Abstract
IMPORTANCE
Chemotherapy response in the majority of patients with ovarian cancer remains unpredictable.
OBJECTIVE
To identify novel molecular markers for predicting chemotherapy response in patients with ovarian cancer.
DESIGN, SETTING, AND PARTICIPANTS
Observational study of genomics and clinical data of high-grade serous ovarian cancer cases with genomic and clinical data made public between 2009 and 2014 via the Cancer Genome Atlas project.
MAIN OUTCOMES AND MEASURES
Chemotherapy response (primary outcome) and overall survival (OS), progression-free survival (PFS), and platinum-free duration (secondary outcome).
RESULTS
In 512 patients with ovarian cancer with available whole-exome sequencing data, mutations from 8 members of the ADAMTS family (ADAMTS mutations) with an overall mutation rate of approximately 10.4% were associated with a significantly higher chemotherapy sensitivity (100% for ADAMTS-mutated vs 64% for ADAMTS wild-type cases; P < .001) and longer platinum-free duration (median platinum-free duration, 21.7 months for ADAMTS-mutated vs 10.1 months for ADAMTS wild-type cases; P = .001). Moreover, ADAMTS mutations were associated with significantly better OS (hazard ratio [HR], 0.54 [95% CI, 0.42–0.89]; P = .01 and median OS, 58.0 months for ADAMTS-mutated vs 41.3 months for ADAMTS wild-type cases) and PFS (HR, 0.42 [95% CI, 0.38–0.70]; P < .001 and median PFS, 31.8 for ADAMTS-mutated vs 15.3 months for ADAMTS wild-type cases). After adjustment by BRCA1 or BRCA2 mutation, surgical stage, residual tumor, and patient age, ADAMTS mutations were significantly associated with better OS (HR, 0.53 [95% CI, 0.32–0.87]; P = .01), PFS (HR, 0.40 [95% CI, 0.25–0.62]; P < .001), and platinum-free survival (HR, 0.45 [95% CI, 0.28–0.73]; P = .001). ADAMTS-mutated cases exhibited a distinct mutation spectrum and were significantly associated with tumors with a higher genome-wide mutation rate than ADAMTS wild-type cases across the whole exome (median mutation number per sample, 121 for ADAMTS-mutated vs 69 for ADAMTS wild-type cases; P < .001).
CONCLUSIONS AND RELEVANCE
ADAMTS mutations may contribute to outcomes in ovarian cancer cases without BRCA1 or BRCA2 mutations and may have important clinical implications.
Ovarian cancer remains the leading cause of mortality from gynecologic cancer.1,2 Despite aggressive surgery and chemotherapy, most patients eventually experience relapse with generally incurable disease mainly due to emergence of chemotherapy resistance.3,4 Early identification and differentiation of patients with chemotherapy-resistant disease could allow enrollment in clinical trials with alternative therapeutics rather than ineffective chemotherapy.
Patients with ovarian cancer with germline or somatic BRCA1 or BRCA2 mutations are recognized to have better response to platinum-based treatment and substantially longer survival than noncarriers.5 Recent analyses showed that BRCA2 mutation demonstrated a stronger association with improved survival and chemotherapy response among women with ovarian cancer than BRCA1 mutation across multiple data sets.6,7
BRCA1 or BRCA2 mutations including both germline and somatic mutations have been found in 20.3% of the Cancer Genome Atlas (TCGA) patients with ovarian cancer,8 which is similar to the mutation rates reported in previous studies.9,10 However, the clinical chemosensitive rates to platinum-based therapy regimens are approximately 70%,11 suggesting that events other than BRCA1 or BRCA2 mutations exist that predict chemotherapy response. In this study, we examined TCGA genomic and clinical data to determine the association between novel gene mutations in ovarian cancer and patient overall survival (OS), progression-free survival (PFS), and chemotherapy response.
Methods
Patients and Study Design
We obtained the whole-exome sequencing data for 512 patients with high-grade serous ovarian cancer from TCGA.8 The specimens were obtained prior to systemic therapy and all patients received platinum-based chemotherapy. The entire TCGA cohort was divided into a discovery set of 210 cases (hereafter referred to as the discovery cohort) and a validation set of 302 cases (hereafter referred to as the validation cohort). The separation of discovery and validation cohorts is described in detail in the eMethods in the Supplement. Details about patient characteristics and study design are described in the eMethods, eFigure 1, and eTables 1, 2, and 3 in the Supplement. Access to TCGA database was approved by the National Cancer Institute (https://tcga-data.nci.nih.gov/tcga). The study was approved by the institutional review board at the University of Texas MD Anderson Cancer Center. The need for consent was waived because of the retrospective nature of the study.
Whole-Exome Sequencing Data Analysis
We analyzed the whole-exome sequencing data for the 210 TCGA cases in the discovery cohort that had explicitly defined response status to chemotherapy (sensitive or resistant). To quantify the association of gene mutation with response status, we calculated for each individual gene the number of mutations in the sensitive (Ns) or resistant (Nr) samples, respectively. We further selected the genes associated with chemosensitivity by applying both of the following criteria: (1) Nr = 0; (2) Ns ≥ 2.
We calculated the mutation frequency in terms of the total number of mutations including single-nucleotide substitution or insertion-deletion (indel) per sample. Fractions of mutations (indels were excluded) in the 6 possible mutation classes (ie, C>T, C>A, C>G, A>G, A>C, and A>T) were calculated for each sample. Details of whole-exome sequencing and chemotherapy response data analyses are provided in the eMethods in the Supplement.
Statistical Analysis
Survival differences were assessed using the log-rank test or Wald test (details are described in the eMethods in the Supplement). Other standard statistical tests were used to analyze the clinical and genomic data, including the Mann-Whitney, Fisher exact, and χ2 tests. All statistical tests were 2 sided, and P < .05 was considered statistically significant. Statistical analyses were performed using scientific software such as Matlab (MathWorks), SPSS version 18 (SPSS Inc), and GraphPad Prism, version 6 (Graphpad Software Inc).
Results
ADAMTS Mutations in TCGA Ovarian Cancer Patients
Whole-exome capture and sequencing of TCGA ovarian cancer samples targeted approximately 180 000 exons from 18 500 genes.8 Of the 210 patients with explicit chemotherapy response status in TCGA discovery cohort (eMethods and eFigure 1 in the Supplement), 141 were designated as sensitive and 69 as resistant. Mutation analysis showed that 2118 genes including BRCA2 were mutated in at least 2 chemosensitive samples (Ns ≥ 2), but not in any of the chemoresistant cases (Nr = 0) (eFigure 2 and eTable 4 in the Supplement). The majority of these genes had small numbers of mutations, which is consistent with the somatic mutation frequency of any gene other than TP53 being relatively low in high-grade serous ovarian cancer.8 ADAMTS16, a member of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) superfamily,12 is one of the most frequently mutated genes (eTable 5 in the Supplement). Because members from the gene family share common protein structural domains and demonstrate functional redundancy,13,14 we next examined whether any other member(s) of the ADAMTS family was associated with chemosensitivity. Interestingly, we found that 6 ADAMTS family members in addition to ADAMTS16 demonstrated a mutation bias in platinum-sensitive patients (eFigure 2 in the Supplement). Furthermore, gene set enrichment analysis of the responder-related genes showed that members of this gene family were significantly enriched in the list (P = .02, χ2 test). The mutated members consisted of ADAMTS16 (~4.3%), ADAMTSL1 (~2.9%), and ADAMTS1, ADAMTS15, ADAMTS6, ADAMTS9, and ADAMTS18 (~1.0% each). To obtain a more comprehensive view of gene mutations from this family, we included ADAMTS13 in the downstream analysis although it was mutated in only 1 chemosensitive sample (Figure 1). We use the term “ADAMTS mutations” to refer to the mutations of these 8 members, unless specified otherwise.
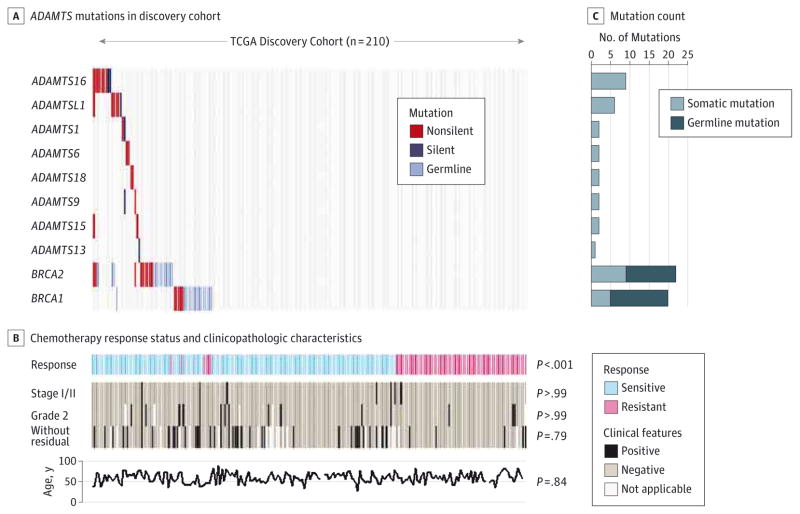
Figure 1. ADAMTS Mutations in the Cancer Genome Atlas (TCGA) Discovery Cohort.
ADAMTS and BRCA1 or BRCA2 mutations that were detected in the 210 TCGA patients with ovarian cancer in the discovery cohort. A, For each gene (row) indicated, tumors (columns) with mutations are labeled with red (nonsilent mutations), dark blue (silent mutations), or light blue (germline mutations) bars. The locations of the residuals altered by ADAMTS mutations are detailed in eFigure 3 in the Supplement. B, Chemotherapy response status and clinicopathologic characteristics for each individual patient. “Without residual” denotes a tumor with no macroscopic disease. The P values show the comparison between the ADAMTS-mutated cases vs ADAMTS wild-type cases. C, Mutation count for each individual gene shown in panel A.
Together, ADAMTS mutations were found in a total of 23 ovarian cancer samples (Figure 1); most of these were missense (eFigure 3 in the Supplement). Forty-two samples harbored BRCA1 or BRCA2 mutations, 66.7% of which were germline mutations (Figure 1). The BRCA and ADAMTS mutations were not correlated with each other (P = .26, Fisher exact test). Except for a significant correlation with chemotherapy response status, ADAMTS mutations were not correlated with age or clinical characteristics such as stage, grade, and residual tumor (Figure 1 and eTable 6 in the Supplement).
Association of ADAMTS Mutations With Patient Survival
We next determined the relationship between the prevalence of ADAMTS mutations and patient outcome (Figure 2). Kaplan-Meier survival analysis revealed that patients with ADAMTS mutations had a 5-year survival rate of approximately 59% and exhibited significantly longer OS than those without (median OS, not reached vs 44.4 months; log-rank P = .007; hazard ratio [HR], 0.37 [95% CI, 0.29–0.82]) (Figure 2A). Similarly, the median PFS of the ADAMTS-mutated cases was almost twice as long as that of the ADAMTS wild-type cases (26.8 vs 14.0 months; log-rank P = .002; HR, 0.47 [95% CI, 0.38–0.80]) (Figure 2B).
Figure 2. Association of ADAMTS Mutations With Clinical Outcome and Chemotherapy Response.
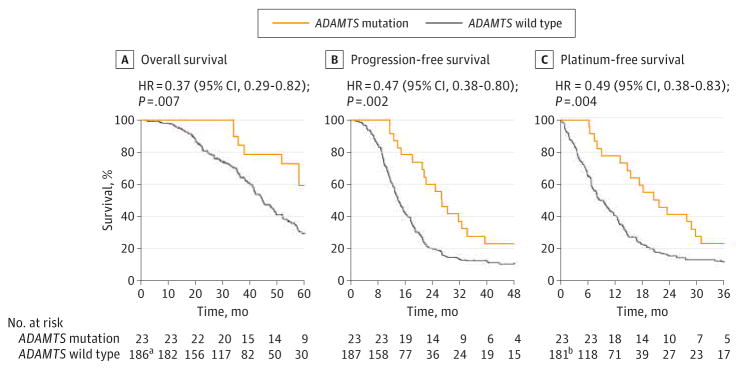
Estimates of clinical outcome and chemotherapy response were performed among patients that were stratified on the basis of ADAMTS mutations. Subgroups were compared with the use of the log-rank test. Kaplan-Meier analyses of overall survival (A), progression-free survival (B), and platinum-free survival (C) of individuals with ovarian cancer in the Cancer Genome Atlas (TCGA) discovery cohort are shown. A and B, The percentage probability is plotted vs time since diagnosis in months. C, The percentage probability is plotted vs time since the end of adjuvant therapy. The number of patients at risk is shown below each curve at various time points.
aOne case is not included in this analysis because of missing overall survival data in TCGA database.
bSix cases are excluded from this analysis because the patients underwent platinum treatment after progression or recurrence.
To test whether this result was independent of known predictive variables such as BRCA1 or BRCA2 mutation status, residual tumor size (eFigures 4 and 5 in the Supplement), stage, orage,15,16 we applied multivariate analysis using a Cox proportional hazards model with ADAMTS mutation status and known predictors as covariates. After adjustment by BRCA1 or BRCA2 mutation, stage, residual tumor, and patient age, ADAMTS mutation was significantly associated with longer OS (HR, 0.32[95% CI, 0.14–0.69]; P = .004) and PFS (HR, 0.42 [95% CI, 0.25–0.71]; P = .001) (Table). Furthermore, we observed no correlation between ADAMTS mutation and these covariates (eTable 6 in the Supplement). These data suggested that ADAMTS mutation is an independent predictor of survival in patients with ovarian cancer.
Table.
Univariate and Multivariate Models for Overall Survival, Progression-Free Survival, and Platinum-Free Survival in Women With Ovarian Cancer in the Cancer Genome Atlas (TCGA) Discovery Cohorta
| Characteristic | Univariate Analysis
|
Multivariate Analysisb
|
||
|---|---|---|---|---|
| HR (95% CI) | P Valuec | HR (95% CI) | P Valuec | |
| Overall Survival | ||||
|
| ||||
| ADAMTS status | ||||
|
| ||||
| Wild type | 1 [Reference] | .01 | 1 [Reference] | .004 |
|
|
|
|||
| Mutation | 0.36 (0.17–0.79) | 0.32 (0.14–0.69) | ||
|
| ||||
| BRCA1 or BRCA2 status | ||||
|
| ||||
| Wild type | 1 [Reference] | .001 | 1 [Reference] | .002 |
|
|
|
|||
| Mutation | 0.36 (0.20–0.64) | 0.40 (0.22–0.72) | ||
|
| ||||
| Tumor stage | ||||
|
| ||||
| II | 1 [Reference] | .24 | 1 [Reference] | .24 |
|
|
|
|||
| III or IV | 1.27 (0.40–4.00) | 2.32 (0.56–9.50) | ||
|
| ||||
| Residual tumor size, mm | ||||
|
| ||||
| 0d | 1 [Reference] | 1 [Reference] | ||
|
| ||||
| 1–20 | 2.07 (1.17–3.66) | .01 | 1.91 (1.08–3.39) | .03 |
|
| ||||
| >20 | 2.17 (1.08–4.37) | .03 | 1.59 (0.79–3.24) | .20 |
|
| ||||
| Age at diagnosis, y | 1.01 (0.99–1.03) | .20 | 1.02 (1.00–1.03) | .09 |
|
| ||||
| Progression-Free Survival | ||||
|
| ||||
| ADAMTS status | ||||
|
| ||||
| Wild type | 1 [Reference] | .003 | 1 [Reference] | .001 |
|
|
|
|||
| Mutation | 0.46 (0.28–0.77) | 0.42 (0.25–0.71) | ||
|
| ||||
| BRCA1 or BRCA2 status | ||||
|
| ||||
| Wild type | 1 [Reference] | .006 | 1 [Reference] | .009 |
|
|
|
|||
| Mutation | 0.59 (0.40–0.86) | 0.58 (0.39–0.88) | ||
|
| ||||
| Tumor stage | ||||
|
| ||||
| II | 1 [Reference] | .26 | 1 [Reference] | .18 |
|
|
|
|||
| III or IV | 1.67 (0.69–4.08) | 2.00 (0.73–5.47) | ||
|
| ||||
| Residual tumor size, mm | ||||
|
| ||||
| 0d | 1 [Reference] | 1 [Reference] | ||
|
| ||||
| 1–20 | 1.89 (1.26–2.83) | .002 | 1.90 (1.26–2.86) | .002 |
|
| ||||
| >20 | 1.82 (1.11–2.98) | .02 | 1.56 (0.94–2.60) | .08 |
|
| ||||
| Age at diagnosis, y | 1.00 (0.98–1.01) | .56 | 1.00 (0.98–1.01) | .47 |
|
| ||||
| Platinum-Free Survival | ||||
|
| ||||
| ADAMTS status | ||||
|
| ||||
| Wild type | 1 [Reference] | .005 | 1 [Reference] | .002 |
|
|
|
|||
| Mutation | 0.48 (0.29–0.80) | 0.43 (0.26–0.73) | ||
|
| ||||
| BRCA1 or BRCA2 status | ||||
|
| ||||
| Wild type | 1 [Reference] | .01 | 1 [Reference] | .01 |
|
|
|
|||
| Mutation | 0.60 (0.41–0.89) | 0.59 (0.39–0.89) | ||
|
| ||||
| Tumor stage | ||||
|
| ||||
| II | 1 [Reference] | .18 | 1 [Reference] | .21 |
|
|
|
|||
| III or IV | 1.83 (0.75–4.47) | 1.91 (0.70–5.23) | ||
|
| ||||
| Residual tumor size, mm | ||||
|
| ||||
| 0d | 1 [Reference] | 1 [Reference] | ||
|
| ||||
| 1–20 | 1.86 (1.24–2.79) | .003 | 1.88 (1.25–2.83) | .03 |
|
| ||||
| >20 | 1.84 (1.11–3.03) | .02 | 1.60 (0.96–2.69) | .07 |
|
| ||||
| Age at diagnosis, y | 1.00 (0.98–1.01) | .63 | 1.00 (0.98–1.01) | .50 |
Abbreviation: HR, hazard ratio.
Included are data from the 210 TCGA patients with ovarian cancer who had an explicitly defined chemotherapy response status. Patient characteristics are detailed in eTable 1 in the Supplement. BRCA mutations include somatic and germline mutations of BRCA1 and BRCA2. Both ADAMTS and BRCA1 and BRCA2 mutations are depicted in Figure 1.
Based on a multivariate Cox proportional hazards model, including all variables in the table.
Wald test.
Patients with no macroscopic disease are categorized as 0 mm.
Association of ADAMTS Mutations With Chemotherapy Response
All ADAMTS-mutated cases in the discovery cohort were designated as chemosensitive. We next determined the association between ADAMTS mutations and platinum-free duration after treatment, a parameter characterizing platinum-based chemotherapy response. As shown in Figure 2C, 41% of patients with ADAMTS mutations had a 2-year platinum-free duration. Patients with ADAMTS mutations exhibited a significantly longer platinum-free duration than those with wild-type ADAMTS (median platinum-free duration, 21.7 vs 8.7 months; log-rank P = .004; HR, 0.49 [95% CI, 0.38–0.83]). Multivariate Cox proportional hazards model analysis showed that ADAMTS mutation had a significant association with platinum-free duration (HR, 0.43 [95% CI, 0.26–0.73]; P = .002), independent of other known predictors such as BRCA1 or BRCA2 mutations, stage, and residual tumor volume (Table).
Association of ADAMTS Mutations With Mutation Spectra
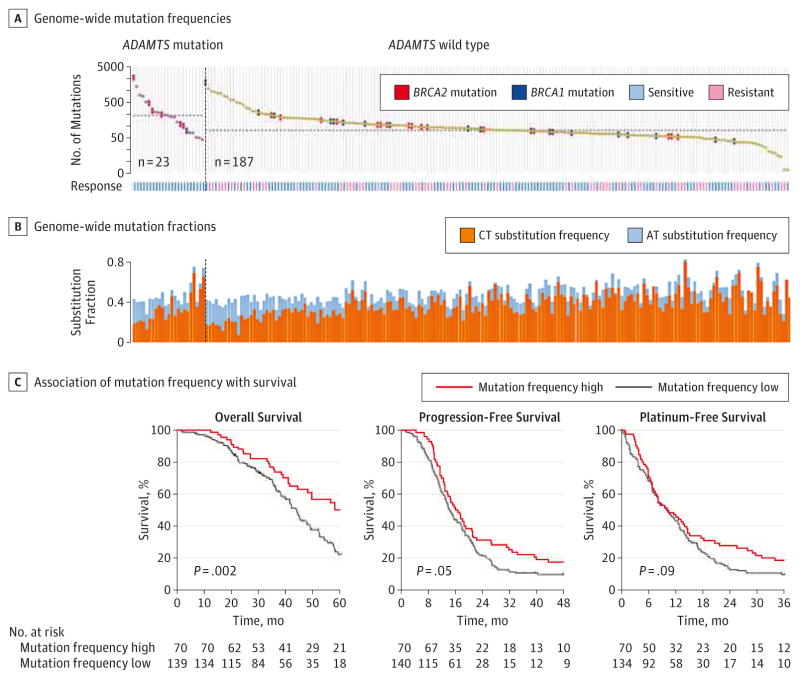
Using whole-exome sequencing data, we further examined the association between ADAMTS mutations with mutation spectra in the ovarian cancer exome. Using the method as previously described,7 we found that the ADAMTS-mutated cases were significantly enriched in the hypermutated samples (P < .01) (eFigure 6 in the Supplement). The median number of mutations was 183 for ADAMTS-mutated vs 69 for ADAMTS wild-type cases (P < .001, Mann-Whitney test) (Figure 3A). In contrast, BRCA2 mutation (P = .02) but not BRCA1 mutation (P = .73) was significantly associated with mutation rate in this cohort (eFigure 7 in the Supplement). Moreover, ADAMTS-mutated cases had a significantly lower percentage of C>T transition (P = .003) but a significantly higher percentage of A>T transversion (P = .03) than ADAMTS wild-type cases (Figure 3B and eFigure 8 in the Supplement). The proportion of C>T transitions was negatively correlated with the mutation rate, whereas the proportion of A>T transversions showed a significant correlation but in the opposite direction (eFigure 9 in the Supplement). Because high mutation rate was previously reported to be associated with better prognosis in endometrial17 and colorectal18 cancer, we hypothesized that the association of ADAMTS mutations with better survival in ovarian cancer could be a consequence of genetic instability. Supporting this notion, we found that patients with higher mutation rates exhibited significantly longer OS (P = .002) and PFS (P = .05) but no significant difference in platinum-free interval (P = .09) as compared with those with lower mutation rates (Figure 3C).
Figure 3. Association of ADAMTS Mutations With Mutation Spectra.
A, Genome-wide mutation frequencies in terms of the number of mutations (vertical axis) detected for each tumor (horizontal axis) in order of descending number of mutations in each patient group stratified according to ADAMTS mutations. The median number of mutations in the ADAMTS-mutated (183) and wild-type groups (69) are indicated by the horizontal dashed lines. Samples with BRCA1 or BRCA2 mutations are also indicated. Patients’ response status to chemotherapy is also shown. B, Fractions (vertical axis) of C>T transition and A>T transversion for each tumor (horizontal axis) in the same order as in A. C, Kaplan-Meier analyses of overall survival, progression-free survival, and platinum-free survival in patients stratified by mutation frequency. The ovarian cancer tumors were dichotomously categorized on the basis of patient mutation rate into 2 groups, high (highest one-third, n = 70) and low (rest of cohort, n = 140) mutation frequency. Subgroups were compared with the use of the log-rank test.
Multifaceted Validation of ADAMTS Mutations
To determine whether the association of ADAMTS mutations with patient outcome is due to chance, we attempted to examine whether any gene combinations from the original 2118 responder-related genes were also significantly associated with patient outcome. We randomly selected 8 genes (to match the 8 ADAMTS genes) from the list and then performed survival analysis between patients stratified by mutation status in the 8 selected genes. This process was repeated 105 times (eMethods and eFigure 10 in the Supplement). We found that mutations in 58.8%, 69.3%, or 64.0% of gene combinations were not significantly associated with OS, PFS, or platinum-free survival, respectively (P > .05). Only 3.4% of gene combinations were simultaneously correlated with OS, PFS, and platinum-free survival (P < .01 for all 3 survival categories), similar to those of ADAMTS mutations (eFigure 11 in the Supplement). The P values of these random selections generated a null distribution for association of the 8-gene combination with outcome, from which we can calculate the nominal P value of association of ADAMTS mutations with outcome relative to this null distribution. This analysis showed that the association of ADAMTS mutations with outcome was statistically significant for PFS (nominal P = .02) and platinum-free survival (nominal P = .05) but not for OS (nominal P = .09), as compared with the background statistical significance levels (eMethods and eFigure 11 in the Supplement).
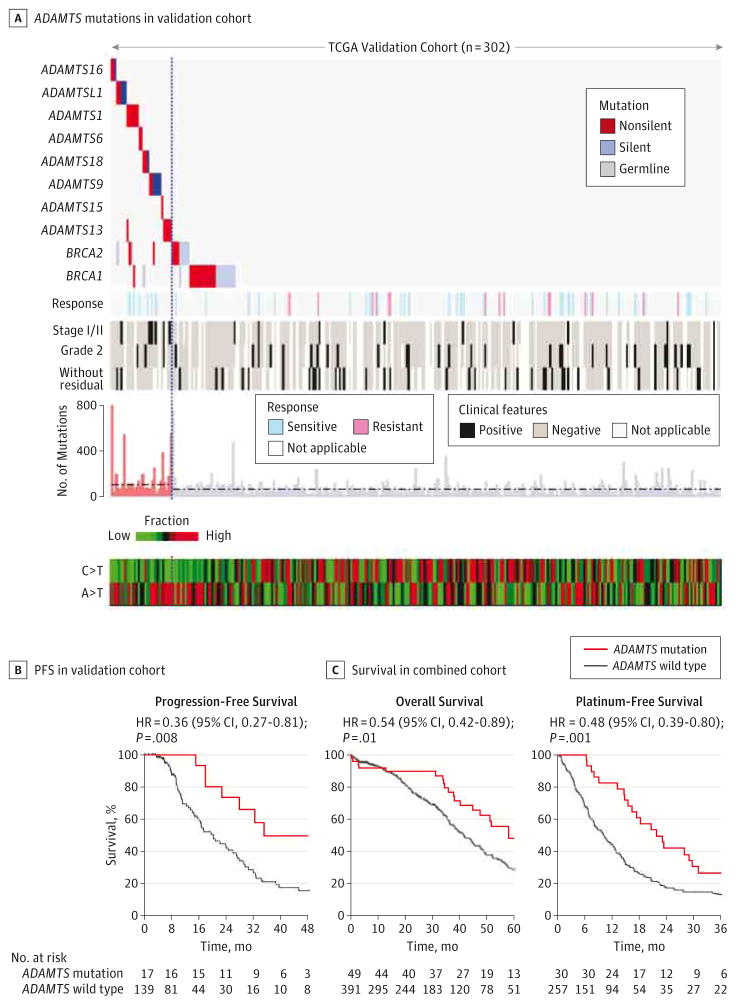
Next we validated the predictive value of ADAMTS mutations in a separate TCGA validation cohort that comprised 302 ovarian cancer samples (eMethods, eFigure 12, and eTable 7 in the Supplement), as evaluated by the associations of ADAMTS mutations with chemosensitivity, survival, and mutation spectra. In this validation cohort, 30 cases had ADAMTS mutations that were not correlated with BRCA1 or BRCA2 mutations (P = .24, Fisher exact test) (eFigures 13 and 14 in the Supplement) (only somatic mutation data were available for the second batch). ADAMTS mutations were significantly associated with hypermutated samples (P < .001, Mann-Whitney test) and had a significantly lower percentage of C>T transition (P < .001) but higher percentage of A>T transversion (P = .003) (Figure 4A and eFigures 15–17 in the Supplement). Among those with known chemotherapy response status, all ADAMTS-mutated cases were sensitive and none were resistant (Figure 4A). These results were consistent with the findings from the discovery cohort. Additionally, patients with ADAMTS mutation had significantly longer PFS than those without (HR, 0.36 [95% CI, 0.27–0.81]; log-rank P = .008) (Figure 4B). ADAMTS mutations exhibited no significant difference in OS and platinum-free interval; this could have resulted from the short OS follow-up duration and smaller size of analyzed samples with platinum-free survival data (eFigure 18 and eTable 8 in the Supplement). Likely for the same reason, the known out come predictor, BRCA1 orBRCA2 mutation status, unexpectedly was not significantly associated with OS and platinum-free survival (eFigure 19 in the Supplement).
Figure 4. Validation of ADAMTS Mutations.
A, Association of ADAMTS mutations with chemotherapy response status, clinicopathologic characteristics, and mutation spectra in the Cancer Genome Atlas (TCGA) validation cohort (n = 302). “Without residual” denotes a tumor with no macroscopic disease. The median number of mutations in the ADAMTS-mutated (111) and wild-type groups (69) are indicated by the horizontal dashed lines. The vertical dashed line highlights the separation of the ADAMTS-mutated samples from the ADAMTS wild-type cases. B, Kaplan-Meier analysis of progression-free survival in patients stratified by ADAMTS mutations in the validation cohort. C, Kaplan-Meier analyses of overall survival and platinum-free survival in patients stratified by ADAMTS mutations in TCGA combined cohort.
We pooled the 2 TCGA cohorts and analyzed ADAMTS mutations in the combined cohort of 512 patients with ovarian cancer. A total of 53 cases had ADAMTS mutations, which corresponded to an overall mutation rate of approximately 10.4%; all were chemotherapy sensitive (eFigure 20 and eTable 9 in the Supplement). Consistently, ADAMTS mutations were significantly associated with hypermutated samples (P < .001, Mann-Whitney test), low C>T transition (P < .001), and high A>T transversion (P < .001) but were not significantly correlated with BRCA1 or BRCA2 mutations (P = .07, Fisher exact test) (eFigures 20 and 21 in the Supplement). With an increased clinical follow-up and more samples included in the platinum-free survival analysis (eTable 8 in the Supplement), BRCA1 or BRCA2 mutations, as anticipated, exhibited a significant correlation with clinical outcome in this combined cohort (eFigure 22 in the Supplement). For the same reason, patients with ADAMTS mutations exhibited significantly longer OS (HR, 0.54 [95% CI, 0.42–0.89]; log-rank P = .01) and platinum-free interval (HR, 0.48 [95% CI, 0.39–0.80]; log-rank P = .001) than did those without (Figure 4C), similar to PFS (HR, 0.42[95% CI, 0.38–0.70]) (eFigure 23 in the Supplement). In an adjusted model, ADAMTS mutation was significantly associated with longer OS (HR, 0.53 [95%CI, 0.32–0.87]; P = .01), PFS (HR, 0.40 [95% CI, 0.25–0.62]; P < .001), and platinum-free survival (HR, 0.45 [95% CI, 0.28–0.73]; P = .001) independent of BRCA1 or BRCA2 mutation, stage, residual tumor, and age (eTable 10 in the Supplement). Moreover, ADAMTS nonsilent mutations consistently exhibited significant association with longer OS (HR, 0.57 [95% CI, 0.41–0.98]; P = .04), PFS (HR, 0.49 [95% CI, 0.40–0.82]; P = .002), and platinum-free survival (HR, 0.52 [95% CI, 0.40–0.89]; P = .01) even when silent mutations were excluded (eFigure 24 in the Supplement).
Discussion
Drug resistance is a major cause of treatment failure in ovarian cancer and primarily contributes to the disease’s high mortality rate. The early identification of patients who are (or are not) benefiting from platinum-based therapy is central to advancing ovarian cancer management and represents an important step toward the goal of personalized treatment. In this study, we found that patients with ADAMTS mutations were significantly correlated with an improved chemotherapy sensitivity and exhibited a significantly longer platinum-free duration than those with ADAMTS wild-type tumors. Moreover, ADAMTS mutation status was an independent predictor of OS and PFS in patients with ovarian cancer regardless of BRCA1 or BRCA2 mutations, stage, residual tumor, and age. Thus, taken together, patients with either ADAMTS or BRCA1 or BRCA2 mutations are more likely to benefit from platinum-based therapy. Nevertheless, additional predictors of sensitivity remain to be detected because many patients without ADAMTS or BRCA1 or BRCA2 mutations are chemosensitive.
Alterations in the ADAMTS genes have been detected in cancers8,13 and other diseases.12 A variant at ADAMTS6 orADAMTS16 was reported to be associated with susceptibility toosteosarcoma19 or with premature ovarian failure.20 ADAMTS13 mutation was recognized to cause thrombotic thrombocytopenic purpura.21 For the first time, to our knowledge, we identified an association between ADAMTS mutations and clinical outcome in patients with ovarian cancer. The ADAMTS genes consist of a protease domain and an ancillary domain, each of which provides substrate-binding or cleavage-site specificity. Similar to BRCA1 orBRCA2, which have no “hot spot” (recurrent) somatic mutations,7,8 there is no common domain that is mutated across the ADAMTS genes. However, unlike BRCA1 or BRCA2, ADAMTS genes are rarely reported to be mutated in the germ line.
Functionally, the ADAMTS proteases are a distinct group of enzymes with broad catalytic activity against a range of substrates22 and have been demonstrated to have important roles in angiogenesis, cell migration, coagulation, andinflammation.23 In particular, ADAMTS1,24 ADAMTS9,25,26 ADAMTS15,27 and ADAMTS1828,29 have been reported to function as tumor suppressor genes and to inhibit angiogenesis26,30,31 in several cancers. ADAMTSL1 was previously shown to be involved in ovary development.32 ADAMTS15 mutations restrained tumor growth and invasion in colorectal cancer. Adamts16-mutant rats exhibited a longer survival rate than did control rats by alteration in the vasculature.33 Collectively, the ADAMTS genes, to a large extent, have been demonstrated to play a critical role in the development of vasculature, which is known to be heavily implicated in ovarian cancer prognosis.34,35 This may explain our observation of a better survival among ADAMTS-mutated patients.
ADAMTS mutations were significantly associated with tumors with a high mutation rate, which was similar to what has been observed for BRCA2 mutations. 7 A recent study showed that some ovarian cancer cases had a mutation signature similar to that found in BRCA1- or BRCA2-mutated cases but did not harbor BRCA1 or BRCA2 mutations, indicating that abnormalities of genes other than BRCA1 or BRCA2 may contribute to this mutation pattern.36 Previous studies suggested that genetic in stability could result in the sensitization to DNA-damaging agents.37 Platinum based treatment induces cross-linking and single-strand or double-strandbreaks.38 Cells that have more mutations in the genome may have compromised DNA repair and altered DNA replication capacities, contributing to an increased sensitivity to apoptosis triggered by platinum-induced DNA damage. In support of this notion, we recently showed that BRCA2 mutations but not BRCA1 mutations were significantly associated with high mutation rate and significantly correlated with an improved chemosensitivity in patients with ovarian cancer, as compared with BRCA1 or BRCA2 wild-type cases.7 Prominently, we further found that the association of mutation rate with ADAMTS mutations is also statistically significant. High mutation rate is associated with better prognosis in ovarian cancer, similar to findings in endometrial17 andcolorectal18 cancer. These data together with a small overlap between tumors with ADAMTS and BRCA1 or BRCA2 mutations suggest that ADAMTS mutations may play a similar role in response to DNA-damaging agents, leading to better survival and improved chemosensitivity in the patients with ovarian cancer whose tumors did not harbor BRCA1 or BRCA2 mutations. However, the molecular mechanism underlying the association of ADAMTS mutations with hypermutated samples remains unclear and requires in-depth studies.
Conclusions
Using whole-exome sequencing, we have, for the first time to our knowledge, reported a novel association of ADAMTS mutations with longer survival and improved chemotherapy sensitivity in patients with ovarian cancer. The finding has important implications for clinical prediction and trial design and may be a useful addition to BRCA mutation assessment for patients with ovarian cancer.
Supplementary Material
At a Glance.
Chemotherapy response in the majority of high-grade serous ovarian cancer patients remains unpredictable.
ADAMTS mutations are significantly associated with improved chemotherapy sensitivity (P < .001) and a longer platinum-free duration (P = .001).
ADAMTS mutations are significantly associated with longer overall survival (P = .01) and progression-free survival (P < .001), independent of BRCA1 or BRCA2 mutations, tumor stage, residual tumor size, and age.
There is no statistically significant correlation between ADAMTS and BRCA1 or BRCA2 mutations.
ADAMTS mutations are significantly associated with patients with ovarian cancer with a higher mutation rate (P < .001).
Acknowledgments
Funding/Support: This study was partially supported by a Sprint for Life Research Award from the Blanton-Davis Ovarian Cancer Research Program (Dr Liu), grants from the Program for Changjiang Scholars and Innovative Research Team in University in China and National Key Scientific and Technological Project (2011ZX0 9307–001–04 to Dr Chen), the Ovarian Cancer SPORE (P50 CA083639 to Dr Sood), U54 CA151668 (Dr Sood), funding for the Genome Data Analysis Centers from the National Institutes of Health (U24 CA143835 to Drs Shmulevich and Zhang), and funding for the Cancer Systems Informatics Center from the National Foundation for Cancer Research (Dr Zhang).
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Supplemental content at jamaoncology.com
Conflict of Interest Disclosures: None reported.
Additional Contributions: We thank Ann Sutton, BA, Department of Scientific Publications, University of Texas MD Anderson Cancer Center, for editing the manuscript. She received no specific compensation for editing this article.
Author Contributions: Dr Zhang had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Liu, Yasukawa, Mills, Sood, Zhang.
Acquisition, analysis, or interpretation of data: Liu, Yasukawa, Chen, Hu, Broaddus, Ding, Mardis, Spellman, Levine, Shmulevich, Sood, Zhang.
Drafting of the manuscript: Liu, Yasukawa, Zhang.
Critical revision of the manuscript for important intellectual content: Liu, Yasukawa, Chen, Hu, Broaddus, Ding, Mardis, Spellman, Levine, Mills, Shmulevich, Sood, Zhang.
Statistical analysis: Liu, Yasukawa, Shmulevich
Obtained funding: Zhang.
Administrative, technical, or material support: Chen, Hu, Broaddus, Ding, Spellman, Levine, Mills, Zhang.
Study supervision: Mardis, Mills, Sood, Zhang.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Spentzos D, Levine DA, Kolia S, et al. Unique gene expression profile based on pathologic response in epithelial ovarian cancer. J Clin Oncol. 2005;23(31):7911–7918. doi: 10.1200/JCO.2005.02.9363. [DOI] [PubMed] [Google Scholar]
- 3.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351(24):2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 4.Selvanayagam ZE, Cheung TH, Wei N, et al. Prediction of chemotherapeutic response in ovarian cancer with DNA microarray expression profiling. Cancer Genet Cytogenet. 2004;154(1):63–66. doi: 10.1016/j.cancergencyto.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Chetrit A, Hirsh-Yechezkel G, Ben-David Y, Lubin F, Friedman E, Sadetzki S. Effect of BRCA1/2 mutations on long-term survival of patients with invasive ovarian cancer: the national Israeli study of ovarian cancer. J Clin Oncol. 2008;26(1):20–25. doi: 10.1200/JCO.2007.11.6905. [DOI] [PubMed] [Google Scholar]
- 6.Bolton KL, Chenevix-Trench G, Goh C, et al. EMBRACE; kConFab Investigators; Cancer Genome Atlas Research Network. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307(4):382–390. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang D, Khan S, Sun Y, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306(14):1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennessy BT, Timms KM, Carey MS, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. 2010;28(22):3570–3576. doi: 10.1200/JCO.2009.27.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal T, Permuth-Wey J, Betts JA, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104(12):2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 11.Rustin GJ, Nelstrop AE, McClean P, et al. Defining response of ovarian carcinoma to initial chemotherapy according to serum CA 125. J Clin Oncol. 1996;14(5):1545–1551. doi: 10.1200/JCO.1996.14.5.1545. [DOI] [PubMed] [Google Scholar]
- 12.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem. 2009;284(46):31493–31497. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 14.Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landrum LM, Java J, Mathews CA, et al. Prognostic factors for stage III epithelial ovarian cancer treated with intraperitoneal chemotherapy: a Gynecologic Oncology Group study. Gynecol Oncol. 2013;130(1):12–18. doi: 10.1016/j.ygyno.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan JK, Tian C, Monk BJ, et al. Gynecologic Oncology Group. Prognostic factors for high-risk early-stage epithelial ovarian cancer: a Gynecologic Oncology Group study. Cancer. 2008;112(10):2202–2210. doi: 10.1002/cncr.23390. [DOI] [PubMed] [Google Scholar]
- 17.Kandoth C, Schultz N, Cherniack AD, et al. Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage SA, Mirabello L, Wang Z, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 2013;45(7):799–803. doi: 10.1038/ng.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyun JA, Kim S, Cha DH, Kwack K. Epistasis between polymorphisms in TSHB and ADAMTS16 is associated with premature ovarian failure. Menopause. 2014;21(8):890–895. doi: 10.1097/GME.0000000000000172. [DOI] [PubMed] [Google Scholar]
- 21.Levy GG, Nichols WC, Lian EC, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413(6855):488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 22.Gao W, Zhu J, Westfield LA, Tuley EA, Anderson PJ, Sadler JE. Rearranging exosites in noncatalytic domains can redirect the substrate specificity of ADAMTS proteases. J Biol Chem. 2012;287(32):26944–26952. doi: 10.1074/jbc.M112.380535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanton H, Melrose J, Little CB, Fosang AJ. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim Biophys Acta. 2011;1812(12):1616–1629. doi: 10.1016/j.bbadis.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Martino-Echarri E, Fernández-Rodríguez R, Rodríguez-Baena FJ, et al. Contribution of ADAMTS1 as a tumor suppressor gene in human breast carcinoma: linking its tumor inhibitory properties to its proteolytic activity on nidogen-1 and nidogen-2. Int J Cancer. 2013;133(10):2315–2324. doi: 10.1002/ijc.28271. [DOI] [PubMed] [Google Scholar]
- 25.Du W, Wang S, Zhou Q, et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32(28):3319–3328. doi: 10.1038/onc.2012.359. [DOI] [PubMed] [Google Scholar]
- 26.Lo PH, Lung HL, Cheung AK, et al. Extracellular protease ADAMTS9 suppresses esophageal and nasopharyngeal carcinoma tumor formation by inhibiting angiogenesis. Cancer Res. 2010;70(13):5567–5576. doi: 10.1158/0008-5472.CAN-09-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter S, Span PN, Sweep FC, et al. ADAMTS8 and ADAMTS15 expression predicts survival in human breast carcinoma. Int J Cancer. 2006;118(5):1241–1247. doi: 10.1002/ijc.21476. [DOI] [PubMed] [Google Scholar]
- 28.Jin H, Wang X, Ying J, et al. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene. 2007;26(53):7490–7498. doi: 10.1038/sj.onc.1210559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordgard SH, Johansen FE, Alnaes GI, et al. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer. 2008;47(8):680–696. doi: 10.1002/gcc.20569. [DOI] [PubMed] [Google Scholar]
- 30.Inagaki J, Takahashi K, Ogawa H, et al. ADAMTS1 inhibits lymphangiogenesis by attenuating phosphorylation of the lymphatic endothelial cell-specific VEGF receptor. Exp Cell Res. 2014;323(2):263–275. doi: 10.1016/j.yexcr.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Kelwick R, Wagstaff L, Decock J, et al. Metalloproteinase-dependent and -independent processes contribute to inhibition of breast cancer cell migration, angiogenesis and liver metastasis by a disintegrin and metalloproteinase with thrombospondin motifs-15. Int J Cancer. 2015;136(4):E14–E26. doi: 10.1002/ijc.29129. [DOI] [PubMed] [Google Scholar]
- 32.Carré GA, Couty I, Hennequet-Antier C, Govoroun MS. Gene expression profiling reveals new potential players of gonad differentiation in the chicken embryo. PLoS One. 2011;6(9):e23959. doi: 10.1371/journal.pone.0023959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gopalakrishnan K, Kumarasamy S, Abdul-Majeed S, et al. Targeted disruption of Adamts16 gene in a rat genetic model of hypertension. Proc Natl Acad Sci U S A. 2012;109(50):20555–20559. doi: 10.1073/pnas.1211290109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown HM, Russell DL. Blood and lymphatic vasculature in the ovary: development, function and disease. Hum Reprod Update. 2014;20(1):29–39. doi: 10.1093/humupd/dmt049. [DOI] [PubMed] [Google Scholar]
- 35.Stone RL, Nick AM, McNeish IA, et al. Paraneoplastic thrombocytosis in ovarian cancer [published correction appears in N Engl J Med. 2012;367(18):1768] N Engl J Med. 2012;366(7):610–618. doi: 10.1056/NEJMoa1110352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer [published correction appears in Nature. 2013;502(7470): 258] Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res. 1989;181(2):305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275(31):23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.