Abstract
Decreases in endoplasmic reticulum calcium content are sensed by resident STIM proteins, which can activate plasma membrane Orai channels to facilitate Ca2+ entry. The role of STIMATE, a previously unidentified component of the store-operated calcium entry complex, has now been identified and defined.
Calcium (Ca2+) is a ubiquitous intracellular signaling ion, responsible for governing a plethora of cellular processes. The most abundant source of Ca2+ within the cell is the endoplasmic reticulum (ER). Engagement of phospholipase C-coupled receptors in the plasma membrane (PM) leads to the production of inositol 1,4,5-trisphosphate (InsP3), a critical second messenger that triggers ER Ca2+ release into the cytosol. The resultant loss of ER Ca2+ content leads to a process termed store-operated calcium entry (SOCE); the ER Ca2+ sensors STIM1 and STIM2 react to decreased Ca2+ by assembling in ER-PM junctions where they can control the function of Orai1, the store-operated Ca2+ channel1, 2. Whereas STIM is capable of mediating these processes independently in vitro3, recent investigations reveal that, in vivo, the assembly and disassembly of STIM proteins is regulated by several critical adaptor proteins4–7. In this issue of Nature Cell Biology, Jing et al. reveal the identity and function of a new member of this family of adaptors termed STIM-activating enhancer (STIMATE)8.
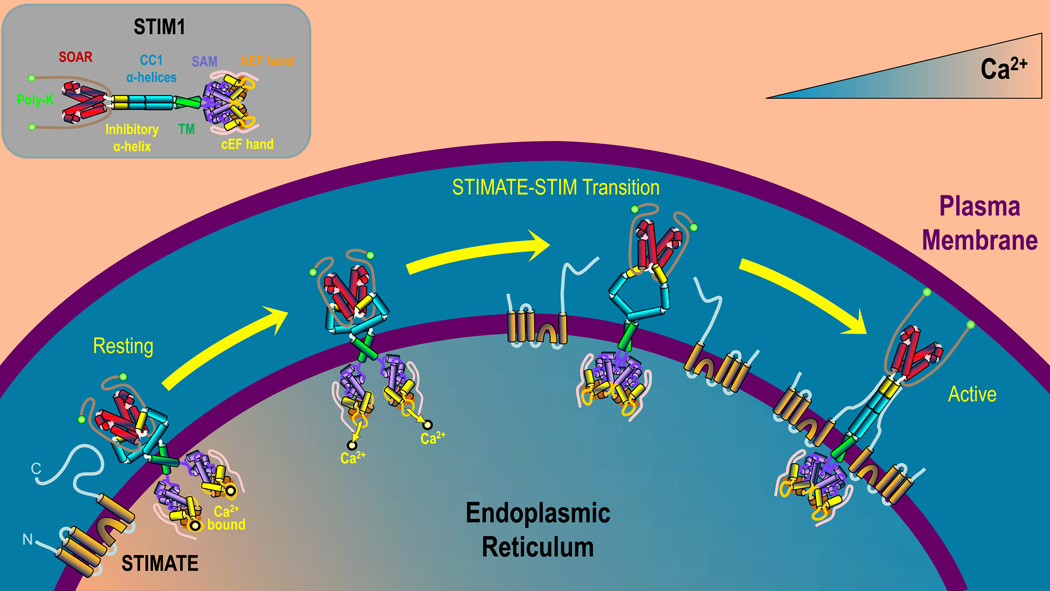
STIMs are highly modular proteins with multiple domains in the cytosol and ER. Critical to its function is the STIM-Orai-Activating-Region (SOAR)9, a 98 amino acid cytosolic region representing the sole domain required to activate Orai (Fig 1; inset). At rest, STIM is a dimer, with SOAR inaccessible due to its association with adjacent inhibitory domains within a juxtamembrane coiled-coil region termed CC110. The luminal Ca2+ - sensing region of STIM contains two EF-hand domains and a sterile α motif (SAM). Following ER Ca2+ depletion, its low affinity EF hand domain releases Ca2+, destabilizing the EF hand/SAM complexes, forcing STIM to aggregate within the ER lumen11. This initiates a series of conformational changes ultimately leading to STIM extension12 and multimerization, SOAR disinhibition and Orai activation. As summarized below and in the accompanying article8, STIMATE serves a critical role in SOAR disinhibition by binding to CC1 when ER Ca2+ is depleted (Fig 1). This intriguing finding reveals the existence of a previously unrecognized transitional state in which Ca2+ has dissociated from STIM1 and the protein has undergone a conformational change on the cytosolic side of the ER, yet SOAR remains inaccessible and non-functional.
Figure 1. Regulation of STIM1 activation by STIMATE.
When the ER is replete with Ca2+ (left) resting STIM1 exists as a dimer with Ca2+ bound to its luminal EF-hands and in a folded conformation with SOAR closely associated with CC1. STIMATE is located in the ER membrane, available to modulate STIM1 activation. As ER luminal Ca2+ levels decrease (middle), Ca2+ dissociates from the EF-hands of STIM, resulting in a conformational change that promotes binding between STIMATE and CC1. This relieves inhibition of SOAR, leading to transformation of STIM1 into fully activated, unfolded conformation (right). STIM1 can then translocate to ER-PM junctions where it can facilitate SOCE. [inset] Identification of STIM1 domains. Poly-K - Lysine-rich domain; SOAR – STIM-Orai1 Activating Region; CC1 – Coiled-coil 1; TM – Transmembrane domain; SAM – Sterile α motif; cEF – canonical EF hand; hEF – homologous EF hand.
STIMATE was identified through a screen of ER-PM junction proteins. This was accomplished by tagging STIM1 with the ascorbate peroxidase 2 enzyme (APEX2); when provided with H2O2, APEX2 generates short-lived phenoxyl radicals that covalently link biotin-phenol to proteins located within a 20 nm radius. Whereas 73 proteins were labelled using this approach, STIMATE was one of the 17 proteins labelled in both store replete and depleted conditions. Although the functional role of STIMATE had not previously been investigated, STIMATE was also identified in a screen for genes contributing to NFAT activation5, implicating STIMATE as a positive regulator of SOCE. The authors firmly established this relationship in this study by showing significantly decreased SOCE after either knockdown or knockout of STIMATE.
Jing et al. provide considerable mechanistic insight into how STIMATE regulates STIM1 activation. One of the first observations made about STIM1 as a regulator of SOCE was that, following ER Ca2+ depletion, it translocates within the ER towards the PM where it forms large clusters or ‘puncta’, easily visible by fluorescence microscopy. Interestingly, this ability was severely compromised in STIMATE-KO cells, while in cells overexpressing STIMATE, STIM1 constitutively formed clusters under ER Ca2+ replete conditions.
The concept that STIMATE facilitates this effect via SOAR disinhibition was built on several key pieces of evidence. The first clue was that the c-terminal domain of STIMATE interacts directly with CC1, the portion of STIM1 responsible for SOAR inhibition. Based on this information, the authors designed an elegant FRET-based experiment to assess association between CC1 and SOAR by truncating STIM1 at the end of CC1, tagging it with CFP and co-expressing it with YFP-tagged SOAR. Remarkably, ER Ca2+ depletion decreased association between these STIM components, demonstrating that ER Ca2+ depletion leads to decreased association between CC1 and SOAR. Co-expressing STIMATE greatly decreased this association. The authors further showed that STIMATE increased FRET between full length STIM1 proteins and between STIM1 and Orai1, consistent with disinhibited SOAR. Considered collectively, these observations provide clear evidence in support of a role for STIMATE in promoting STIM1 activation through regulation of intramolecular interactions between SOAR and CC1.
Given that STIMATE was found to be a regulator of STIM1 activation, it is somewhat surprising that association between STIMATE and STIM1 was detected under both store replete and depleted conditions. One possibility is that, even under store replete conditions, not all STIM1 exists in its resting state. Indeed, whereas the authors observe FRET between STIM1 and STIMATE under resting conditions, they see a considerable increase in STIMATE-STIM1 association upon store depletion. Hence, loss of Ca2+ from STIM1 must increase the availability of CC1, the STIM1 domain that STIMATE binds to. However, the fact that this change is insufficient to facilitate cluster formation by STIM1 in the absence of STIMATE reveals the existence of a transitional state in which STIM1 is not bound Ca2+ and a conformational change has occurred, but SOAR remains bound to its inhibitory domain (Fig 1). Future investigations may shed new insight into the molecular dynamics of this transition and the identities of any additional players involved in its regulation.
An alternative explanation for the ability of STIMATE to affect STIM1 localization in ER-PM junctions, would be regulation of ER-PM junction formation itself. However, whereas the data provided by the authors in this study makes this possibility seem unlikely, the authors did not entirely rule this possibility out. Hence, the authors used 2 novel tools to measure the amount of cortical ER in STIMATE-KO cells, revealing an ~10% decrease. This does not seem sufficient to account for the effect of STIMATE on STIM1 puncta formation, but it is curious that any difference was observed at all. Recent investigations have implicated extended synaptotagmins (E-Syts) as the primary regulators of cortical ER formation13. Since E-Syts have C2 domains and are Ca2+ sensitive14, a significant possibility is that decreased Ca2+ entry in STIMATE-KO cells decreases E-Syt activity, thereby indirectly leading to a loss of cortical ER.
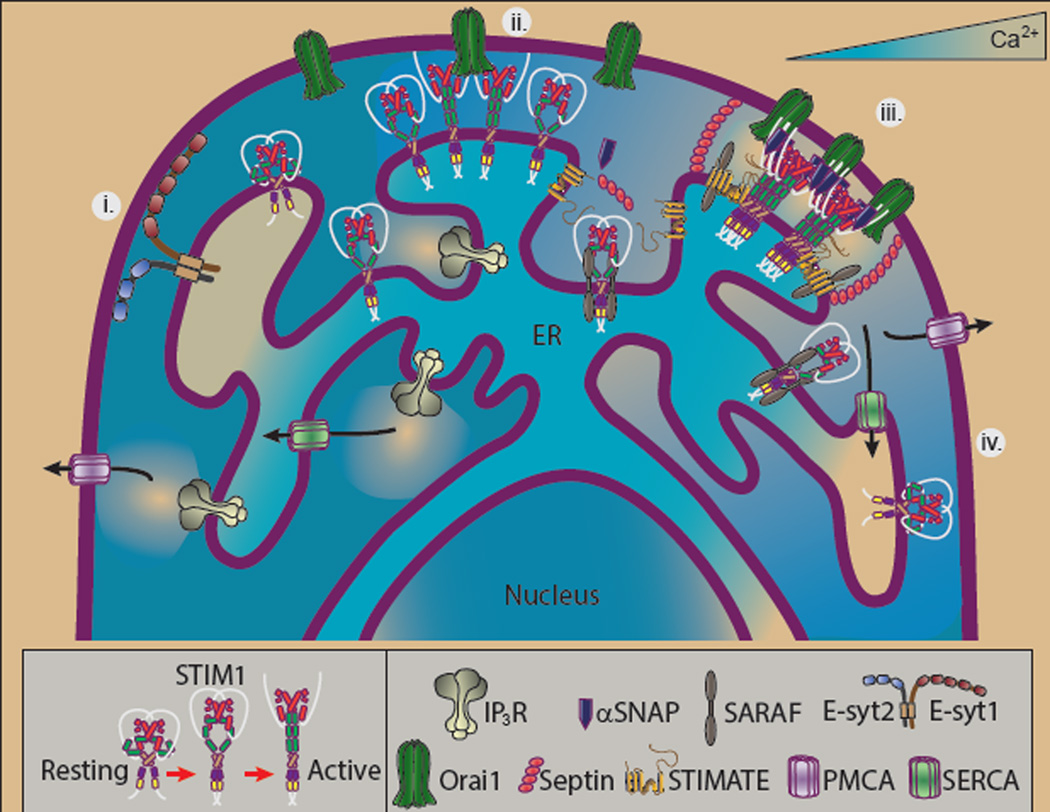
In conclusion, Jing et al. reveal STIMATE as the newest member of a rapidly forming class of STIM adaptor proteins. Over the last 10 years, our understanding of how STIM1 forms puncta has advanced considerably. It has become increasingly clear that formation of STIM1 puncta is a tightly regulated process involved a surprisingly wide field of players. Indeed, since early work by Yousang Gwack and colleagues revealed junctate and CRACR2A as STIM1 interacting proteins4, the field has expanded to include septins5, αSNAP6, SARAF7 and now STIMATE. Significantly, these investigations have revealed distinct inter-dependent roles for each of these adaptor proteins (Fig 2). Hence, the current investigation reveals a critical role for STIMATE as a regulator of STIM1 activation. In order for activated STIM1 to associate with Orai1, septins facilitate its recruitment and trapping into ER-PM junctions. To define optimal stoichiometry within these junctions, αSNAP is recruited. Finally, the dissociation of STIM1 puncta as ER Ca2+ content is restored is promoted by SARAF. Future investigations may lead to the identification of additional players in this increasingly complex and exciting new field.
Figure 2. STIM-Orai interaction is regulated by the actions and interactions of multiple adaptors.
i) ER-PM junctions exist in cells independent of SOCE function. The extended synaptotagmins are (E-syts) are key proteins in regulating such sites by binding to Pi(4,5)P2-enriched regions of the plasma membrane. ii) STIM1 translocates to ER-PM junctions in response to store depletion even in the absence of adaptor proteins, although clusters fail to form properly and Orai1 is activated with poor efficiency. iii) In the presence of a full array of adaptor proteins STIM1-Orai association occurs with optimal efficiency. When ER luminal Ca2+ decreases, SARAF dissociates from SOAR while STIMATE interacts with CC1, promoting the STIM1 transition to its active conformation. Septins target Orai1 to ER-PM junctions where αSNAP regulates STIM-Orai stoichiometry. iv) When luminal ER Ca2+ content rises, SARAF again binds STIM1, mediating Ca2+-dependent inactivation of SOCE, promoting disassembly of STIM1 clusters.
References
- 1.Fahrner M, Derler I, Jardin I, Romanin C. Channels (Austin) 2013;7:330–343. doi: 10.4161/chan.26742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soboloff J, Rothberg BS, Madesh M, Gill DL. Nat Rev Mol Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Y, et al. Nat Struct Mol Biol. 2010;17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srikanth S, Gwack Y. J Physiol. 2012;590:4169–4177. doi: 10.1113/jphysiol.2012.231522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S, et al. Nature. 2013;499:238–242. doi: 10.1038/nature12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao Y, et al. eLife. 2013;2:e00802. doi: 10.7554/eLife.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palty R, Raveh A, Kaminsky I, Meller R, Reuveny E. Cell. 2012;149:425–438. doi: 10.1016/j.cell.2012.01.055. [DOI] [PubMed] [Google Scholar]
- 8.Jing J, et al. Nature Cell Biology. 2015 in press. [Google Scholar]
- 9.Yuan JP, et al. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Jin H, Cai X, Li S, Shen Y. Proc Natl Acad Sci U S A. 2012;109:5657–5662. doi: 10.1073/pnas.1118947109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Muik M, et al. EMBO J. 2011;30:1678–1689. doi: 10.1038/emboj.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giordano F, et al. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idevall-Hagren O, Lu A, Xie B, De Camilli P. EMBO J. 2015 doi: 10.15252/embj.201591565. [DOI] [PMC free article] [PubMed] [Google Scholar]