 5) genetic risk categories, a significant gradient of risk for incident or recurrent CHD was demonstrated with the multivariable-adjusted HRs (95% CI) for CHD for the intermediate and high genetic risk categories vs. low genetic risk category being 1.32 (1.20-1.46, P<0.0001) and 1.71 (1.54-1.91, P<0.0001), respectively. In terms of the benefit of statin therapy in the four randomized trials, there was a significant gradient of increasing relative risk reduction across the low, intermediate, and high genetic risk categories (13%, 29%, and 48%, P=0.0277). Similarly, greater absolute risk reductions were seen in those individuals in higher genetic risk categories (P=0.0101), resulting in an approximate three-fold gradient in the number needed to treat (NNT) in the primary prevention trials. Specifically, in the primary prevention trials, the NNT to prevent one MACE over 10 years for the low, intermediate, and high GRS individuals was 66, 42, and 25 in JUPITER and 57, 47, and 20 in ASCOT.
Interpretation
A genetic risk score identified individuals at increased risk for both incident and recurrent CHD events. Individuals with the highest burden of genetic risk derived the largest relative and absolute clinical benefit with statin therapy.
INTRODUCTION
The risk of developing coronary heart disease (CHD) depends on a number of factors related both to lifestyle and genetics. Heritable factors account for as much as 30-60% of the variation in risk,1, 2 and large-scale studies have identified genetic variants associated with CHD at stringent levels of statistical significance.3, 4 Prior studies have demonstrated that an assessment of genetic risk burden using multiple loci can identify individuals at increased risk for incident CHD in population-based epidemiological cohorts.5-12 Additionally, whereas some individual variants have been evaluated in isolated studies for an association with recurrent events, an independent association between a multilocus genetic risk score and recurrent CHD events has not been clearly demonstrated.13-16
The clinical benefit from therapies that reduce the likelihood of CHD events might vary by the degree of risk at baseline. As such, in addition to identifying risk, a genetic risk score consisting of validated CHD-risk SNPs might also distinguish individuals who enjoy greater clinical benefit from statin therapy, a hypothesis that has not been validated to date in randomized controlled trials of statin therapy. Such a finding would be of particular interest in the setting of primary prevention. Therefore, the goals of the present study were two-fold: (1) to test if a multilocus genetic risk score based on a combination of 27 loci might predict not only incident CHD in an epidemiologic cohort but also incident or recurrent CHD events in a clinical trial setting; and (2) to evaluate whether the clinical benefit of statin therapy varies by genetic risk score in four randomized controlled trials of statin therapy.
METHODS
Primary Prevention Populations
Baseline characteristics from each study are provided in Supplemental Table 1. In brief, the Malmo Diet and Cancer Study (MDCS) is a community-based prospective epidemiologic cohort of middle-aged individuals from Southern Sweden.17 Genetic samples were available in 27,817 individuals without documented CHD at baseline. JUPITER was a primary prevention trial that tested rosuvastatin 20 mg daily versus placebo in individuals with low-density lipoprotein (LDL) cholesterol <130 mg/dl and hs-CRP ≥2 mg/L and no known cardiovascular disease.18 Genetic samples were available in 8,749 individuals for this analysis. Another primary prevention trial, ASCOT, tested the clinical benefit of different classes of antihypertensive therapy in high-risk individuals without documented coronary heart disease. The lipid lowering arm (ASCOT-LLA) randomized individuals from the main trial with a total cholesterol ≤251 mg/dL to either atorvastatin 10 mg daily or placebo.19 In total, 4,219 individuals from ASCOT-LLA were included in the treatment-related analyses. Additionally, 2,759 individuals from the blood pressure lowering arm of ASCOT not taking a statin medication were assessed in the non-treatment related analyses.
Secondary Prevention Populations
CARE was a secondary-prevention trial that investigated the clinical benefit of pravastatin 40 mg daily versus placebo in individuals with prior MI who had total cholesterol ≤240 mg/dL and LDL cholesterol between 115 and 174 mg/dL.20 Genetic samples were available for 2,878 individuals. Another secondary prevention trial, PROVE-IT TIMI 22, investigated the clinical benefit of moderate statin therapy (pravastatin 40 mg daily) versus intensive statin therapy (atorvastatin 80 mg daily) in individuals after an acute coronary syndrome (ACS) and who had total cholesterol ≤ 240 mg/dL.21 A genetic substudy included 1,999 individuals.
Genetic Risk Score
A genetic risk score was derived based on 27 single nucleotide polymorphisms (SNPs) that were significantly associated with CHD at a genome-wide level in prior analyses (see Supplemental Methods).22 The loci, lead SNP, effect sizes, risk allele, and risk allele frequency are shown in Table 1, with the specifics for each study provided in Supplemental Table 2. Each individual received a score equal to the sum of the number of risk alleles for each SNP weighted by the log of the odds ratio observed with the SNP in the original report (Supplemental Figure 1).
Table 1.
Components of the Genetic Risk Score
| Locus | Lead SNP | OR for CHD | Risk Allele | Risk Allele Frequency |
|---|---|---|---|---|
| 1p13.3 (SORT1) | rs646776 | 1.19 | T | 0.77 |
| 1p32.2 (PPAP2B) | rs17114036 | 1.17 | A | 0.92 |
| 1p32.3 (PCSK9) | rs11206510 | 1.15 | T | 0.82 |
| 1q41 (MIA3) | rs17465637 | 1.14 | C | 0.75 |
| 2q33.1 (WDR12) | rs6725887 | 1.17 | C | 0.13 |
| 3q22.3 (MRAS) | rs9818870 | 1.15 | T | 0.15 |
| 6p21.31 (ANKS1A) | rs17609940 | 1.07 | G | 0.79 |
| 6p24.1 (PHACTR1) | rs9349379 | 1.12 | G | 0.43 |
| 6q23.2 (TCF21) | rs12190287 | 1.08 | C | 0.63 |
| 6q25.3 (LPA) | rs3798220 | 1.47 | C | 0.01 |
| 6q25.3 (LPA) | rs10455872 | 1.70 | G | 0.07 |
| 7q32.2 (ZC3HC1) | rs11556924 | 1.09 | C | 0.64 |
| 9p21.3 (CDKN2A) | rs4977574 | 1.29 | G | 0.55 |
| 9q34.2 (ABO) | rs9411489 | 1.10 | T | 0.21 |
| 10q11.21 (CXCL12) | rs1746048 | 1.17 | C | 0.86 |
| 10q24.32 (CYP17A1) | rs12413409 | 1.12 | G | 0.90 |
| 11q23.3 (APOA5) | rs964184 | 1.13 | G | 0.13 |
| 12q24 (HNF1A) | rs2259816 | 1.08 | T | 0.35 |
| 12q24.12 (SH2B3) | rs3184504 | 1.13 | T | 0.48 |
| 13q34 (COL4A1) | rs4773144 | 1.07 | G | 0.41 |
| 14q32.2 (HHIPL1) | rs2895811 | 1.07 | C | 0.45 |
| 15q25.1 (ADAMTS7) | rs3825807 | 1.08 | T | 0.57 |
| 17p11.2 (RASD1) | rs12936587 | 1.07 | G | 0.53 |
| 17p13.3 (SMG6) | rs216172 | 1.07 | C | 0.64 |
| 17q21.32 (UBE2Z) | rs46522 | 1.06 | T | 0.48 |
| 19p13.2 (LDLR) | rs1122608 | 1.15 | G | 0.77 |
| 21q22.11 (KCNE2) | rs9982601 | 1.20 | T | 0.13 |
The material in this table is from MDCS. CHD indicates coronary heart disease.
Statistical Analysis
The outcome of interest was CHD given that the SNPs were originally reported to be associated with coronary events. CHD reflected the available endpoints, and an effort was made to harmonize the definitions across studies. In JUPITER, ASCOT, and PROVE IT-TIMI 22, CHD was defined as a composite of coronary heart death, MI, or unstable angina. In MDCS, CHD represented a composite of fatal or nonfatal MI, coronary artery bypass grafting, or PCI; for CARE, CHD was coronary heart death or MI.
Using Cox proportional hazard models, the risk of CHD was assessed for each quintile of genetic risk using the first quintile as the reference group; additionally, the risk for categories [low (quintile 1), intermediate (quintiles 2-4), and high (quintile 5)] and per 1-standard deviation was calculated. These analyses were conducted among participants in MDCS and in the placebo or lower intensity statin treatment arms of the applicable trials. The models were adjusted for age, sex, diabetes status, smoking, race where applicable, family history of CHD, high-density lipoprotein (HDL) cholesterol, LDL cholesterol, and hypertension. In a meta-analysis, the estimates were combined from each study using a random-effects model to account for possible differences in study populations. Heterogeneity across studies and types of studies was assessed, and analyses were stratified based on the primary and secondary prevention populations.
The treatment-specific analyses were conducted in the JUPITER, ASCOT, CARE, and PROVE IT-TIMI 22 trials. The effect of statin versus placebo (or high-intensity versus moderate-intensity statin in the case of PROVE-IT TIMI 22) was evaluated, and the number of events and event rates in the statin and placebo groups were analyzed based on the genetic risk score quintiles and aforementioned categories. Hazard ratios and 95% confidence intervals were generated, and absolute risk reductions were calculated. For the primary prevention trials, JUPITER and ASCOT, ten-year event rates were extrapolated and the numbers needed to treat were calculated for each study.
The relative risk ratios for the benefit of statin therapy within each genetic risk score category were combined across the trials using meta-analytic techniques, with separate analyses conducted for the primary and secondary prevention populations. The resulting meta-analytic risk ratios across the genetic risk score categories were evaluated using meta-regression. In terms of absolute risk reductions with statins, within each trial and within each genetic risk score category, the absolute risk difference for statin versus placebo and corresponding standard errors and 95% confidence interval (CI) were generated. Notably, the fact that the trials had populations with different absolute event rates (due to different cardiovascular risk and different durations of follow-up) precluded a clinically interpretable result from simply combining the raw absolute risk differences across the 4 trials. Therefore, to normalize across the trials, a scaling factor was applied to the data (see Supplement for further details). Then within each trial, meta-regression was performed across the genetic risk score categories to determine how the relative magnitude of absolute risk reduction with statin therapy varied by genetic risk score category. Meta-analysis was then performed combining the regression coefficients from the 4 trials, again stratified by primary and secondary prevention populations. For this analysis, the funders did not assist in analysis, interpretation, or writing of the manuscript. Investigators associated with each study had access to the data, and JLM, NOS, SK, and MSS were responsible for the decision to submit the manuscript.
RESULTS
Genetic Risk Score and Cardiovascular Risk
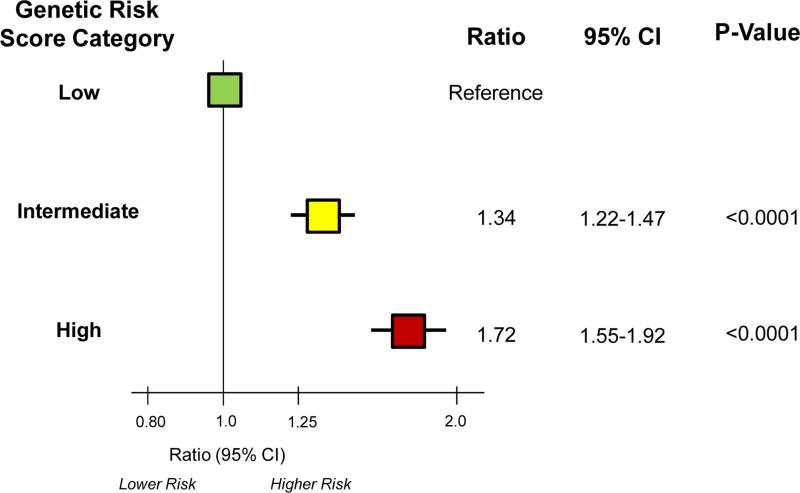
Higher genetic risk scores were associated with a higher risk of CHD, adjusting for established clinical predictors. Specifically, dividing participants into low (quintile 1), intermediate (quintiles 2-4), and high (quintile 5) genetic risk score categories, a gradient of risk for CHD was observed in the studies (Table 2). Combining the data from the primary prevention cohorts, the multivariable-adjusted HRs (95% CI) for incident CHD for the intermediate and high genetic risk categories versus low genetic risk category were 1.31 (1.19-1.45, P<0.0001) and 1.72 (1.53-1.92, P<0.0001), respectively (Table 2). Similarly, the multivariable-adjusted HRs (95% CI) for recurrent CHD in the secondary prevention cohorts were 1.65 (1.19-2.30, P=0.0030) and 1.81 (1.22-2.67, P=0.0029, respectively (Table 2). Overall, the multivariable-adjusted HRs (95% CI) were 1.34 (1.22-1.47, P<0.0001) and 1.72 (1.55-1.92, P<0.0001), respectively (Figure 1). Data for individual quintiles were similar and are presented in Supplemental Table 3.
Table 2.
Risk of Coronary Heart Disease Across Genetic Risk Score Categories
| Study | Low Genetic Risk Category | Intermediate Genetic Risk Category | High Genetic Risk Category | P Value (Intermediate vs. Low) | P Value (High vs. Low) |
|---|---|---|---|---|---|
| Primary Prevention Populations | |||||
| MDCS | 1.00 | 1.30 (1.17-1.44) | 1.70 (1.51-1.91) | 2×10−6 | 2×10−18 |
| JUPITER | 1.00 | 1.23 (0.61-2.44) | 1.32 (0.58-2.98) | 0.56 | 0.51 |
| ASCOT | 1.00 | 1.58 (1.06-2.34) | 2.10 (1.35-3.25) | 0.0236 | 0.0009 |
| Meta-Analysis | 1.00 | 1.31 (1.19-1.45) | 1.72 (1.53-1.92) | <0.0001 | <0.0001 |
| Secondary Prevention Populations | |||||
| CARE | 1.00 | 1.52 (0.99-2.33) | 1.67 (1.01-2.76) | 0.0575 | 0.0482 |
| PROVE IT-TIMI 22 | 1.00 | 1.87 (1.11-3.16) | 2.04 (1.10-3.79) | 0.0190 | 0.0239 |
| Meta-Analysis | 1.00 | 1.65 (1.19-2.30) | 1.81 (1.22-2.67) | 0.0030 | 0.0029 |
These analyses were conducted among participants in MDCS and in the placebo or lower intensity statin treatment arms (PROVE IT-TIMI 22) of the applicable trials.
Figure 1. Summary of Risk of Coronary Heart Disease Across Genetic Risk Score Categories in Primary and Secondary Prevention Populations.
The boxes indicate the point estimates and the horizontal lines the 95% confidence intervals.
Benefit of Statins across Genetic Risk Categories
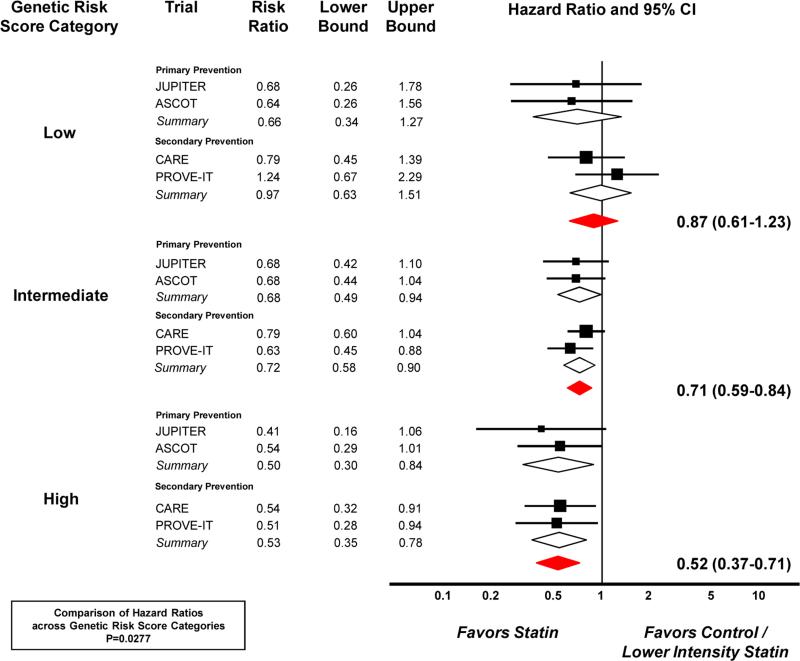
Baseline LDL cholesterol and HDL cholesterol were similar across genetic risk score categories within each trial, as were the absolute and percentage changes with statin therapy (Supplemental Tables 4 and 5). Analyses were conducted to investigate the clinical benefit of statin therapy across genetic risk score. The benefit of statin versus placebo in the primary and secondary prevention trials with a total of 806 events is presented across genetic risk score categories in Table 3 and genetic risk score quintiles in Supplemental Table 6. The relative risk reductions across low, intermediate, and high genetic risk score categories were 34%, 32% and 50%, respectively, in the primary prevention trials and 3%, 28%, and 47% in the secondary prevention trials. When combining the data, the gradient of relative risk reductions with statin therapy across low, intermediate, and high genetic risk score categories were 13%, 29%, and 48% (P for trend=0.0277, Figure 2).
Table 3.
Risk of Coronary Heart Disease Associated with Statin Therapy Across Genetic Risk Score Categories
| # of Events/Yrs of Followup/Person Yrs of Followup | Genetic Risk Group | # of Events, Control | # of Individuals, Control | Event Rate, Control (%) | # of Events, Statin | # of Individuals, Statin | Event Rate, Statin (%) | Hazard Ratio | ARR (%) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Primary Prevention Populations | ||||||||||
| JUPITER | 108 | Low | 10 | 865 | 1.16 | 7 | 878 | 0.80 | 0.68 | 0.36 |
| 2.37 | Intermediate | 43 | 2621 | 1.64 | 28 | 2605 | 1.07 | 0.68 | 0.57 | |
| 20,612 | High | 14 | 864 | 1.62 | 6 | 878 | 0.68 | 0.41 | 0.94 | |
| ASCOT | 149 | Low | 13 | 432 | 3.00 | 8 | 412 | 1.94 | 0.64 | 1.06 |
| 6.07 | Intermediate | 48 | 1187 | 4.04 | 37 | 1344 | 2.75 | 0.68 | 1.29 | |
| 25,609 | High | 28 | 426 | 6.57 | 15 | 418 | 3.59 | 0.54 | 2.98 | |
|
Secondary Prevention Populations | ||||||||||
| CARE | 320 | Low | 26 | 278 | 9.35 | 22 | 297 | 7.41 | 0.79 | 1.94 |
| 4.94 | Intermediate | 119 | 877 | 13.57 | 92 | 850 | 10.82 | 0.79 | 2.75 | |
| 13,623 | High | 38 | 277 | 13.72 | 23 | 299 | 7.69 | 0.54 | 6.03 | |
| PROVE IT-TIMI 22* | 229 | Low | 20 | 213 | 9.39 | 21 | 186 | 11.29 | 1.24 | −1.90 |
| 2.03 | Intermediate | 88 | 605 | 14.55 | 55 | 595 | 9.24 | 0.63 | 5.31 | |
| 3,769 | High | 28 | 188 | 14.89 | 17 | 212 | 8.02 | 0.51 | 6.87 | |
In PROVE IT-TIMI 22, the control group is moderate intensity statin therapy (pravastatin 40 mg) and the statin group is high intensity statin therapy (atorvastatin 80 mg).
ARR indicates absolute risk reduction; yrs, years.
Figure 2. Risk Ratios for Coronary Heart Disease with Statin Therapy across Genetic Risk Score Categories.
The boxes indicate the point estimates, and the size of each box reflects the weight of a trial's data within that subgroup. The horizontal lines display the 95% confidence intervals. The diamonds provide summary data. In PROVE IT-TIMI 22, the control group is moderate intensity statin therapy (pravastatin 40 mg) and the statin group is high intensity statin therapy (atorvastatin 80 mg).
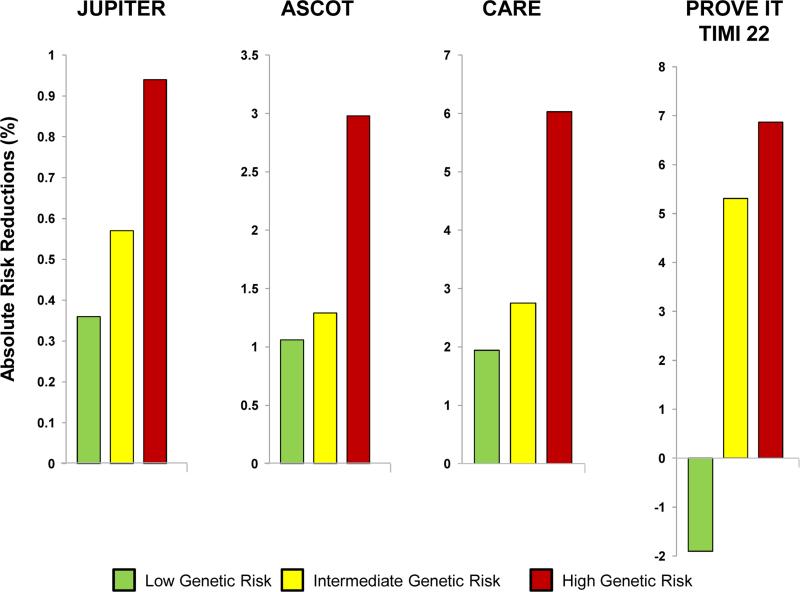
Likewise, in terms of the absolute risk reductions, a graded increase in the benefit of statin therapy across the genetic risk score categories was evident in both the primary prevention (JUPITER and ASCOT) and the secondary prevention trials (CARE and PROVE IT-TIMI 22) (Table 3 and Figure 3). Correspondingly, the number needed to treat (NNT) to reduce CHD events with statin therapy differed according to genetic risk score. Focusing on the primary prevention trials, in JUPITER, the NNT to prevent one CHD event over 10 years for those individuals with low, intermediate, and high genetic risk score was 66, 42, and 25. In ASCOT, the NNT to prevent one CHD event over 10 years was 57, 47, and 20 across the three genetic risk score categories.
Figure 3. Absolute Risk Reductions of Coronary Heart Disease with Statin Therapy across Genetic Risk Score Categories.
In PROVE IT-TIMI 22, the control group is moderate intensity statin therapy (pravastatin 40 mg) and the statin group is high intensity statin therapy (atorvastatin 80 mg).
Calculating the difference in absolute risk reduction in each trial as a function of genetic risk score category and combining the data from the trials demonstrated a consistent and significant gradient, with greater absolute risk reductions seen in those individuals in higher genetic risk score categories (P=0.0101, Supplemental Figure 2). The regression coefficient of 0.71 indicates that, compared with the absolute risk reduction seen in the intermediate risk category, statin therapy would result in an absolute risk difference that is 71% lower in the low genetic risk score category and 71% higher in the high genetic risk score category (i.e., if the absolute risk reduction was 1% in the intermediate risk genetic risk score category, it would be 0.29% and 1.71% in the low and high genetic risk score categories, respectively).
DISCUSSION
Large-scale genetic association studies have identified a number of genetic variants that are individually associated with the risk of CHD. When combined into a 27-variant risk score, we found, in multivariable adjusted analyses, that these variants were able to identify individuals at increased degrees of risk of CHD events, including incident CHD in primary prevention populations and recurrent CHD events in secondary prevention populations. Furthermore, when compared with individuals at low genetic risk, those with the highest genetic risk scores derived greater relative risk reduction and absolute risk reduction with statin therapy. Notably, among the primary prevention statin studies, there was an approximate three-fold gradient in the number needed to treat to prevent one CHD event.
Clinical, biochemical, and imaging parameters have been used to stratify cardiovascular risk and potentially to tailor therapy. The present analysis suggests that genetics might also play such a role. Prior data for genetic variants predicting recurrent CHD events independent of traditional risk factors is inconclusive,13-16 perhaps a function of varying definitions of prevalent CHD (e.g., angina vs documented MI) and inclusion of less specific outcomes (e.g., non-cardiovascular death) in a composite endpoint. With regard to treatment options, the decision to prescribe any drug depends on weighing several factors including efficacy, safety, and cost. In the case of statins, substantial relative risk reductions in major cardiovascular outcomes have been demonstrated across the spectrum of primary and secondary prevention.23 Absolute risk reductions can depend on the risk profile of the population, but even in lower risk individuals, statins provide clinical benefit.24
Nonetheless, there remains debate about the use of statins in relatively lower risk individuals and particularly primary prevention populations,25 driven by concerns about safety and cost-effectiveness in an extremely broad population. For that reason, an understanding of the absolute risk reductions achieved with statin therapy in different subgroups may be useful in some circumstances. Moreover, there has been interest in considering lifetime risk of CHD and information about genetic risk could be obtained early in life.26 As such, there have been discussions about considering statin therapy earlier in individuals who do not currently meet practice guidelines, but who may still be at elevated risk of events. Defining the best approach for maximizing the benefit of statin therapy in such a population is a complex challenge that requires further study. A genetic risk score offers a unique window onto future risk and may aid in selecting populations for clinical trials that, so enriched, would be better positioned to test the clinical benefit of early initiation of statin therapy in primary prevention; specifically, the role of statin therapy in individuals with apparently low clinical risk but with high genetic risk could be tested.27
Prior analyses have evaluated genetics and CHD events in the setting of statin therapy.28-39 However, such approaches have been limited by either examining only a single SNP, using SNPs whose association with CHD has not been well validated, not testing the SNP in a randomized trial of statin therapy, and/or not consistently validating any observed interactions. In contrast, the present analysis used a multi-locus genetic risk score compromised of well-validated CHD-risk SNPs and tested the score in 4 randomized controlled trials of statin therapy.
There are some potential limitations to these analyses. First, the data from several studies were utilized in this analysis, and each study has its own entry criteria, treatment allocation, and follow-up. As such, the hazards associated with the genetic risk categories differed somewhat, with the genetic-based risk in JUPITER appearing to be the lowest and possibly related to the patient population or the lower event rates with wider confidence intervals. However, having access to data from a large community cohort study, as well as primary and secondary prevention clinical trials, allowed us to test the generalizability of the genetic risk score across the various populations. Second, the NNT data were calculated by extrapolating the effect of statin therapy over 10 years, and the treatment effect could vary over time. Nonetheless, studies of statin therapy suggest a relatively linear relationship with coronary event reduction over the long-term.40 Third, these analyses were conducted within completed clinical trials, and the genetic risk score was not specifically used as an enrollment criterion. Dedicated clinical trials using a genetic risk score to triage statin therapy would further add to the knowledge base. Fourth, although we focused on the ability of genetics to offer insight into risk of CHD and benefit of statin therapy, optimal tailoring of therapy may require a combination of several factors. Fifth, the present analysis focuses on genetic variants that were associated with the risk of CHD. Other variants have been described that are associated with LDL cholesterol levels. However, these variants primarily affect baseline LDL cholesterol, which is already routinely measured. Moreover, as prior studies show, they have little impact on the change in LDL cholesterol with statins and/or the clinical response to statin therapy.41, 42 Therefore, the current analyses do not explore such variants unless there was a prior association with CHD. Nonetheless, such a line of inquiry might also be informative. Finally, the gradient of relative risk reduction across genetic risk score categories seen in the present study was unexpected. Data from other studies suggest that patients with a higher burden of CHD-risk SNPs have more extensive atherosclerosis.8 Thus, it is plausible that such patients experience a relatively greater benefit with statin therapy because there are more plaques that can be stabilized. However, this concept deserves additional study to elucidate the underlying mechanism.
In conclusion, we found that a genetic risk score identified individuals at increased risk of CHD across primary and secondary prevention populations. Furthermore, individuals with high genetic risk scores demonstrated the numerically largest relative and absolute risk reductions with statin therapy. In situations where optimizing the number needed to treat is relevant, such as in the primary prevention setting, tools such as genetics may prove useful.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic Review
We searched PubMed for original research pertaining to this analysis. The following combination of keywords were used: “genetic,” “risk score,” and “coronary disease,” and identified studies that describe the association of genetic variants and the risk of CHD, including several analyses examining the prognostic significance of multiple genetic variants, although these studies were largely confined to epidemiological cohorts and prediction of first manifestation of CHD. We searched “genetic,” “risk score,” “coronary disease,” and “statins” without identifying studies that directly tested this specific concept. Therefore, in the present study we aimed to: (1) test if a multi-locus genetic risk score predicts not only incident CHD in an epidemiologic cohort but also recurrent CHD events in a clinical trial setting; and (2) evaluate whether the clinical benefit of statin therapy varies by genetic risk score.
Interpretation
A genetic risk score was derived based on 27 single nucleotide polymorphisms (SNPs) that have been significantly associated with CHD at a genome-wide level in prior analyses. First, in an epidemiologic cohort, individuals with higher genetic risk scores were determined to have a higher risk of CHD, even after adjusting for established clinical predictors. We then evaluated the association between the genetic risk score and CHD in primary and secondary prevention trials of statin therapy, and validated a gradient of risk for incident as well as recurrent CHD. In terms of the benefit of statin therapy, we tested the genetic risk score in four clinical trials and identified a significant gradient of increasing relative risk reduction across the low, intermediate, and high genetic risk categories. Similarly, in each trial, greater absolute risk reductions were seen in those individuals in higher genetic risk categories, resulting in an approximate three-fold gradient in the number needed to treat in the primary prevention trials. Thus, genetics could aid in selecting populations for clinical trials that, so enriched, would be better positioned to test the clinical benefit of early initiation of statin therapy. Moreover, in situations where optimizing the number needed to treat is relevant, genetics could provide useful information.
Acknowledgments
ADDITIONAL FUNDING SOURCES
JUPITER was supported by AstraZeneca; ASCOT was supported by Pfizer; CARE was supported by Bristol-Myers Squibb; and, PROVE IT-TIMI 22 was supported by Bristol-Myers Squibb and Sankyo.
Mega JL, Kathiresan S, and Sabatine MS received funding from the National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute (NHLBI) R01 HL098082.
Stitziel NO was supported, in part, by a career development award from the NIH/NHLBI K08 HL114642.
The Malmö Diet and Cancer Study was funded by The Swedish Research Council, The Swedish Heart and Lung Foundation, the European Research Council and the Novo Nordisk Foundation.
This work was facilitated by the National Institutes for Health Research (NIHR) Biomedical Research Unit at Barts and the NIHR Biomedical Research Centre at Imperial College. Caulfield M, Poulter N, and Sever P are recipients of NIHR Senior Investigator Awards.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONTRIBUTIONS
JLM, NOS, SK, and MSS contributed to the study design, data analysis, data interpretation, and manuscript writing. JGS, DIC, MC, JJD, FN, CH, CPC, FS, NP, PS, RMR, EB, and OM contributed to the study design, data analysis, and data interpretation of either the present analysis or the associated clinical trials, and they each reviewed and/or revised the manuscript.
DISCLOSURES
Mega JL: Research grant support through Brigham and Women's Hospital from Bristol-Myers Squibb and Sankyo, during the conduct of the study; grant support through Brigham and Women's Hospital from Janssen, Bayer, Bristol-Myers Squibb/Sanofi, Daiichi Sankyo, and AstraZeneca, outside the submitted work: personal fees from Janssen, American Genomics, and Boehringer-Ingelheim, outside the submitted work. Stitziel NO: Grants from the National Institutes of Health, during the conduct of the study; personal fees from American Genomics, outside the submitted work. Smith JG: None. Chasman DI: Research support for genetic analysis in the JUPITER population from AstraZeneca. Caulfield M: Reports non-financial support (member of the board): Genomics England. Devlin J: Reports employment at Quest Diagnostics, a company that offers tests for cardiovascular disease; personal fees from Quest Diagnostics, outside the submitted work. Nordio F: None. Hyde C: Reports employment at Pfizer; reports personal fees from Pfizer, outside the submitted work. Cannon CP: Research grant support through Brigham and Women's Hospital from Accumetrics, Arisaph, AstraZeneca, Boehringer-Ingelheim, Glaxo Smith Kline, Janssen, Merck, Regeneron, Sanofi, and Takeda; personal fees from Accumetrics, CSL Behring, Essentialis, Merck, Regeneron, Sanofi, Takeda, Bristol-Myers Squibb, Lipimedix, Pfizer: advisory board for, Bristol-Myers Squibb, Lipimedix, and Pfizer, outside the submitted work. Sacks F: Reports grants from Bristol Myers Squibb during the conduct of the study: other from Pfizer, outside the submitted work. Poulter N: Reports personal fees for Pfizer; grants from Servier, Pfizer, and Amgen, outside the submitted work. Sever P: Reports grant support through Imperial College from Pfizer; personal fees (Honoraria) from Pfizer for lectures, outside the submitted work. Ridker PM: Research grant through Brigham and Women's Hospital from AstraZeneca and Amgen. For outside the submitted work he received grants through the Brigham and Women's Hospital from Novartis and Pfizer, and is listed as a co-inventor on patents held by the Brigham and Women's Hospital that relate to the use of inflammatory biomarkers in cardiovascular disease and diabetes that have been licensed to AstraZeneca and Seimens. Braunwald E: For the work under consideration; grants through Brigham and Women's Hospital from Merck, and Bristol Myers Squibb and uncompensated consultancies and lectures from Merck. For outside the submitted work; grants from Daiichi Sankyo, Duke University, Astra Zeneca, Glaxo Smith Kline, Johnson and Johnson, and Sanofi Aventis; personal fees for consultancies from Genzyme, The Medicines Company, and Sanofi Aventis; personal fees for lectures from Menari International, Medscape, Bayer, and Daiichi Sankyo. Melander O: None. Kathiresan S: Grants from National Institutes of Health, during the conduct of the study; Research grant support from Merck, Celera, outside the submitted work; personal fees from, Catabasis, Regeneron, Amgen, Amarin, and Eli Lilly, outside the submitted work : grants from National Institutes of Health during the conduct of the study. Sabatine MS: Research grant support through Brigham and Women's Hospital from Abbott Laboratories, Accumetrics, Amgen, AstraZeneca, AstraZeneca / Bristol-Myers Squibb Alliance, BRAHMS, Bristol-Myers Squibb / Sanofi-aventis Joint Venture, Critical Diagnostics, Daiichi-Sankyo, diaDexus, Eisai, Genzyme, Glaxo Smith Kline, Intarcia, Merck, Nanosphere, Ortho-Clinical Diagnostics, Roche Diagnostics, Sanofi-aventis, Singulex, and Takeda; consultant to Aegerion, Amgen, AstraZeneca, Bristol-Myers Squibb, Daiichi-Sankyo / Eli Lilly, Glaxo Smith Kline, Intarcia, Merck, MyoKardia, Pfizer, Sanofi-Aventis, Vertex, Zeus, Cubist, and Quest Diagnostics.
REFERENCES
- 1.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330(15):1041–6. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones DM, Nam BH, D'Agostino RB, Sr., Levy D, Murabito JM, Wang TJ, et al. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. Jama. 2004;291(18):2204–11. doi: 10.1001/jama.291.18.2204. [DOI] [PubMed] [Google Scholar]
- 3.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148(6):1242–57. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45(1):25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphries SE, Cooper JA, Talmud PJ, Miller GJ. Candidate gene genotypes, along with conventional risk factor assessment, improve estimation of coronary heart disease risk in healthy UK men. Clinical chemistry. 2007;53(1):8–16. doi: 10.1373/clinchem.2006.074591. [DOI] [PubMed] [Google Scholar]
- 6.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358(12):1240–9. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 7.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376(9750):1393–400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, et al. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5(1):113–21. doi: 10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brautbar A, Pompeii LA, Dehghan A, Ngwa JS, Nambi V, Virani SS, et al. A genetic risk score based on direct associations with coronary heart disease improves coronary heart disease risk prediction in the Atherosclerosis Risk in Communities (ARIC), but not in the Rotterdam and Framingham Offspring, Studies. Atherosclerosis. 2012;223(2):421–6. doi: 10.1016/j.atherosclerosis.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes MF, Saarela O, Stritzke J, Kee F, Silander K, Klopp N, et al. Genetic markers enhance coronary risk prediction in men: the MORGAM prospective cohorts. PLoS One. 2012;7(7):e40922. doi: 10.1371/journal.pone.0040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tikkanen E, Havulinna AS, Palotie A, Salomaa V, Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33(9):2261–6. doi: 10.1161/ATVBAHA.112.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganna A, Magnusson PK, Pedersen NL, de Faire U, Reilly M, Arnlov J, et al. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol. 2013;33(9):2267–72. doi: 10.1161/ATVBAHA.113.301218. [DOI] [PubMed] [Google Scholar]
- 13.Buysschaert I, Carruthers KF, Dunbar DR, Peuteman G, Rietzschel E, Belmans A, et al. A variant at chromosome 9p21 is associated with recurrent myocardial infarction and cardiac death after acute coronary syndrome: the GRACE Genetics Study. Eur Heart J. 2010;31(9):1132–41. doi: 10.1093/eurheartj/ehq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wauters E, Carruthers KF, Buysschaert I, Dunbar DR, Peuteman G, Belmans A, et al. Influence of 23 coronary artery disease variants on recurrent myocardial infarction or cardiac death: the GRACE Genetics Study. Eur Heart J. 2013;34(13):993–1001. doi: 10.1093/eurheartj/ehs389. [DOI] [PubMed] [Google Scholar]
- 15.Tragante V, Doevendans PA, Nathoe HM, van der Graaf Y, Spiering W, Algra A, et al. The impact of susceptibility loci for coronary artery disease on other vascular domains and recurrence risk. Eur Heart J. 2013;34(37):2896–904. doi: 10.1093/eurheartj/eht222. [DOI] [PubMed] [Google Scholar]
- 16.Patel RS, Asselbergs FW, Quyyumi AA, Palmer TM, Finan CI, Tragante V, et al. Genetic variants at chromosome 9p21 and risk of first versus subsequent coronary heart disease events: a systematic review and meta-analysis. J Am Coll Cardiol. 2014;63(21):2234–45. doi: 10.1016/j.jacc.2014.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berglund G, Elmstahl S, Janzon L, Larsson SA. The Malmo Diet and Cancer Study. Design and feasibility. Journal of internal medicine. 1993;233(1):45–51. doi: 10.1111/j.1365-2796.1993.tb00647.x. [DOI] [PubMed] [Google Scholar]
- 18.Ridker PM, Danielson E, Fonseca FA, Genest J, Gotto AM, Jr., Kastelein JJ, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359(21):2195–207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 19.Sever PS, Dahlof B, Poulter NR, Wedel H, Beevers G, Caulfield M, et al. Prevention of coronary and stroke events with atorvastatin in hypertensive patients who have average or lower-than-average cholesterol concentrations, in the Anglo-Scandinavian Cardiac Outcomes Trial--Lipid Lowering Arm (ASCOT-LLA): a multicentre randomised controlled trial. Lancet. 2003;361(9364):1149–58. doi: 10.1016/S0140-6736(03)12948-0. [DOI] [PubMed] [Google Scholar]
- 20.Sacks FM, Pfeffer MA, Moye LA, Rouleau JL, Rutherford JD, Cole TG, et al. The effect of pravastatin on coronary events after myocardial infarction in patients with average cholesterol levels. Cholesterol and Recurrent Events Trial investigators. N Engl J Med. 1996;335(14):1001–9. doi: 10.1056/NEJM199610033351401. [DOI] [PubMed] [Google Scholar]
- 21.Cannon CP, Braunwald E, McCabe CH, Rader DJ, Rouleau JL, Belder R, et al. Intensive versus moderate lipid lowering with statins after acute coronary syndromes. N Engl J Med. 2004;350(15):1495–504. doi: 10.1056/NEJMoa040583. [DOI] [PubMed] [Google Scholar]
- 22.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baigent C, Blackwell L, Emberson J, Holland LE, Reith C, Bhala N, et al. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376(9753):1670–81. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mihaylova B, Emberson J, Blackwell L, Keech A, Simes J, Barnes EH, et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet. 2012;380(9841):581–90. doi: 10.1016/S0140-6736(12)60367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor F, Huffman MD, Macedo AF, Moore TH, Burke M, Davey Smith G, et al. Statins for the primary prevention of cardiovascular disease. The Cochrane database of systematic reviews. 2013;1:CD004816. doi: 10.1002/14651858.CD004816.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry JD, Dyer A, Cai X, Garside DB, Ning H, Thomas A, et al. Lifetime risks of cardiovascular disease. N Engl J Med. 2012;366(4):321–9. doi: 10.1056/NEJMoa1012848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laufs U, Weintraub WS, Packard CJ. Beyond statins: what to expect from add-on lipid regulating therapy? Eur Heart J. 2013;34(34):2660–5. doi: 10.1093/eurheartj/eht213. [DOI] [PubMed] [Google Scholar]
- 28.Basso F, Lowe GD, Rumley A, McMahon AD, Humphries SE. Interleukin-6 -174G>C polymorphism and risk of coronary heart disease in West of Scotland coronary prevention study (WOSCOPS). Arterioscler Thromb Vasc Biol. 2002;22(4):599–604. doi: 10.1161/01.atv.0000013283.84306.1a. [DOI] [PubMed] [Google Scholar]
- 29.Iakoubova OA, Tong CH, Chokkalingam AP, Rowland CM, Kirchgessner TG, Louie JZ, et al. Asp92Asn polymorphism in the myeloid IgA Fc receptor is associated with myocardial infarction in two disparate populations: CARE and WOSCOPS. Arterioscler Thromb Vasc Biol. 2006;26(12):2763–8. doi: 10.1161/01.ATV.0000247248.76409.8b. [DOI] [PubMed] [Google Scholar]
- 30.Chiodini BD, Franzosi MG, Barlera S, Signorini S, Lewis CM, D'Orazio A, et al. Apolipoprotein E polymorphisms influence effect of pravastatin on survival after myocardial infarction in a Mediterranean population: the GISSI-Prevenzione study. Eur Heart J. 2007;28(16):1977–83. doi: 10.1093/eurheartj/ehm196. [DOI] [PubMed] [Google Scholar]
- 31.Maitland-van der Zee AH, Lynch A, Boerwinkle E, Arnett DK, Davis BR, Leiendecker-Foster C, et al. Interactions between the single nucleotide polymorphisms in the homocysteine pathway (MTHFR 677C>T, MTHFR 1298 A>C, and CBSins) and the efficacy of HMG-CoA reductase inhibitors in preventing cardiovascular disease in high-risk patients of hypertension: the GenHAT study. Pharmacogenet Genomics. 2008;18(8):651–6. doi: 10.1097/FPC.0b013e3282fe1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabatine MS, Ploughman L, Simonsen KL, Iakoubova OA, Kirchgessner TG, Ranade K, et al. Association between ADAMTS1 matrix metalloproteinase gene variation, coronary heart disease, and benefit of statin therapy. Arterioscler Thromb Vasc Biol. 2008;28(3):562–7. doi: 10.1161/ATVBAHA.107.156653. [DOI] [PubMed] [Google Scholar]
- 33.Iakoubova OA, Sabatine MS, Rowland CM, Tong CH, Catanese JJ, Ranade K, et al. Polymorphism in KIF6 Gene and Benefit From Statins After Acute Coronary Syndromes: Results From the PROVE IT-TIMI 22 Study. J Am Coll Cardiol. 2008;51(4):449–55. doi: 10.1016/j.jacc.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Maitland-van der Zee AH, Peters BJ, Lynch AI, Boerwinkle E, Arnett DK, Cheng S, et al. The effect of nine common polymorphisms in coagulation factor genes (F2, F5, F7, F12 and F13 ) on the effectiveness of statins: the GenHAT study. Pharmacogenet Genomics. 2009;19(5):338–44. doi: 10.1097/fpc.0b013e32832933b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iakoubova OA, Robertson M, Tong CH, Rowland CM, Catanese JJ, Blauw GJ, et al. KIF6 Trp719Arg polymorphism and the effect of statin therapy in elderly patients: results from the PROSPER study. Eur J Cardiovasc Prev Rehabil. 2010;17(4):455–61. doi: 10.1097/HJR.0b013e328336a0dd. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Sabatine MS, Tong CH, Ford I, Kirchgessner TG, Packard CJ, et al. Genetic variants in the KIF6 region and coronary event reduction from statin therapy. Human genetics. 2011;129(1):17–23. doi: 10.1007/s00439-010-0892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brautbar A, Covarrubias D, Belmont J, Lara-Garduno F, Virani SS, Jones PH, et al. Variants in the APOA5 gene region and the response to combination therapy with statins and fenofibric acid in a randomized clinical trial of individuals with mixed dyslipidemia. Atherosclerosis. 2011;219(2):737–42. doi: 10.1016/j.atherosclerosis.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akao H, Polisecki E, Kajinami K, Trompet S, Robertson M, Ford I, et al. KIF6, LPA, TAS2R50, and VAMP8 genetic variation, low density lipoprotein cholesterol lowering response to pravastatin, and heart disease risk reduction in the elderly. Atherosclerosis. 2012;220(2):456–62. doi: 10.1016/j.atherosclerosis.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 39.Shiffman D, Trompet S, Louie JZ, Rowland CM, Catanese JJ, Iakoubova OA, et al. Genome-wide study of gene variants associated with differential cardiovascular event reduction by pravastatin therapy. PLoS One. 2012;7(5):e38240. doi: 10.1371/journal.pone.0038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MRC/BHF Heart Protection Study of cholesterol lowering with simvastatin in 20,536 high-risk individuals: a randomised placebo-controlled trial. Lancet. 2002;360(9326):7–22. doi: 10.1016/S0140-6736(02)09327-3. [DOI] [PubMed] [Google Scholar]
- 41.Mega JL, Morrow DA, Brown A, Cannon CP, Sabatine MS. Identification of genetic variants associated with response to statin therapy. Arterioscler Thromb Vasc Biol. 2009;29(9):1310–5. doi: 10.1161/ATVBAHA.109.188474. [DOI] [PubMed] [Google Scholar]
- 42.Hopewell JC, Parish S, Offer A, Link E, Clarke R, Lathrop M, et al. Impact of common genetic variation on response to simvastatin therapy among 18 705 participants in the Heart Protection Study. Eur Heart J. 2013;34(13):982–92. doi: 10.1093/eurheartj/ehs344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.