Abstract
Inflammatory phenotypes of asthma are associated with differences in disease characteristics. It is unknown whether these inflammatory phenotypes are reflected by the activation status of neutrophils in blood and sputum. We obtained peripheral blood and induced sputum from 21 asthma patients and stratified our samples based on sputum eosinophilia resulting in two groups (>3% eosinophils: n = 13, <3%: n = 8). Eosinophils and neutrophils from blood and sputum were analysed for expression of activation and degranulation markers by flow cytometry. Data were analysed by both classical, non-parametric statistics and a multi-dimensional approach, using principal component analysis (PCA). Patients with sputum eosinophilia were characterized by increased asthma control questionnaire (ACQ) scores and blood eosinophil counts. Both sputum neutrophils and eosinophils displayed an activated and degranulated phenotype compared to cells obtained from blood. Specifically, degranulation of all granule types was detected in sputum cells, combined with an increased expression of the activation markers (activated) Mac-1 (CD11b), programmed death ligand 1 (PD-L1) (CD274) and a decreased expression of CD62L. CD69 expression was only increased on sputum eosinophils. Surface marker expression of neutrophils was similar in the presence or absence of eosinophilia, either by single or multi-dimensional analysis. Sputum neutrophils were highly activated and degranulated irrespective of sputum eosinophilia. Therefore, we conclude that differences in granulocyte activation in sputum and/or blood are not associated with clinical differences in the two groups of asthma patients. The finding of PD-L1 expression on sputum granulocytes suggests an immunomodulatory role of these cells in the tissue.
Keywords: cell surface molecules, lung, neutrophils
Introduction
An estimated 300 million individuals worldwide are affected by asthma 1. Asthma is characterized by airway inflammation, bronchial hyperresponsiveness and reversible airway obstruction. Several different inflammatory phenotypes have been identified in asthma, which are accompanied by different clinical characteristics 2. Sputum eosinophilia is associated with bronchial hyperresponsiveness, high forced exhaled nitric oxide (FeNO) and (specific) immunoglobulin (Ig)E levels. In addition, the presence of either neutrophils or eosinophils in sputum is associated with a decreased forced expiratory volume in 1 s (FEV1) 3,4. These inflammatory phenotypes are identified by microscopically evaluating the percentages of neutrophils and eosinophils in sputum or by transcriptomic profiling of whole sputum samples 2,5,6. In addition to microscopic evaluation, cells obtained by sputum induction can be analysed by flow cytometry 7–11. Sputum granulocytes analysed by this technique have been shown to display an activated phenotype, with up-regulation of markers for activation and degranulation on either neutrophils 11, eosinophils 12 or both 13. However, to date there are no reports on in-depth analysis of activation markers comparing blood and sputum granulocytes using multi-dimensional analysis. In addition, no studies have been published that compare sputum granulocyte expression profiles adequately between patients with different asthma phenotypes. A single study compared the expression levels of two classical activation markers between patients with moderate and severe asthma, but did not find a correlation between expression and disease severity 13. Transcriptomic profiling of whole blood and sputum samples showed up-regulation of neutrophil defensins and proteases in the blood of neutrophilic asthma patients, and significant differences between sputum samples of patients with different asthma phenotypes 6,14. It is unknown, however, whether these differences reflect differences in expression of the granulocytes themselves or differences in cell numbers.
Traditionally, granulocyte activation markers include adhesion receptors such as the integrin Mac-1 (CD11b/CD18) and L-selectin (CD62L). Other activation markers described to be up-regulated on circulating or sputum granulocytes from asthma patients are the CD11b activation epitope CBRM1/5 15 and the intercellular adhesion molecule-1 (ICAM-1, CD54); the latter adhesion receptor was proposed as a therapeutic target for antigen-induced acute airway inflammation 16. The activation marker CD69, which is well known as a T cell activation marker 17, was shown to be up-regulated on eosinophils from broncheoalveolar lavage (BAL) when compared to blood cells 15.
Neutrophils and eosinophils possess different types of granules containing anti-microbial and proinflammatory proteins. For neutrophils, release of these proteins to the outside of the cell (degranulation) occurs sequentially in response to increasing strength of activation signals, with secretory vesicles degranulating by the most mild stimulus, followed by tertiary, specific and azurophilic granules 18–20. The marker for neutrophil tertiary and eosinophil secretory granules (CD11b) was shown to be up-regulated on cells from both BAL and sputum, compared to blood granulocytes 15. Markers for specific and azurophilic/crystalloid granules were shown to be up-regulated on BAL and on sputum granulocytes in several diseases, but so far not on sputum cells from asthma patients 15,21,22. Degranulation of neutrophil secretory vesicles was shown to occur already in the blood of asthma patients 23,24.
Another activation marker of interest, programmed death ligand 1 (PD-L1, CD274), has been implicated in a murine model of airway hyperresponsiveness 25. The immunomodulatory protein CD274 is not as well characterized in granulocytes as in lymphocytes, but was shown to be responsible for suppression of T cell responses by interferon (IFN)-γ-stimulated neutrophils 26 and is highly expressed in granulomas in the lungs of sarcoidosis patients, although its expression on granulocytes was not tested by flow cytometry 27.
Until the present, all immunophenotyping of sputum cells has been performed by analysis of single-dimensional marker expression, which ignores the interaction between multiple markers. To overcome this issue, marker expressions and interactions can be studied using principal component analysis (PCA). PCA is a multivariate analysis method that detects systematic variability within multiple parameters and explores correlations between these parameters 28. It transforms data sets with a large number of measured parameters into a smaller number of parameters, called principal components. As the resulting smaller number of components is interpreted more easily, it is a preferred technique for the analysis of large data sets and forms the basis of cluster approaches used to identify asthma phenotypes 29,30.
In this study we investigated differences in granulocyte activation and degranulation by flow cytometric evaluation of neutrophils and eosinophils isolated from blood and sputum of asthma patients with and without sputum eosinophilia, both by single and multi-dimensional analysis.
Methods
Study population and ethics
This study was approved by the local medical ethics committee. Patients were recruited at the pulmonary out-patient clinic of the University Medical Centre Utrecht (see Table1 for patient baseline characteristics) and gave written informed consent in accordance to the Declaration of Helsinki (seventh revision, Fortaleza, 2013).
Table 1.
Patient baseline characteristics at time of sputum induction grouped for sputum eosinophilia.
| >3% sputum eo | <3% sputum eo | P-value* | |
|---|---|---|---|
| Number | 13 | 8 | n.a. |
| Gender, male/female | 9/4 | 4/4 | 0·88 |
| Age (years) | 45 (19–66) | 47 (25–54) | 0·45 |
| BMI (kg/m2) | 27 (22–35) | 27 (22–32) | 0·885 |
| FeNO (ppb) | 32 (16–215) | 23 (12–42) | 0·09 |
| ACQ score | 2·3 (0·0–4·4) | 1·3 (0·14–2·0) | 0·035 |
| Blood eosinophil count (109/ml) | 0·68 (0·1–1·2) | 0·18 (0·0–0·5) | 0·007 |
| Medication† | 4 (3–5) | 3·5 (0–4) | 0·09 |
| FEV1 (l) | 2·4 (1·5–3·9) | 3·5 (2·3–4·0) | 0·053 |
| FEV1 (% predicted) | 76 (61–105) | 89 (61–113) | 0·16 |
| Reversibility FEV1‡ | 0·47 (0·0–16·6) | 0·43 (0·0–4·9) | 0·46 |
| Sputum eosinophils (%)§ | 29 (3–82) | 0 (0–1) | n.a. |
| Sputum neutrophils (%)§ | 39 (10–77) | 49 (19–83) | 0·41 |
Values are medians ± range unless indicated otherwise.
Based on Wilcoxon–Mann–Whitney test or Fisher's exact test where appropriate.
5-point ordinal scale based on guidelines of the British Thoracic Society, with (0) no medication, (1) inhaled short-acting beta agonist (SABA) when required, (2) low-dose ICS + SABA, (3) low/medium-dose inhaled corticosteroids (ICS) + long-acting beta agonist (LABA), or medium-dose ICS + LABA, (4) high-dose ICS ± LABA and leucotriene receptor antagonist test and (5) high-dose ICS + oral corticosteroids (OCS) ± LABA.
Percentage reversibility in FEV1 (corrected for age, gender, length and body weight) after inhalation of 2 × 100 μg salbutamol.
As determined by microscopic evaluation. BMI = body mass index; FeNO = forced exhaled nitric oxide; ACQ = asthma control questionnaire; FEV1 = forced expiratory volume in 1 s; n.a. = not applicable; eo = eosinophil; ppb = parts per billion.
Patients were included according to the following criteria: age 18–75 years, having adult asthma defined by the Global Initiative for Asthma (GINA) guidelines on clinical features (episodic shortness of breath, particularly at night and often accompanied by cough and wheezing) and reversibility of FEV1 upon inhalation of 400 μg salbutamol (≥12% predicted or ≥200 ml) and/or airway hyperresponsiveness to histamine (PC20 < 8 mg/ml) 1. Patient numbers were calculated to be able to distinguish medium effect sizes with a power of 0·8 and an α of 0·05 using G*Power version 3·1·3 31.
Exclusion criteria for the study were smoking in the last 12 months, a smoking history ≥10 pack-years, treatment with antibiotics <4 weeks ago or confirmed allergic bronchopulmonary aspergillosis.
Blood and sputum samples were stratified on the presence or absence of sputum eosinophilia based on a cut-off value of 3% sputum eosinophils, as determined by differential count (described below). All samples were collected between 9 a.m. and 1 p.m. to minimize the effects of diurnal variation such as found in FeNO and haematological parameters 32,33.
Lung function and clinical parameters
After inclusion of patients, the asthma control questionnaire (ACQ) and medication adherence report scale (MARS) were completed. FEV1 measurements were performed by peak flow measurement using the PiKo-1 (nSpire Health, Longmont, CO, USA) and FeNO was determined using NIOX MINO® (Aerocrine, Solna, Sweden), with an expiration time of 10 s. Absolute blood eosinophil counts were calculated from percentages obtained by flow cytometric (FACS) analysis (see below) and white blood cell (WBC) counts obtained from a Cell-Dyn haematology analyser (Abbot Diagnostics, North Chicago, IL, USA).
Blood processing
Erythrocytes were lysed from sodium heparin blood using lysis buffer consisting of 150 mM NH4Cl, 10 mM KHCO3 and 0·1 mM Na2 ethylenediaminetetraacetic acid (EDTA) dissolved in ddH2O. The resulting total leucocyte preparations were washed and resuspended in staining buffer consisting of phosphate-buffered saline (PBS) supplemented with trisodium citrate (0·32% w/v) (both prepared by the UMCU pharmacy) and human pasteurized plasma solution (10% w/v; Sanquin, Amsterdam, the Netherlands).
Sputum induction and processing
Sputum was induced by inhalation of 0·9–5% saline aerosols and processed as published previously using Sputolysin prepared from a ×10 stock (Merck Millipore, Darmstadt, Germany) and supplemented with NaCl to reach an osmolarity of 280–290 mOsm 34,35. Cytospin slides of sputum cells were stained with May–Grünwald–Giemsa for differential cell count. Samples containing > 80% squamous epithelial cells were excluded from analysis. The cell percentages in sputum were calculated after exclusion of squamous epithelial cells 36. Remaining cells were resuspended in staining buffer for FACS staining procedure.
Flow cytometry
Samples were stained with antibodies for 30 min on ice at a maximum concentration of 5 × 106 cells/ml and washed twice before analysis on a Gallios flow cytometer (Beckman Coulter, Pasadena, CA, USA).
Granulocytes in blood were identified based on forward-/side-scatter (FSC/SSC) and CD16 was used to differentiate into neutrophils (CD16+) and eosinophils (CD16–). A different sorting strategy was chosen for sputum samples due to the presence of epithelial cells and alveolar macrophages with a high SSC (Fig. 1). Epithelial cells do not express CD11b and alveolar macrophages are highly autofluorescent at emission wavelengths of around 450 nm when excited by a 405 nm laser. Therefore, after exclusion of debris on FSC/SSC, granulocytes were identified as CD11b+ and 405/450 nm-autofluorescencelow. Subsequently, they were differentiated into neutrophils and eosinophils based on CD16 expression and SSC. Analysis was performed on at least 250 neutrophils/eosinophils. Degranulation of neutrophils was measured using CR1 (CD35), integrin αM (CD11b), carcinoembryonic antigen-related cell adhesion molecule-8 (CEACAM-8) (CD66b) and lysosome-associated membrane protein 3 (LAMP-3) (CD63) antibodies for secretory, tertiary, specific and azurophilic granules, respectively 19. For eosinophils, LAMP-3 was used as a marker for crystalloid (specific/secondary) granule degranulation 37 and CD11b as a marker for secretory (sombrero) vesicles 20. As CD63 is also expressed on activated platelets, its expression was only determined on cells negative for platelet marker CD41 38.
Figure 1.
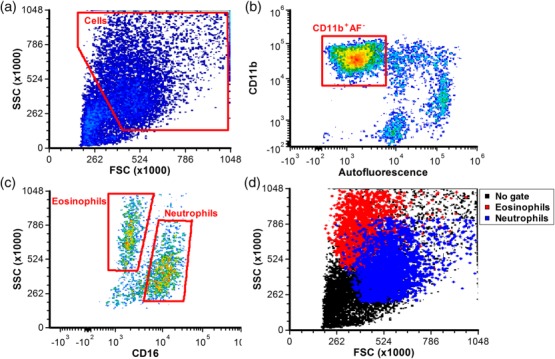
Gating strategy to identify sputum granulocytes. (a) A forward-/side-scatter (FSC/SSC) gate is used to exclude most debris and lymphocytes. (b) Subsequently, cells positive for CD11b and negative for ∼450 nm autofluorescence were gated to exclude residual lymphocytes and alveolar macrophages (which are highly autofluorescent). (c) The resulting granulocytes were subdivided into neutrophils and eosinophils based on SSC and expression of CD16. (d) The resulting neutrophil and eosinophil population show FSC/SSC patterns similar to those in blood, with a higher SSC for eosinophils and higher FSC for neutrophils.
Antibodies used were CD11b-allophycocyanin (APC)-Alexa750 (clone Bear1), CD16-Krome Orange (3G8), CD274-phycoerythrin-cyanin 7 (PeCy7) (PDL1.3.1) and CD62L-endothelial cell density (ECD) (DREG56) from Beckman Coulter (Pasadena, CA, USA), CD35-fluorescein isothiocyanate (FITC) (E11) from Biolegend (San Diego, CA, USA). CD63-PE (H5C6), CD69-PeCy7 (FN50), IgG1 isotype control-PE (X40), IgG2a isotype control-FITC (X39) and the annexin-V PE apoptosis kit I from BD (San Jose, CA, USA), active CD11b-Alexa700 (CBRM1/5) from eBiosciences (San Diego, CA, USA), CD54-PE (MEM-111) from EXBIO Praha (Vestec, Czech Republic) and CD41-FITC (VIPL3) from Life Technologies (Carlsbad, CA, USA).
Data analysis and representation
FCS express 4·0 (De Novo Software, Los Angeles, CA, USA) was used for evaluation of flow cytometric data and determination of median fluorescence intensities. Data were plotted as boxes representing median ± IQR with error bars plotted according to Tukey's method in Prism version 6·04 (GraphPad Software, La Jolla, CA, USA). Statistical analysis was performed in spss statistics version 22 (IBM, Armonk, NY, USA). Samples were compared using multiple Wilcoxon–Mann–Whitney tests or related-samples Wilcoxon signed-rank tests where applicable. Correction for multiple testing was performed using the Bonferroni method. Correlations between parameters were determined using Spearman's correlation coefficient.
PCA was performed as described extensively elsewhere 28. In short, PCA was performed on the median fluorescence intensities (MFIs) of all measured parameters (except isotype controls) using spss statistics version 22. MFIs were classified as numerical data, missing values were mode imputed and discretization was used. Scree plots (not shown) were used to determine the required amount of components. Data points with object scores of >3·5 or <3·5 were considered as outliers and excluded from the analysis. Graphs of object scores and loading plots were made in Prism version 6·04.
Results
Patient characteristics
Sputum and blood samples were obtained from 21 patients (Table1 patient baseline characteristics). Thirteen patients (68%) had more than 3% eosinophils in their sputum samples. This sputum eosinophilia was accompanied with higher eosinophil counts in peripheral blood (P = 0·007), less well-controlled asthma (P = 0·035) and trends towards higher FeNO, more medication use and lower absolute FEV1 (P = 0·09, 0·09 and 0·053, respectively).
Viability of sputum granulocytes
After processing of sputum samples, the viability of the cells in the sample was determined by annexin-V and 7-aminoactinomycin D (7-AAD) staining 39. Sputum neutrophils and eosinophils were typically 90–95% alive (Fig. 2) and 5–10% necrotic or late apoptotic. Fewer than 1% of sputum cells were annexin-V single positive and, thus, early apoptotic.
Figure 2.
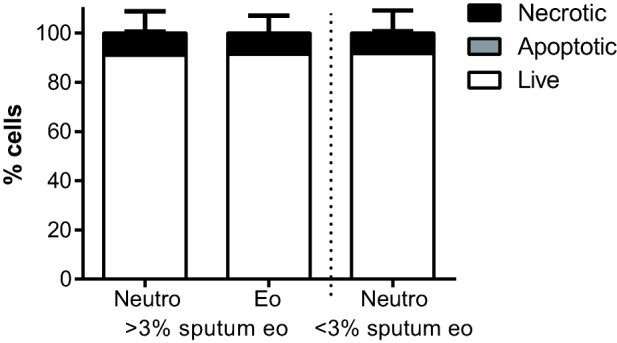
Viability of sputum granulocytes. Sputum cells are typically >90% viable, with <1% early apoptotic annexin-V single positive cells and the remainder being necrotic or late apoptotic. Bars depict means with standard deviation of patients with (n = 13) and without (n = 8) sputum eosinophilia.
Expression of activation markers in blood and sputum
Both neutrophils and eosinophils showed an activated phenotype in sputum compared to blood (Fig. 3 and Supporting information, Fig. S1). CD62L was shed from sputum neutrophils and eosinophils. An increased CD69 expression was found only on sputum eosinophils, whereas CD11b, CBRM1/5 and CD274 were up-regulated on both cell types. Expression of CD54 was not detected, irrespective of cell or sampling location. Increased expression of markers for tertiary (CD11b), specific (CD66b) and azurophilic/crystalloid (CD63) granules indicated a highly degranulated state for sputum granulocytes.
Figure 3.
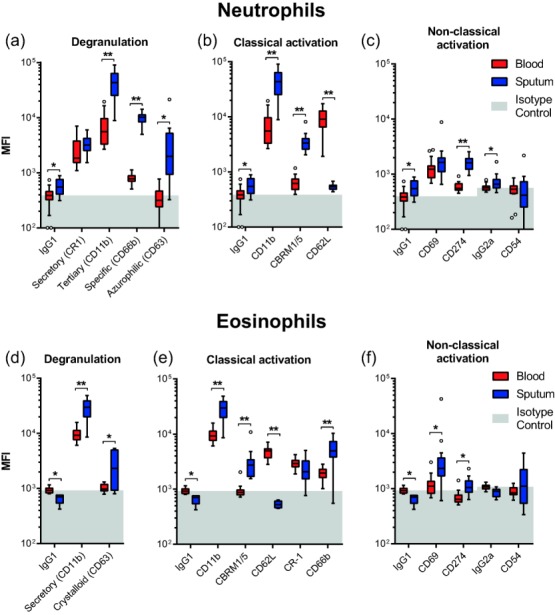
Expression profiles of blood granulocytes compared to sputum. Expression of activation markers on blood and sputum neutrophils (a–c, n = 21) and eosinophils (e–f, n = 13). As CD11b is both a classical activation marker and a marker for degranulation of tertiary granules, it is displayed twice. Boxes represent medians ± interquartile range (IQR) with whiskers of 1·5 IQR according to Tukey's method. Light grey fills represent data points below the isotype control median measured on blood granulocytes. *P < 0·05, **P < 0·001 significant differences between blood and sputum as determined by multiple Wilcoxon's signed-rank test corrected for multiplicity by Bonferroni correction.
In contrast to the marked differences in expression patterns between blood and sputum granulocytes, no significant differences were detected when comparing expression levels of markers on blood and sputum cells isolated from individuals with high and low eosinophil counts in sputum (Fig. 4). In addition, no significant correlations were found between ACQ score, FEV1 (percentage predicted and/or absolute value) and expression of any of the granulocyte markers (data not shown). Due to the absence or small numbers of eosinophils in patients with <3% sputum eosinophils, surface marker expressions of these cells could not be compared sufficiently between patient groups.
Figure 4.
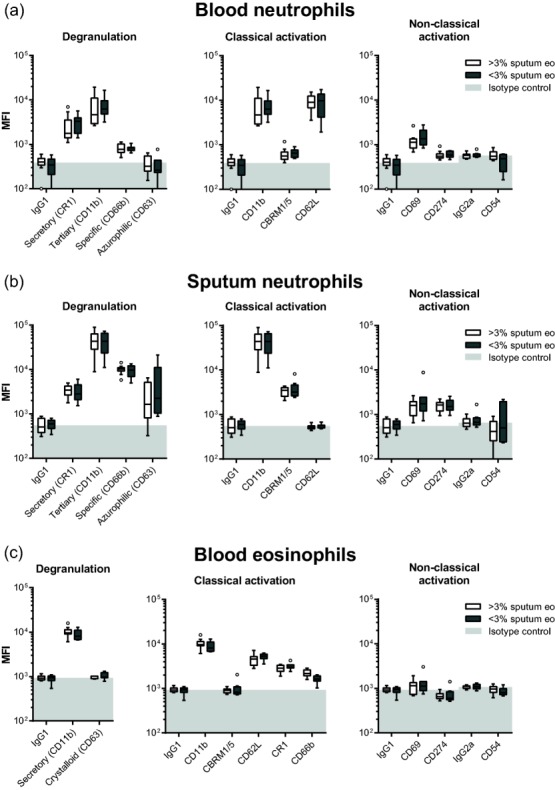
Expression profiles of sputum granulocytes in patients with presence of absence of sputum eosinophilia. Expression profiles of (a) blood neutrophils, (b) blood eosinophils and (c) sputum neutrophils are similar in patients with the presence or absence of sputum eosinophilia. Boxes represent medians ± interquartile range (IQR) with whiskers of 1·5 IQR, according to Tukey's method. Light grey fills indicate values below the median isotype control. No statistically significant differences were found as determined by multiple Wilcoxon–Mann–Whitney tests with Bonferroni correction (P < 0·05 after multiplicity correction).
PCA
PCA was performed on the neutrophil data and eosinophil data for the patients with adequate numbers of sputum eosinophils (Fig. 5). Scree plots (not shown) indicated that all PCAs comparing blood and sputum samples required two components and the PCA comparing the two asthma phenotypes required four components.
Figure 5.
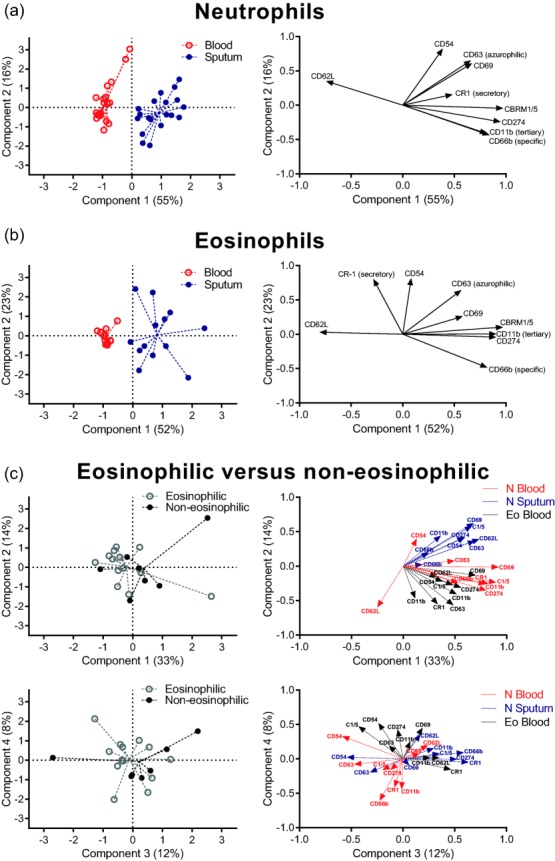
Multi-dimensional analysis of flow cytometric data. Object scores of each patient (left) and loading plots of each marker (right) of principal component analysis (PCA) of receptor expression levels on blood and sputum neutrophils (a) and eosinophils (b). Dashed lines originate from the mean object scores of a group. Component 1 showed a clear separation between blood and sputum cell types with some residual variation explained by component 2. Loading plots indicate the contribution of each marker in a component, with a large distance from 0 indicating a large influence. Markers which are not (differently) expressed remain close to zero on the x-axis, so play little role in the component 1. Markers up-regulated in sputum have positive values for component 1 (e.g. CD11b), whereas down-regulated markers (CD62L) have negative values. PCA analysis did not discriminate between patient groups (c) in any of the four components. Group sizes were 21 (a), 13 (b) and 21 (c).
Principal component 1 separates samples from blood and sputum for both neutrophils (Fig. 5a) and eosinophils (Fig. 5b) accurately, demonstrating that receptor expression profiles differ between sampling locations. The first principal component accounted for a total of 55 and 52% of the variation in receptor expression for neutrophils and eosinophils, respectively. Principal component 2 described a further 16 and 23% of the variation.
In contrast, none of the four principal components distinguished between patients with or without eosinophils in their sputum after a PCA on the combined data of blood eosinophils, blood neutrophils and sputum neutrophils (Fig. 5c). Separate PCA of surface marker data for blood eosinophils, blood neutrophils and sputum neutrophils did also not distinguish between patient groups (Supporting information, Fig. S2).
Discussion
In our study, asthma patients (n = 21) were characterized by the presence or absence of sputum eosinophilia (> 3% eosinophils: n = 13, < 3%: n = 8) at the time of inclusion. At inclusion, the two patient groups showed clear differences in clinical parameters. Patients with sputum eosinophilia had more uncontrolled asthma, blood eosinophilia and trends towards higher FeNO, increased medication use and lower FEV1.
However, there were no differences in the expression of activation and degranulation markers between the two patient groups. No correlations were found between surface expression markers and markers of disease severity such as ACQ or FEV1. Post-hoc analysis of the effect size for each marker revealed a median effect sizes r of 0·146 (range 0·032–0·345), indicating that any effects missed due to the small sample size would have been small. Therefore, we conclude that activation or degranulation of granulocytes in both blood and sputum are not associated with clinical differences in eosinophilic and non-eosinophilic asthma, defined by sputum analysis. The absence of differences in marker expressions on sputum neutrophils is in line with the finding of an activated phenotype of neutrophils in the BAL of both healthy volunteers and chronic obstructive pulmonary disease (COPD) patients 40 and supports the hypothesis that the process of recruitment leads per se to granulocyte activation.
Our results confirm that sputum granulocytes have increased expression of the classical activation marker CD11b and decreased CD62L expression 8,10–13. Both neutrophils and eosinophils display a highly degranulated phenotype, with up-regulation of markers for tertiary, specific and azurophilic granules on neutrophils and crystalloid and secretory granules on eosinophils. Interestingly, the expression of secretory granule marker CR1 (CD35) was already high on blood cells and did not increase further on sputum cells. This can be explained by the fact that secretory vesicles are the first to fuse with the membrane and that in the peripheral blood of asthma patients this process has already taken place 23,24. Alternatively, CR1 may have been shed from the cell surface, as described in other diseases 41.
CD54 has been described as an eosinophil activation marker 42,43, which is expressed on sputum eosinophils 12. However, in line with another study 8, we did not detect CD54 expression on sputum granulocytes, while using the same antibody clone (MEM-111). The reason for this discrepancy remains to be elucidated. We show CD69 to be an activation marker expressed on sputum eosinophils, just as found on BAL eosinophils 15.
Another important finding is the high up-regulation of immune-regulatory protein CD274 on sputum neutrophils, and to a lesser extent on eosinophils (see Fig. 3). Immune suppressive blood neutrophils have been shown to up-regulate mRNA for this marker during acute inflammation and employ it for suppression of T cell proliferation 26. These cells cannot be identified as easily in the lungs, as cells have shed CD62L after leaving the bloodstream, but the expression of CD274 supports the view that sputum neutrophils might have a suppressive phenotype. Interestingly, high expression has also been found in sarcoid lung granulomas, even though the expression of CD274 on neutrophils was not studied specifically. In addition, blockade of the PD-1 (the receptor for CD274) pathway restores T cell functioning in vitro 27. Taken together, the expression of CD274 in the lungs of asthmatic patients favours the hypothesis that sputum cells can modulate inflammation in asthma rather than merely causing tissue damage and perpetuation of the inflammatory response.
In conclusion, granulocytes in sputum display a highly activated and degranulated phenotype compared to granulocytes in peripheral blood. However, sputum granulocytes receptor profiles do not differ in presence or absence of sputum eosinophilia in patients with asthma. Furthermore, we found the immune-inhibitory protein CD274 to be expressed specifically on sputum cells, supporting the hypothesis that sputum granulocytes can have an immune-modulatory instead of a detrimental role in asthma.
Acknowledgments
B.H. recruited patients, B.H. and T.T. performed flow-cytometric measurements. K.T. and L.K. participated in the design and coordination of the study. T.T. performed statistical analyses and wrote the draft of the manuscript, which was revised with the help of all other authors. All authors read and approved the final manuscript. We would like to thank Dr Caroline van Baal from the Julius Centre for her advice on the statistics. Our gratitude goes to the UMCU pulmonary function test department in general and to research nurse Simone Sluis-Eising in particular, without whom this study would not have been possible. This work was supported by the Dutch Lung Foundation (grant no. 3.2.10.052).
Disclosure
The authors declare no competing interests.
Supporting Information
Additional Supporting information may be found in the online version of this article at the publisher's web-site:
Fig. S1. Representative histograms of all measured markers in blood (red) and sputum (blue). Filled solid lines represent the relevant antibody, dashed lines indicate isotype controls. Histograms are binned and normalized for peak values.
Fig. S2. Object scores of principle components analysis (PCA) on marker expression for each celltype and location. PCA was performed on (a) blood neutrophils, (b) sputum neutrophils and (c) blood eosinophils separately. None of these analyses was able to discriminate between patient groups. Dashed lines originate from median object scores of patient groups; n = 13 (>3% sputum eosinophils) and 8 (<3% sputum eosinophils).
References
- Global Initiative for Asthma (GINA) 2012. Gina report, global strategy for asthma management and prevention. Available at: http://www.ginasthma.org/. Accessed February 2 2014.
- Simpson JL, Scott R, Boyle MJ, Gibson PG. Inflammatory subtypes in asthma: assessment and identification using induced sputum. Respirology. 2006;11:54–61. doi: 10.1111/j.1440-1843.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- Woodruff PG, Khashayar R, Lazarus SC, et al. Relationship between airway inflammation, hyperresponsiveness, and obstruction in asthma. J Allergy Clin Immunol. 2001;108:753–8. doi: 10.1067/mai.2001.119411. [DOI] [PubMed] [Google Scholar]
- Wagener AH, de Nijs SB, Lutter R, et al. External validation of blood eosinophils, FENO and serum periostin as surrogates for sputum eosinophils in asthma. Thorax. 2015;70:115–20. doi: 10.1136/thoraxjnl-2014-205634. [DOI] [PubMed] [Google Scholar]
- Pavord ID, Pizzichini MM, Pizzichini E, Hargreave FE. The use of induced sputum to investigate airway inflammation. Thorax. 1997;52:498–501. doi: 10.1136/thx.52.6.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines KJ, Simpson JL, Wood LG, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol. 2014;133:997–1007. doi: 10.1016/j.jaci.2013.12.1091. [DOI] [PubMed] [Google Scholar]
- Brooks CR, van Dalen CJ, Hermans IF, Douwes J. Identifying leukocyte populations in fresh and cryopreserved sputum using flow cytometry. Cytometry B Clin Cytom. 2013;84:104–13. doi: 10.1002/cyto.b.21069. [DOI] [PubMed] [Google Scholar]
- Dominguez OJ, Leon F, Martinez Alonso JC, et al. Fluorocytometric analysis of induced sputum cells in an asthmatic population. J Investig Allergol Clin Immunol. 2004;14:108–13. [PubMed] [Google Scholar]
- Lay JC, Peden DB, Alexis NE. Flow cytometry of sputum: assessing inflammation and immune response elements in the bronchial airways. Inhal Toxicol. 2011;23:392–406. doi: 10.3109/08958378.2011.575568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S, Bellido-Casado J, Granel C, Crespo A, Plaza V, Juarez C. Flow cytometry analysis of leukocytes in induced sputum from asthmatic patients. Immunobiology. 2012;217:692–7. doi: 10.1016/j.imbio.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Alexis N, Soukup J, Ghio A, Becker S. Sputum phagocytes from healthy individuals are functional and activated: a flow cytometric comparison with cells in bronchoalveolar lavage and peripheral blood. Clin Immunol. 2000;97:21–32. doi: 10.1006/clim.2000.4911. [DOI] [PubMed] [Google Scholar]
- Hansel TT, Braunstein JB, Walker C, et al. Sputum eosinophils from asthmatics express ICAM-1 and HLA-DR. Clin Exp Immunol. 1991;86:271–7. doi: 10.1111/j.1365-2249.1991.tb05809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veen JC, Grootendorst DC, Bel EH, et al. CD11b and L-selectin expression on eosinophils and neutrophils in blood and induced sputum of patients with asthma compared with normal subjects. 1998;28:606–15. doi: 10.1046/j.1365-2222.1998.00279.x. Clin Exp Allergy. [DOI] [PubMed] [Google Scholar]
- Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Systemic upregulation of neutrophil alpha-defensins and serine proteases in neutrophilic asthma. Thorax. 2011;66:942–7. doi: 10.1136/thx.2010.157719. [DOI] [PubMed] [Google Scholar]
- Johansson MW. Activation states of blood eosinophils in asthma. Clin Exp Allergy. 2014;44:482–98. doi: 10.1111/cea.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundel RH, Wegner CD, Torcellini CA, et al. Endothelial leukocyte adhesion molecule-1 mediates antigen-induced acute airway inflammation and late-phase airway obstruction in monkeys. J Clin Invest. 1991;88:1407–11. doi: 10.1172/JCI115447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso A, Cosulich ME, Rubartelli A, Mazza MR, Bargellesi A. MLR3 molecule is an activation antigen shared by human B, T lymphocytes and T cell precursors. Eur J Immunol. 1989;19:323–8. doi: 10.1002/eji.1830190216. [DOI] [PubMed] [Google Scholar]
- Gullberg U, Bengtsson N, Bulow E, Garwicz D, Lindmark A, Olsson I. Processing and targeting of granule proteins in human neutrophils. J Immunol Methods. 1999;232:201–10. doi: 10.1016/s0022-1759(99)00177-5. [DOI] [PubMed] [Google Scholar]
- Borregaard N, Cowland JB. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 1997;89:3503–21. [PubMed] [Google Scholar]
- Adkinson NF, Bochner BS, Burks W, Busse WW, Holgate ST. Middleton's allergy: principles and practice. Elsevier-Health Sciences Division; 2013. Philadelphia, PA: [Google Scholar]
- Mallia P, Message SD, Contoli M, et al. Neutrophil adhesion molecules in experimental rhinovirus infection in COPD. Respir Res. 2013;14:72. doi: 10.1186/1465-9921-14-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirouvanziam R, Gernez Y, Conrad CK, et al. Profound functional and signaling changes in viable inflammatory neutrophils homing to cystic fibrosis airways. Proc Natl Acad Sci USA. 2008;105:4335–9. doi: 10.1073/pnas.0712386105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends C, Hoekstra MO, Dijkhuizen B, de Monchy JG, Gerritsen J, Kauffman HF. Expression of CD35 (CR1) and CD11b (CR3) on circulating neutrophils and eosinophils from allergic asthmatic children. Clin Exp Allergy. 1993;23:926–33. doi: 10.1111/j.1365-2222.1993.tb00277.x. [DOI] [PubMed] [Google Scholar]
- Lundahl J, Skedinger M, Hed J, Johansson SG, Zetterstrom O. Lability in complement receptor mobilization of granulocytes in patients with bronchial hyperreactivity. Clin Exp Allergy. 1992;22:834–8. doi: 10.1111/j.1365-2222.1992.tb02828.x. [DOI] [PubMed] [Google Scholar]
- Akbari O, Stock P, Singh AK, et al. PD-L1 and PD-L2 modulate airway inflammation and iNKT-cell-dependent airway hyperreactivity in opposing directions. Mucosal Immunol. 2010;3:81–91. doi: 10.1038/mi.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kleijn S, Langereis JD, Leentjens J, et al. Stimulated neutrophils suppress lymphocyte proliferation through expression of PD-L1. PLOS ONE. 2013;8:e72249. doi: 10.1371/journal.pone.0072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun NA, Celada LJ, Herazo-Maya JD, et al. Blockade of the programmed death-1 pathway restores sarcoidosis CD4+ T cells proliferative capacity. Am J Respir Crit Care Med. 2014;190:560–71. doi: 10.1164/rccm.201401-0188OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linting M, van der Kooij A. Nonlinear principal components analysis with CATPCA: a tutorial. J Pers Assess. 2012;94:12–25. doi: 10.1080/00223891.2011.627965. [DOI] [PubMed] [Google Scholar]
- Wu W, Bleecker E, Moore W, et al. Unsupervised phenotyping of Severe Asthma Research Program participants using expanded lung data. J Allergy Clin Immunol. 2014;133:1280–8. doi: 10.1016/j.jaci.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar P, Pavord ID, Shaw DE, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–24. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–91. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- Sennels HP, Jorgensen HL, Goetze JP, Fahrenkrug J. Rhythmic 24-hour variations of frequently used clinical biochemical parameters in healthy young males – the Bispebjerg study of diurnal variations. Scand J Clin Lab Invest. 2012;72:287–95. doi: 10.3109/00365513.2012.662281. [DOI] [PubMed] [Google Scholar]
- Mattes J, Storm van's Gravesande K, Moeller C, Moseler M, Brandis M, Kuehr J. Circadian variation of exhaled nitric oxide and urinary eosinophil protein X in asthmatic and healthy children. Pediatr Res. 2002;51:190–4. doi: 10.1203/00006450-200202000-00011. [DOI] [PubMed] [Google Scholar]
- Rutgers SR, Timens W, Kaufmann HF, van der Mark TW, Koeter GH, Postma DS. Comparison of induced sputum with bronchial wash, bronchoalveolar lavage and bronchial biopsies in COPD. Eur Respir J. 2000;15:109–15. doi: 10.1183/09031936.00.15110900. [DOI] [PubMed] [Google Scholar]
- Efthimiadis A, Spanevello A, Hamid Q, et al. Methods of sputum processing for cell counts, immunocytochemistry and in situ hybridisation. Eur Respir J Suppl. 2002;37:19s–23s. doi: 10.1183/09031936.02.00001902. [DOI] [PubMed] [Google Scholar]
- Simpson JL, McElduff P, Gibson PG. Assessment and reproducibility of non-eosinophilic asthma using induced sputum. Respiration. 2010;79:147–51. doi: 10.1159/000245899. [DOI] [PubMed] [Google Scholar]
- Mahmudi-Azer S, Downey GP, Moqbel R. Translocation of the tetraspanin CD63 in association with human eosinophil mediator release. Blood. 2002;99:4039–47. doi: 10.1182/blood.v99.11.4039. [DOI] [PubMed] [Google Scholar]
- van Velzen JF, Laros-van Gorkom BA, Pop GA, van Heerde WL. Multicolor flow cytometry for evaluation of platelet surface antigens and activation markers. Thromb Res. 2012;130:92–8. doi: 10.1016/j.thromres.2012.02.041. [DOI] [PubMed] [Google Scholar]
- Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- Fortunati E, Kazemier KM, Grutters JC, Koenderman L, Van den Bosch vJ. Human neutrophils switch to an activated phenotype after homing to the lung irrespective of inflammatory disease. Clin Exp Immunol. 2009;155:559–66. doi: 10.1111/j.1365-2249.2008.03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson C, Pascual M, French L, Steiger G, Schifferli JA. Soluble complement receptor type 1 (CD35) is released from leukocytes by surface cleavage. Eur J Immunol. 1994;24:2725–31. doi: 10.1002/eji.1830241123. [DOI] [PubMed] [Google Scholar]
- Walker C, Rihs S, Braun RK, Betz S, Bruijnzeel PL. Increased expression of CD11b and functional changes in eosinophils after migration across endothelial cell monolayers. J Immunol. 1993;150:4061–71. [PubMed] [Google Scholar]
- Yamamoto H, Sedgwick JB, Vrtis RF, Busse WW. The effect of transendothelial migration on eosinophil function. Am J Respir Cell Mol Biol. 2000;23:379–88. doi: 10.1165/ajrcmb.23.3.3707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Representative histograms of all measured markers in blood (red) and sputum (blue). Filled solid lines represent the relevant antibody, dashed lines indicate isotype controls. Histograms are binned and normalized for peak values.
Fig. S2. Object scores of principle components analysis (PCA) on marker expression for each celltype and location. PCA was performed on (a) blood neutrophils, (b) sputum neutrophils and (c) blood eosinophils separately. None of these analyses was able to discriminate between patient groups. Dashed lines originate from median object scores of patient groups; n = 13 (>3% sputum eosinophils) and 8 (<3% sputum eosinophils).


