Abstract
The photoactivated toxin, cercosporin, produced by Cercospora species, plays an important role in pathogenesis of this fungus to host plants. Cercosporin has almost universal toxicity to cells due to its production of reactive oxygen species including singlet oxygen. For that reason, Cercospora species, which are highly resistant to their own toxin, are good candidates to identify genes for resistance to cercosporin and to the reactive oxygen species it produces. In previous research, the zinc cluster transcription factor CRG1 (cercosporin resistance gene 1) was found to be crucial for Cercospora species’ resistance against cercosporin, and subtractive hybridization analysis identified 185 genes differentially expressed between Cercospora nicotianae wild type (wt) and a crg1 mutant. The focus of this work was to identify and characterize the hypothetical proteins that were identified in the Cercospora nicotianae subtractive library as potential resistance factors. Quantitative RT-PCR analysis of the 20 genes encoding hypothetical proteins showed that two, 24cF and 71cR, were induced under conditions of cercosporin toxicity, suggesting a role in resistance. Transformation and expression of 24cF and 71cR in the cercosporin-sensitive fungus, Neurospora crassa, showed that 71cR provided increased resistance to cercosporin toxicity, whereas no significant increase was observed in 24cF transformants. Gene disruption was used to generate C. nicotianae 71cR mutants; these mutants did not differ from wt C. nicotianae in cercosporin resistance or production. Quantitative RT-PCR analysis showed induction of other resistance genes in the 71cR mutant that may compensate for the loss of 71cR. Analysis of 71cR conserved domains and secondary and tertiary structure identify the protein as having an NTF2-like superfamily DUF1348 domain with unknown function, to be intracellular and localized in the cytosol, and to have similarities to proteins in the steroid delta-isomerase family.
Introduction
The purpose of this work is to identify genes encoding hypothetical proteins that may play a role in resistance of Cercospora fungi to the photoactivated toxin cercosporin produced by these fungi for infection of host plants. Cercospora species cause leaf spot and blight diseases on many major crops including corn, soybean, sugar beet, and coffee, leading to significant crop losses world-wide. One of the reasons for the success of these pathogens is their production of cercosporin, a photoactivated perylenequinone toxin [1]. Mutants deficient in cercosporin production are significantly less virulent on their host plants, thus engineering cercosporin-resistant crop plants via expression of cercosporin resistance genes may be an ecologically friendly method for controlling these damaging diseases.
Our work has focused on identifying cercosporin-autoresistance genes from Cercospora species, as cercosporin is almost universally toxic to cells due to its production of reactive oxygen species (ROS) [2]. In the light, cercosporin is converted to an energetically activated triplet state, which reacts with oxygen, generating ROS including singlet oxygen (1O2), a highly toxic species of ROS. Unlike free-radical forms of ROS that are components of cellular metabolism and for which defense mechanisms are understood, cellular resistance to 1O2 is not well characterized [2], and cercosporin is toxic to mice, bacteria, and many fungi in addition to host and non-host plants. During disease development, production of 1O2 and other ROS leads to peroxidation of cell membrane lipids in host plants and can also damage nucleic acids, proteins and lipids in the target cells [1]. Cercospora fungi are immune to cercosporin toxicity, thus they may be a source of genes for engineering crop resistance [3], and also serve as a model for understanding cellular resistance to 1O2.
Characterization of C. nicotianae mutants selected for sensitivity to cercosporin [4,5] led to the discovery of the zinc cluster transcription factor CRG1 (cercosporin resistance gene 1) [6] required for autoresistance to cercosporin. To identify putative resistance genes, a subtractive cDNA library was generated between the C. nicotianae wild type and a crg1 mutant [7]. From the library, 185 differentially regulated expressed sequence tags (ESTs) were found that are candidate resistance genes. The ESTs were classified into functional categories based on their homology to known sequences. These functional categories include ones known to be involved in cercosporin resistance including reductases [8], antioxidants and quenchers of ROS [9], and membrane transporters [10–13]. Several of the library genes have been characterized for their role in cercosporin resistance. For example, two genes encoding transporters in the library, CnATR1 and CnCFP, were determined to have an important role in resistance [10,12] as disruption of either of these genes in Cercospora species caused the disruptant strains to be sensitive to cercosporin. In addition, tobacco transformed to express CFP showed significantly reduced lesion size after inoculation with C. nicotianae, confirming a role in resistance and suggesting that CFP might be useful for engineering plants for disease resistance [14]. In the case of ROS quenchers, vitamin B6 was shown to quench 1O2 and be involved in defense against cercosporin [9,15]. Studies to engineer tobacco to constitutively express C. nicotianae B6 biosynthetic genes (PDX1 and PDX2), however, resulted in no statistically significant increase in levels of the B6 vitamers, due to heavy regulation of these genes in plants [16]. Although antioxidants have been implicated in cercosporin resistance [2], a recent study of a gene encoding glutathione S-transferase from the library failed to demonstrate a link with cercosporin resistance [17].
In addition to genes that were categorized into defined functional categories, the library contains genes encoding hypothetical proteins whose functions are not yet characterized. Most studies of cellular resistance to 1O2 have been done with photosynthetic organisms, as 1O2 is a byproduct of photosynthesis [2]. In Rhodobacter sphaeroides, a model for studies of 1O2 resistance, resistance is associated with protein synthesis and turnover, amino acid metabolism, and glutathione-dependent and–independent detoxification pathways, among others [2]. A recent study comparing putative resistance genes from our Cercospora subtractive library with orthologs identified in transcriptional studies of Rhodobacter 1O2 resistance, however, showed little commonality between the two species [17]. Thus Cercospora fungi must have unique mechanisms not common to photosynthetic organisms. We thus chose to investigate the library genes encoding hypothetical proteins as possible novel sources of cercosporin and 1O2 resistance.
Materials and Methods
Culture conditions, fungal strains, and plasmids
Potato dextrose agar (PDA; Difco, Sparks, MD) was used for growth and maintenance of all C. nicotianae strains including the wild type (wt) strain ATCC18366. Neurospora crassa strains including the wt strain ORS-6a (Fungal Genetics Stock Center) were grown on Vogel’s medium [18].
DNA plasmid isolation, cloning, and ligation used standard molecular techniques [19]. iProof High-Fidelity DNA Polymerase from Bio-Rad Laboratories and OneTaq® DNA Polymerase from New England Biosciences were used along with the gene-specific primers (Tables 1 and 2) for standard PCR work. The plasmid pGEM-T Easy (Promega, Madison, WI) was used for cloning and sequencing. Plasmid pCB1636 [20] and pTxA-1 [12], were used for cloning, and fungal transformation. Escherichia coli strain DH5-α was used to maintain all plasmids.
Table 1. Primer sets used for RT-qPCR analysis for screening C. nicotianae hypothetical protein genes for induction under cercosporin toxicity conditions.
| Gene | Forward Primer | Reverse Primer | Efficiency 1 |
|---|---|---|---|
| 11sf | AGTGGCTAATCTGCTCTGG | CACTGCTATTCCTGTTGG | 100.08 |
| 15cF | ACATGCCGCACTCTGTTCATTTGG | GAAGCCCACAACCTGCAACCATTT | 104.48 |
| 1cF | CCGTTCCTGCAGAGCTCAAG | GCCATTGACCATGTTGAGCATGTC | 109.33 |
| 200sF | TGCTGATCCGCTAAATGCTAAAAC | GCCGATGGACAAGGGTATAAGATC | 100.21 |
| 207sF | ATGCTCGATCCTCCGAACCA | CGGCAGCTTTGAGCGTCTTT | 101.78 |
| 214sR3 | ATCAGAAGAGACAGCATAAAGC | AAGTGGTGGCTCAGCGTGG | 89.57 |
| 24cF | TGCTTTCACCTTCAAGTTCGAC | TGCCCTTGCCAAAGCTAGG | 106.51 |
| 40cR | TTGGTCCATCCCTGATCCTGTTGT | TCAGACTCAGCGAGCGAAGGATTT | 99.25 |
| 55cR | GGAGACAGCCAAGCAAGAAGTC | GGGAAGAAGGCGATTGAGGA | 100.16 |
| 56cR | ATCGTTCAAGACCGAGAGGCTCAA | ATGCCGAGAGATCAATGTCCCGAT | 105.38 |
| 71cR | TCAAGCCACCCTACAATGCCTCAA | TTATTTGGTCGGTGCCTTGGACGA | 100.09 |
| 77sR3 | TGAGTGGTCGCTTGATTCG | ATCTGGACCCGAAATCGTGC | 97.62 |
| 84sF | AGTGGGAGTGGGAGTCTTGG | AACACTGGAGAACGAATCAACG | 91.29 |
| Actin | TGACGATGCGCCACGAGCTGT | TTGATTGGAGCCTCGGTGAGC | 101.20 |
1 Calculated efficiencies for primers for RT-qPCR analysis
Table 2. Primer sequences for gene cloning and for screening transformants 1 .
| Purpose | Primer Name | Sequence |
|---|---|---|
| ToxA plasmid cloning | 71cR-F-EcoRI | TTTAATGAATTCAGTGAACACGAACGACTAGGATG |
| ToxA plasmid cloning | 71cR-R-HindIII | TTTAATAAGCTTTCCACGATACGAACTAATGCTCACC |
| ToxA plasmid cloning | 24cF-F-EcoRI | TTTAATGAATTCTCGTAACATCGTTGGGTCAG |
| ToxA plasmid cloning | 24cF-R-HindIII | TTTAATAAGCTTTCGTAACATCGTTGGGTCAG |
| N. crassa transformant screening | 71cR-F | GAGGAGGAGAGGTGGTTCAAGGA |
| N. crassa transformant screening | 24cF-F | TTCCAATCTACGTTTTCGACCCTG |
| N. crassa transformant screening | Hyg-R | TGTCGGGCGTACACAAATCG |
| N. crassa transformant screening | ToxA-Rev1 | ATAAAGGGCTAAGGTGTCCGTCC |
| N. crassa transformant screening | 71cR-R | TTATTTGGTCGGTGCCTTGGACGA |
| N. crassa transformant screening | 24cF-R | ATCAACTGGTAGGCGACTGTGAC |
| 71cR disruption construct | 71cF-5'-F ApaI | attaatGGGCCCTGCTCGTCATCTTGCTCATCG |
| 71cR disruption construct | 71cF-5'-R ApaI | attaatGGGCCCAGAGACATTTTGAGATGGAATTCG |
| 71cR disruption construct | Hyg-split3S | CGTTGCAAGACCTGCCTGAA |
| 71cR disruption construct | Hyg-split5A | GGATGCCTCCGCTCGAAGTA |
| 71cR disruption construct | 71cf-3'-R Sac1 | attaatGAGCTCCAAGTCAAATCCAC |
| 71cR disruption construct | 71cF-3'-F Sac1 | attaatGAGCTCGCCACCCTACAATGCCTCAAC |
| 71cR disruption construct | 71cF-5'-F | TGCTCGTCATCTTGCTCATCG |
| 71cR disruption construct | 71cf-3'-R | GAGCTCCAAGTCAAATCCAC |
| C. nicotianae disruption screening | 71cR-Rev3 | TCTTCCACTTCATCCCATCATTCTTA |
| C. nicotianae disruption screening | 71cR-inv5’ | CCTAGCATCTCAATCTCACCAACTAAC |
| C. nicotianae disruption screening | 71cR-Forw10 | CTAGATGAGACGACGCCTGATC |
| C. nicotianae disruption screening | Hyg-F2 | TGAACCATCTTGTCAAACGACAC |
| C. nicotianae disruption screening | HygR | TGTCGGGCGTACACAAATCG |
| C. nicotianae disruption screening | 71cR-DisR | GATGAAGTCGAGAGCACAACAAG |
1 Bold and underlined sequences represent restriction enzyme targets
Domains of hypothetical proteins in the library
The EST sequences recovered in the subtractive library between C. nicotianae wt and the crg1 mutant [6] were used for finding orthologs of these EST sequences with full length sequence information using tBLASTx analysis against the NCBI (the US National Center for Biotechnology Information) protein database (11/21/2013 update). The full-length gene sequences of the orthologs were then used to find conserved domains using the Conserved Domain Database by NCBI.
Gene expression under conditions of cercosporin toxicity
A CnATR1 (atr1)-disrupted C. nicotianae mutant [12] and two 71cR-disrupted C. nicotianae mutants (#16 and 18) were used to quantify gene expression under conditions of cercosporin toxicity as previously described [13]. RT-qPCR reactions were carried out as described [13]. The primers used to amplify each gene are shown in Table 1. Each sample was normalized against the C. nicotianae-specific actin reference gene, and fold-change relative to no-cercosporin (acetone control) was calculated according to the 2−ΔΔCT method [21–23].
Sequencing of full length gene sequences of 24cR and 71cR
Previously described methods [13] were used to obtain the full-length C. nicotianae genomic sequences of the two hypothetical protein genes, 71cR and 24cF, based on their partial EST sequences from the subtractive library [7]. Nucleotide and amino acid sequences of 71cR and 24cF are in the GenBank database with the accession numbers KJ126714 and KJ126715, respectively.
Expression of hypothetical genes in N. crassa
The fungal transformation plasmid pTxA-1 [12], containing a hygromycin B (Hyg) resistance cassette and an ampicillin (Amp) cassette for selection in fungi and E. coli, respectively, was used to insert the two hypothetical protein genes, 71cR and 24cF, under the control of the fungal constitutive promoter ToxA from Pyrenophora tritici-repentis. Full-length copies of 71cR and 24cF were amplified, respectively, using primers 71cR-F-EcoRI and 71cR-R-HindIII (containing EcoRI and HindIII sites, respectively) for 71cR amplification (512 bp) and 24cF-F-EcoRI and 24cF-R-HindIII (containing EcoRI and HindIII sites, respectively) for 24cF amplification (1224 bp) (Table 2). PCR fragments were digested with EcoRI and HindIII, and ligated to the pTxA-1. The resulting plasmids, pTxA -71cR and pTxA -24cF, were sequenced to confirm the presence of an intact 71cR or 24cF sequence.
The two plasmids were transformed into N. crassa as previously described [17]. Transformation and presence of 71cR or 24cF was confirmed in hyg-resistant colonies by PCR screening using gene specific primers (ToxA-Rev1 and 71cR-R, ToxA-Rev1 and 24cF-R, HYG-R and 71cR-F, HYG-R and 24cF-F) (Table 2). An NaOH DNA extraction method [24] was used to extract gDNA for PCR analysis.
Screening of N. crassa 71cR and 24cF transformants for cercosporin resistance
N. crassa 71cR and 24cF transformants were tested for cercosporin resistance as previously described [17]. Percent growth on medium containing 10 μM cercosporin was calculated relative to the acetone control on the same plate by measuring radial growth of colonies at 21 hours after inoculation. Total RNA was extracted from 4 randomly chosen transformants for gene expression analysis, and RT-qPCR analysis was conducted as previously described [17]. The N. crassa-specific tubulin control was used as the reference gene.
Disruption of 71cr in C. nicotianae
The 71cR gene was disrupted in wt C. nicotianae using a split-marker recombination technique previously described [25]. The 5’ (from 1517 bp upstream to 5 bp downstream of the start codon [1.5 kb]) and 3’ (from 443bp upstream to 43 bp downstream of the stop codon [0.5 kb]) 71cR sequences were amplified from C. nicotianae gDNA by PCR with 71cR-specific primers containing restriction enzyme linkers (Table 2). The two primer sets used to amplify the 5’ and 3’ ends of 71cR were: 71cR-5’-F-ApaI, 71cR-5’-R-ApaI and 71cR-3’-F-SacI, 71cR-3’-R-SacI. The PCR products were digested with the appropriate restriction enzymes (71cR-5’ digested with ApaI; 71cR-3’ digested with SacI) for cloning into the receptor plasmid pCB1636 [20]. Plasmid pCB1636 was first digested with ApaI and ligated with ApaI digested 71cR-5’. Recombinant plasmids isolated from this step were selected based on the insert’s correct orientation, and subsequently digested with SacI and ligated with SacI digested 71cR-3’ to obtain a 71cR disruption construct. Using this construct as a template, two different overlapping PCR fragments were amplified using primers specific to the 71cR sequence and the HygR cassette (split marker 1: 71cR-5’-F and HYG-split 5A [2.6 kb]; split marker 2: 71cR-3’-R and HYG-split 3S [1.3kb]) (Table 2). The identity of each of the split marker PCR fragments was confirmed by sequencing.
The methods used for isolation and transformation of the protoplasts of C. nicotianae were as previously described [13]. After transformation, colonies were transferred at least 5 times to PDA amended with 125 μg/ml hyg to ensure stability of transformation. 71cR disruption was confirmed by PCR analysis using primer sequences shown in Table 2.
Screening of 71cR disruption mutants for cercosporin sensitivity and production
71cR disruptants confirmed with the PCR analysis described above were screened for cercosporin sensitivity as previously described [13]. Data shown are the result of two independent experiments with 4 replications each. Cercosporin production by each disruptant was measured at 480 nm as previously described [13,26].
Phylogenetic analysis and species distribution of 71cR orthologs
Phylogenetic analysis of protein sequences was done with representatives from the DUF1348 family and each of the characterized families within the NTF2-like superfamily. The conserved NTF2-like domain from each sequence was identified using the Conserved Domain Database, and the domain sequences were aligned using the MUSCLE [27] algorithm in Mesquite v3.0.1 [28]. The protein substitution model WAG+G+F [29] was selected using ModelGenerator v0.85 [30]. The alignment was analyzed using maximum likelihood with raxmlGUI v. 1.3.1 [31] with no outgroup, using bootstrap analysis with the autoMRE function.
Blastp and tblastn searches were performed on the NCBI database (11/21/2013 update) to identify 130 of the closest orthologs of 71cR. The Cercospora canescens whole genome sequence (Accession number: ANSM00000000) was downloaded from NCBI, and tblastn in BLAST+ [32] was used to identify its 71cR ortholog (S1 Table). To further characterize the species distribution of 71cR orthologs, tblastn searches were done of the 71cR protein sequence against filtered model transcripts of each fungal genome available on the JGI Mycocosm portal [33,34].
Secondary and tertiary structure prediction of 71cR
The secondary and tertiary structure of the 71cR amino acid sequence was analyzed using Raptor software (8/12/2015 update) [35]. Raptor software was also used for the ligand binding predictions. The Dali server (2/20/2014 update) [36] was used to find orthologs based on the tertiary structure similarities. The crystal structure of 71cR is not available, thus the crystal structure of the protein PFL_3262 from Pseudomonas fluorescens (PDB ID: 2IMJ) (the closest ortholog with a crystal structure), was used for the Dali analysis. Predict Protein server [37] was used to identify transmembrane helices or disulphide bridges and localization predictions for 71cR.
Conserved regions of 71cR were found by aligning 71cR with the closest 130 orthologs in S1 Table, using MUSCLE in Mesquite as described previously, and Chimera 1.10.1 [38] was used to display the degree of conservation for each residue within the conserved domain. These conserved residues were then compared with the ligand binding predictions from Raptor software.
Assays for cercosporin modification or degradation
To test the activity of the 71cR protein to degrade or modify cercosporin, two methods were used. To assay for possible degradation of cercosporin, 10 mg of freshly growing mycelia of wt N. crassa and two 71cR N. crassa transformants (# 379, 380) were collected and grown in 20 ml of PDB medium for 5 days in the dark at room temperature. Cercosporin was added to final concentration of 10 μM and incubated in the dark for 5 hours. To extract cercosporin from the mycelium, it was filtered and ground in liquid nitrogen, and cercosporin was extracted in acetone. The amount of cercosporin in the growth medium and in the mycelial extracts was analyzed by measuring the absorbance at 460 nm using a spectrophotometer. To assay for possible modification of cercosporin, mycelium of wt N. crassa and 71cR transformants #379 and 380 was grown in PDB, and cultures were incubated for 5 hours with cercosporin at a final concentration of 10 μM. We imaged the mycelia using Zeiss LSM 710 confocal microscopy with a 488 nm argon laser reflecting off a 488 nm main beam splitter. A series of images with the same field of view at different emission wavelengths (λ-stack) was captured between 504 and 728 nm in 10 nm intervals. Images were analyzed offline using Zeiss ZEN software to generate the fluorescence emission spectra for the hyphae.
Results
Identification of genes encoding hypothetical proteins and domain analysis
To identify genes encoding resistance to the toxin cercosporin and to 1O2, we have been characterizing genes in a subtractive library [7] between the C. nicotianae cercosporin-resistant wt and the sensitive crg1 mutant deficient for the CRG1 transcription factor [6]. In previous studies we characterized genes encoding membrane transporters [12,13] as well as enzymes that generate reducing power [17], as both toxin transport and toxin reduction have been identified as important mechanisms of cercosporin resistance [2]. Our focus in this study is on genes encoding hypothetical proteins, as resistance to 1O2 is poorly understood. We screened 13 ESTs from the library that are homologous to genes encoding hypothetical proteins. A genome sequence for C. nicotianae is not available. Therefore, tBLASTx searches of the EST sequences were performed against NCBI’s protein database to identify the closest orthologs. Functional domains of these orthologs were then identified using NCBI’s conserved domain database [39]. With the exception of 56cR, 40cR, and 200sF EST sequences, all EST sequences were aligned with their orthologs with high similarity (p-value of e-10 or lower).
The functional domains of the orthologs of the hypothetical EST sequences are listed in Table 3. No functional domains were found for the orthologs of 40cR or 77sR3 EST sequences with the p-value threshold of e-5. Orthologs of the remaining 11 hypothetical proteins had domains with characterized functions, some of which are consistent with previously identified mechanisms of resistance to cercosporin or ROS damage including reducing activity, amino acid turn-over, and transporters [2, 8, 12, 13, 14]. ESTs with these functions include 56cR, which has a hydrolase domain, 1cF and 200sF with domains involved, respectively, in tryptophan and tyrosine biosynthesis, and 15cF that has a permease transporter domain. Other identified domains include a phospholipid methyltransferase domain (24cR and 84cR), a peptidoglycan binding domain (11sf), domains for proteins triggering apoptosis (55cR), and ones involved in signal transduction (207sF).
Table 3. Conserved domains of the orthologs of hypothetical proteins in the subtractive library.
| EST 1 | Domain Name 2 | Accession 3 | Description 4 | Interval 5 | E-value 6 |
|---|---|---|---|---|---|
| 11sF | LysM | cd00118 | Lysine Motif | 130–258 | 6.18E-11 |
| LytE | COG1388 | FOG: LysM repeat (found in outer membranes) | 382–657 | 1.11E-08 | |
| FAM53 super family | cl21101 | Family of FAM53 | 170–514 | 4.44E-05 | |
| 1cF | RHOD | cd00158 | Rhodanese Homology Domain | 1057–1404 | 3.08E-09 |
| Trp-synth-beta_II super family | cl00342 | Tryptophan synthase beta superfamily (fold type II) | 85–969 | 9.42E-40 | |
| 56cR | Abhydrolase_6 | pfam12697 | Alpha/beta hydrolase family | 241–1038 | 4.14E-16 |
| TT_ORF1 super family | cl20238 | Torque teno viral orf 1 | 797–916 | 2.67E-05 | |
| 15cF | RhaT | COG0697 | Permeases of the drug/metabolite transporter (DMT) superfamily | 461–1243 | 2.35E-14 |
| 24cF | PEMT super family | cl21511 | Phospholipid methyltransferase | 85–675 | 6.50E-38 |
| 55cR | EI24 | pfam07264 | Etoposide-induced protein 2.4 | 247–807 | 3.53E-10 |
| 71cR | NTF2_like super family | cl09109 | Nuclear transport factor 2 (NTF2-like) superfamily | 90–455 | 2.19E-54 |
| 84cR | PEMT super family | cl21511 | Phospholipid methyltransferase | 163–1020 | 1.05E-44 |
| 200sF | Tyrosinase | pfam00264 | Common central domain of tyrosinase | 295–984 | 2.98E-71 |
| 207sF | LDB19 super family | cl15231 | Arrestin N terminal like family | 578–1117 | 9.96E-39 |
| 214sR3 | R3H-assoc | pfam13902 | R3H-associated N-terminal domain | 247–582 | 1.63E-34 |
| 40cR | no domains | ||||
| 77sR3 | no domains |
1 The library names of the EST sequences in the subtractive library [7]; F = forward library (down-regulated in crg1 mutant); R = reverse library (upregulated in crg1 mutant)
2 The domain name given in The Conserved Domain Database in the NCBI website [39]
3 Accession numbers of the conserved domains
4 A brief description of the domains
5 The positions within the C. nicotianae genes that the domains are aligning.
6 E-value indicates statistical significance for the similarity between the hypothetical protein sequence and the sequence of the characterized domain.
Expression of hypothetical genes under conditions of cercosporin toxicity
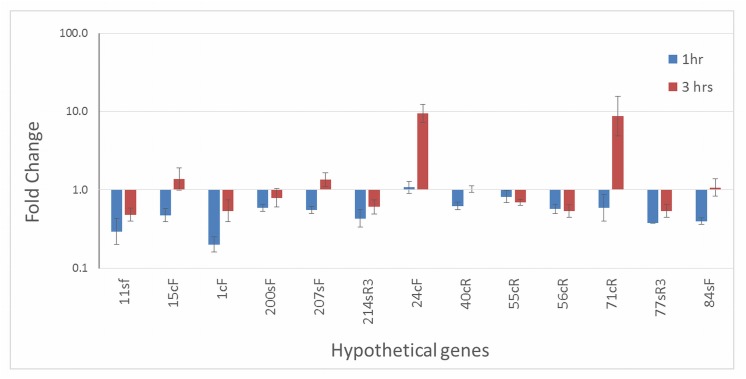
In order to define a possible role in cercosporin resistance, the 13 genes encoding hypothetical proteins were assayed for changes in expression under conditions of cercosporin toxicity. The cercosporin-sensitive C. nicotianae atr1 mutant, deficient in the ATR1 ABC transporter involved in cercosporin resistance [12], was treated with cercosporin under high-light conditions to induce toxicity, and gene expression was assayed by RT-qPCR. This mutant was used because it is highly sensitive to cercosporin and has previously been shown to be useful for identifying putative cercosporin-resistance genes that are upregulated under cercosporin toxicity conditions [13]. Genes 24cF and 71cR were upregulated 9.4- and 8.7-fold, respectively, three hours after cercosporin toxicity was induced (Fig 1). None of the other 11 genes were upregulated under these conditions. Based on these gene expression results, 24cF and 71cR were chosen for further characterization.
Fig 1. Quantitative RT-PCR analysis of gene expression of hypothetical protein genes in the C. nicotianae atr1 mutant treated with cercosporin in the light.
Each sample was normalized against the actin reference gene, and fold-change relative to no-cercosporin was calculated according to the 2(-ΔΔC(T)) method [21]. Data represent the mean of two independent experiments with three replications each. Error bars represent 95% confidence intervals.
Ability of 24cF and 71cR to impart resistance in N. crassa
The cercosporin-sensitive fungus N. crassa was transformed with 24cF and 71cR to test the ability of these genes to impart cercosporin resistance. N. crassa was chosen for this assay as it is highly sensitive to cercosporin, easily transformable, and has been shown in previous studies as an excellent system for screening genes that impart cercosporin resistance [13, 17]. Hyg-resistant colonies after transformation were screened for the presence of the genes by PCR. Out of a total of 84 and 93 hyg-resistant colonies, respectively, of 71cR and 24cF transformants, 60 and 12, respectively, were confirmed to contain the intact transgene by PCR screening (data not shown). All 12 of the 24cF and 27 of the 71cR PCR-positive strains were assayed for resistance to cercosporin by measuring radial growth on cercosporin-containing medium relative to growth on control medium. Cercosporin at 10 μM inhibits radial growth of N. crassa, resulting in approximately 30% of the radial growth on medium lacking cercosporin (Fig 2 and S1 Fig). Of the randomly chosen 27 PCR-positive 71cR transformants tested, eleven were found to be significantly more resistant to cercosporin than wt (P < 0.05) (Fig 2). However, none of the 12 PCR-positive 24cF transformants tested had significantly greater resistance to cercosporin than wt (P < 0.05) (Fig 2).
Fig 2. Cercosporin resistance of Neurospora crassa 71cR-transformed strains.
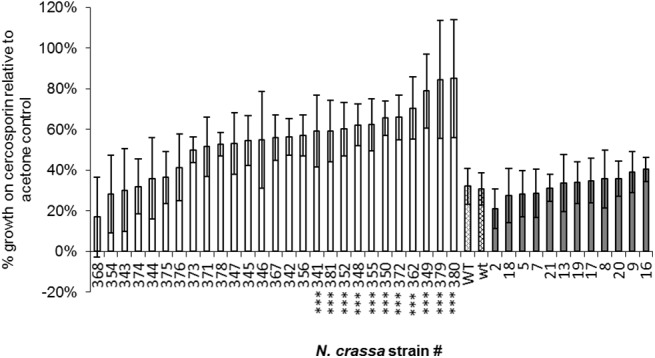
White bars: 71cR transformed strains; patterned bars: wild type N. crassa (WT = 71cR control; wt = 24cF control); grey bars: 24cF-transformed strains. Data are the result of two independent experiments with 5 replications each. Strains marked with *** have significantly greater resistance than wild type (P < 0.05). Error bars represent 95% confidence intervals.
Expression of the transgenes was assayed in selected transformants screened for resistance. RT-qPCR analysis of 71cR expression in the two resistant 71cR transformants assayed (#349 and 350) showed high levels of expression (15,973- and 9,006- fold increase respectively), compared to the lowest expresser, a non-resistant 71cR transformant (#344) (Fig 3). Expression of the other non-resistant 71cR transformant was much lower than the two resistant 71cR transformants (199-fold change increase relative to #344). Thus 71cR expression correlated with cercosporin resistance. For 24cF, all four transformants tested had similar CT values as the high-expressing 71cR transformants, indicating expression of 24cF, and there was little difference in expression between the four 24cF transformants (data not shown). From these results we concluded that the 71cR protein can provide resistance to cercosporin toxicity, but that 24cF cannot.
Fig 3. Quantitative RT-PCR analysis of gene expression of the 71cR gene in selected Neurospora crassa transformants.
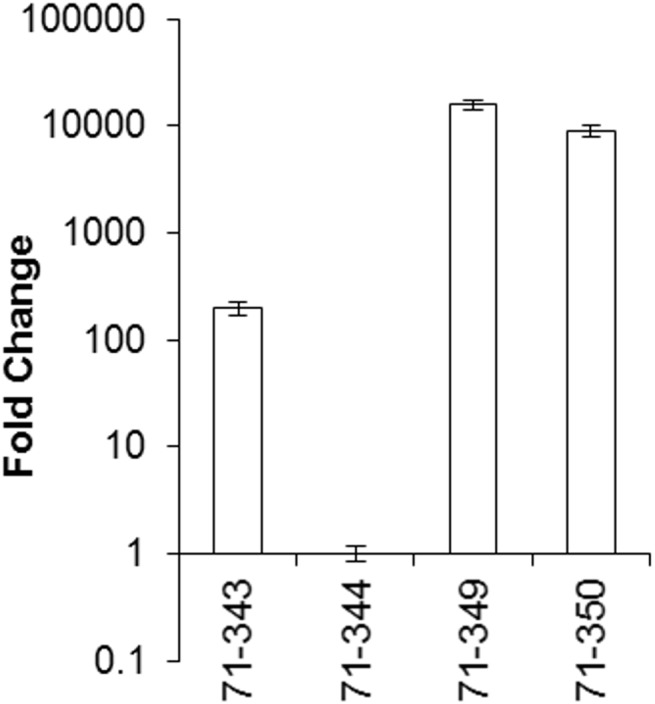
Each sample was normalized against the tubulin reference gene, and fold-change relative to expression of the lowest expressing Neurospora crassa transformant was calculated according to the 2(-ΔΔC(T)) method. Error bars represent 95% confidence intervals.
Disruption of 71cR in C. nicotianae and phenotype of disruption mutants
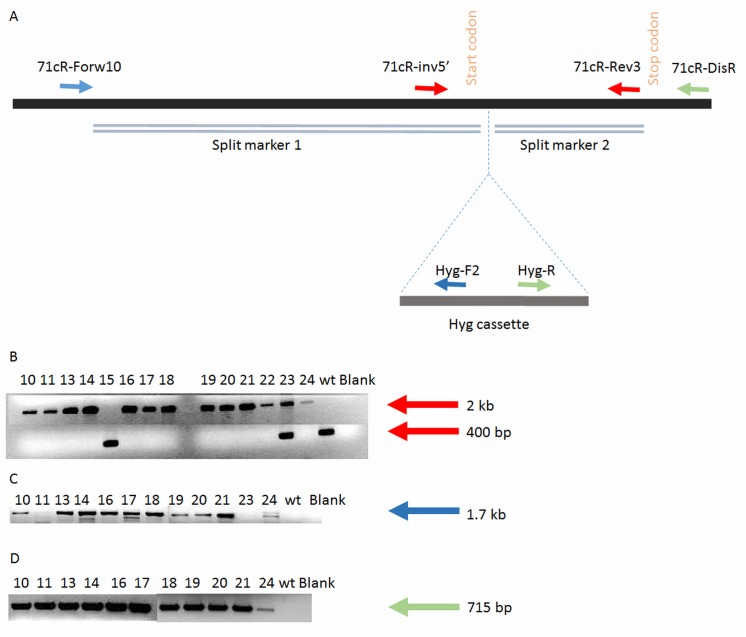
A split marker strategy was used to generate C. nicotianae disruption mutants by homologous recombination. A total of 14 hyg-resistant colonies were screened to confirm 71cR disruption by PCR analysis using primer sequences shown in Table 2. Primers 71cR-Rev3 and 71cR-inv5’ that span the hyg-resistance cassette were used to screen the disruptants; with these primers the wt band is 400 bp and the band resulting from disruption is 2 kb (Fig 4A and 4B). A total of 12 transformants were confirmed as lacking the 400 bp wild type band. One had an ectopic integration of the split markers (#23) and one lacked evidence for integration of the split marker (#15) (Fig 4B). Eleven of the disruptants and the strain with ectopic integration of the split markers (#23) were then screened using a 71cR locus-specific primer for the 5’ end outside the split marker sequence (71cR-Forw10) and a primer specific to hyg cassette sequence (Hyg-F2). Presence of the 1.7 kb fragment confirmed the correct location of split marker integration for 10 of the putative disruptants, and no band was seen on amplification of strain #23 (transformant with ectopic integration of the split markers) (Fig 4C). Finally, we screened transformants using a 71cR locus-specific primer for the 3’ end outside the split marker sequence (71cR-DisR) and a second primer specific to the hyg cassette sequence (Hyg-R) (Fig 4D); all of the transformants were also confirmed to have a 715 bp band again confirming the correct location of the split marker integration. Through this analysis, 10 transformants (# 10, 13, 14, 16, 17, 18, 19, 20, 21, 24) were confirmed as being disrupted for 71cR.
Fig 4. Disruption of 71cR.
A. The gDNA of wt C. nicotianae is shown as a black bar with the start and stop codons of 71cR indicated. In disrupted transformants the hyg cassette is predicted to integrate in the location specified by dotted lines. The homologous regions of each split marker are represented as double lines. Location of primers used in PCR screening is also shown. B. Gel image of screening putative 71cR disruptants with primers that span the hyg cassette integration site (71cR-Rev3 and 71cR-inv5’; red arrows in panel A). Integration results in a 2 kb band; wt band is 400bp (indication of non-disruptant genotype). Wild type C. nicotianae and transformant #15 showed only the wild type band; transformant #23 has both bands indicating an ectopic integration of the split markers. C. Bands represent the 1.7 kb amplification of the 5’ integration site by using a 71cR locus-specific forward primer outside the split marker 1 sequence (71cR-Forw10) and a reverse primer specific to the hyg cassette sequence (Hyg-F2) (blue arrows in panel A). D. Bands represent the 715 bp amplification of the 3’ integration site (Hyg-R and 71cR-DisR; green arrows in panel A).
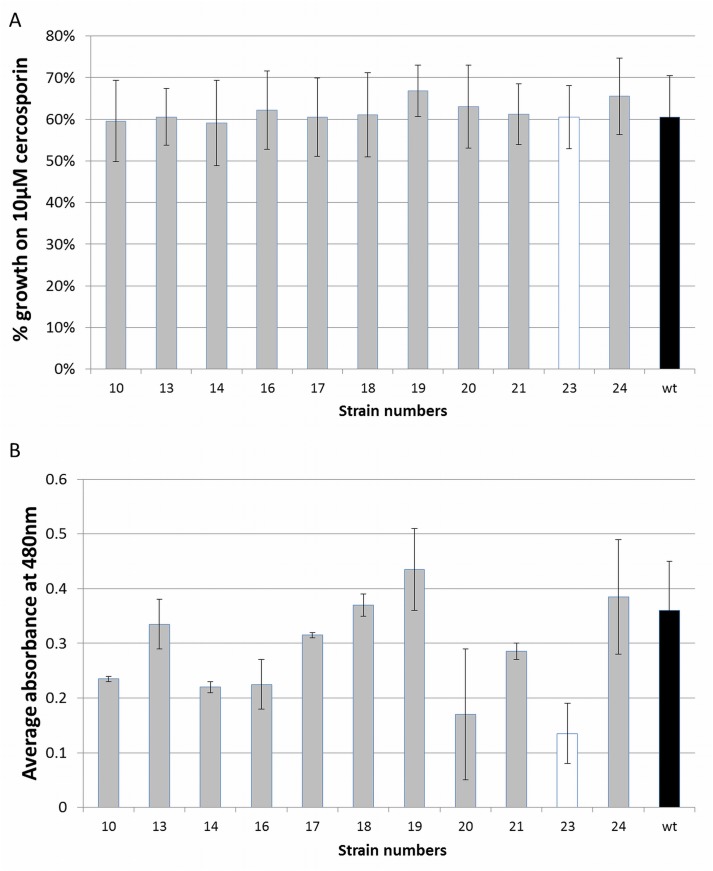
The ten 71cR-disruption strains were tested for cercosporin sensitivity by growing them on cercosporin-containing medium in the light. The wt, a transformed but non-disrupted strain (#23), and the cercosporin-sensitive crg1 mutant [6] were used as controls. Results are shown in Fig 5A. Under the conditions of the assay, the crg1 mutant does not grow on cercosporin (0% growth relative to the control [not shown]). There was no statistically significant difference in cercosporin resistance between the wt, the non-disrupted transformant (#23), and 71cR disruptants in radial growth on cercosporin. As other resistance proteins such as ATR1 and CFP have shown a dual role in both cercosporin resistance and production [10, 12], 71cR disruptants were also assayed for cercosporin production (Fig 5B) as compared to wt and the non-disrupted transformant #23. Cercosporin production varied between the different strains. However none of the 71cR disruptants produced significantly less cercosporin than wt.
Fig 5. Phenotypic characterization of the 71cR disruptants.
A. Cercosporin resistance of Cercospora nicotianae wild type (black bar), 71cR-disruptant strains (grey bars) and 71cR-transformed, but non-disruptant (white bar). The cercosporin sensitive crg1 mutant had 0% growth on cercosporin relative to control growth (not shown). Data are the result of two independent experiments with 4 replications each. Error bars represent standard error. B. Cercosporin production by Cercospora nicotianae wild type (black bar), 71cR-disruptant strains (grey bars) and 71cR-transformed, but non-disruptant strain (white bar). Cercosporin production was measured by extraction of mycelium with 5N KOH and measuring absorbance at 480 nm. Data are the results of two independent experiments with 2 replications each. Error bars represent standard error.
Expression of transporters and cercosporin biosynthetic genes in 71cR disrupted mutants under cercosporin toxicity
Expression of 71cR in N. crassa demonstrated that 71cR can provide cercosporin resistance, however, 71cR disruption mutants of C. nicotianae were not more sensitive to cercosporin or produce less cercosporin than wt. Previous studies have suggested that C. nicotianae upregulates other resistance genes to compensate for resistance gene mutations [13]. We thus assayed for expression of additional library genes previously shown to impart cercosporin resistance (CnATR1, CnATR2 and CnCFP) to determine if they are up-regulated in the 71cR mutant background (Fig 6A). Two 71cR disruptants (disruptant #16 and 18) were tested. Expression of CnATR1 and CnATR2 was not increased significantly in either of the disruptants. By contrast, CnCFP expression was strongly increased: 285 and 20 fold at 1 hour, and 40- and 3- fold at 3 hours in disruptants #16 and 18, respectively.
Fig 6. Quantitative RT-PCR analysis of gene expression of genes in the C. nicotianae 71cR mutant treated with cercosporin in the light.
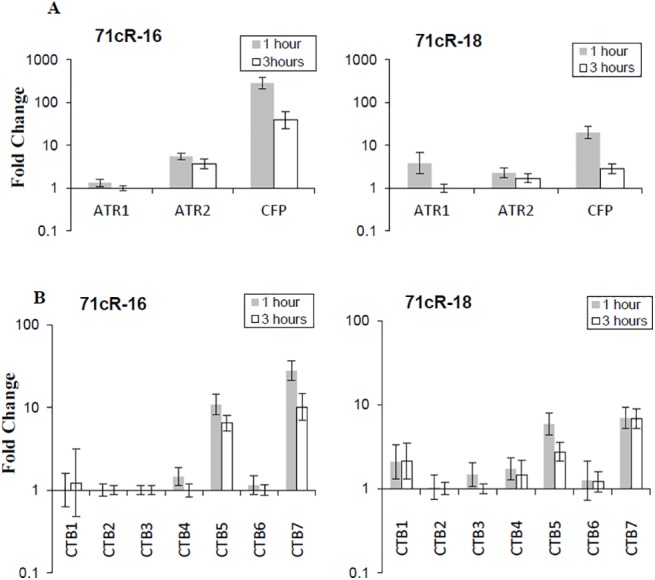
Two 71cR disrupted strains, 71cR-16 and 71cR-18, were tested. Each sample was normalized against the C. nicotianae-specific actin reference gene, and fold-change relative to the no-cercosporin control was calculated according to the 2(-ΔΔC(T)) method [21]. Data represent the mean of two independent experiments. Error bars represent 95% confidence intervals. A. Expression of 3 transporters in 71cR disruptants #16 and #18. B. Expression of CTB genes in 71cR disruptants #16 and #18.
The subtractive library of putative cercosporin resistance genes also includes two genes from the cercosporin biosynthetic pathway: CTB2 (cercosporin toxin biosynthesis gene 2), encoding an O-methyltransferase, and CTB5, encoding an O2, FAD/FMN-dependent oxidoreductase [7,40]. Previous studies to test the role of each of the 8 biosynthetic pathway (CTB) gene products did not document a role in resistance [2], however we previously documented up-regulation of two of the CTB genes in response to cercosporin toxicity in C. nicotianae mutants deficient for a membrane transporter (ATR2) shown to play a role in cercosporin resistance [13]. Thus we assayed expression of all genes in the cercosporin biosynthetic cluster in the 71cR disruption mutants under cercosporin toxicity. Two genes were significantly up-regulated: CTB5 (found in the library) as well as CTB7, encoding a second FAD/FMN-dependent oxidoreductase (Fig 6B). Expression of CTB2 and the other biosynthetic genes including the CTB4 transporter gene were not altered. Up-regulation of CTB5 and CTB7 was less than that of CFP in both 71cR disruptants induced with cercosporin toxicity. CTB5 was induced 10- and 6-fold in disruptants #16 and 18, respectively, at 1 hour, and 6- and 3-fold, respectively, at 3 hours. For CTB7, fold-increase in disruptants #16 and 18 was, respectively, 28- and 10-fold at 1 hour and 6- and 7-fold at 3 hours. Interestingly, CTB5 and CTB7 are the same two CTB genes that are upregulated in response to cercosporin toxicity in the membrane transporter atr2 mutants [13].
Phylogenetic analysis of 71cR
71cR is an intronless 457 bp sequence. Blastp and tblastn searches on the NCBI database and tblastx with BLAST+ on the Cercospora canescens genome sequence revealed high homology to hypothetical proteins from many different fungi. The closest ortholog is in Cercospora canescens (ANSM00000000.1, 92% similar in 139 amino acids), with orthologs present in a wide range of fungi (S1 Table). Orthologs with high homology include ones from Coniosporium apollinis Sterfl. (XM_007783477.1, 84% similar in 151 amino acids), Talaromyces stipitatus (XM_002479737.1, 83% similar in 153 amino acids), and Pyrenophora teres (XM_003300623.1, 81% similar in 151 amino acids). None of the 130 closest relatives to the 71cR protein from different fungal strains found by the BLASTP search (the least similar one having 68% similarity in 133 amino acids) (S1 Table) have been characterized for function.
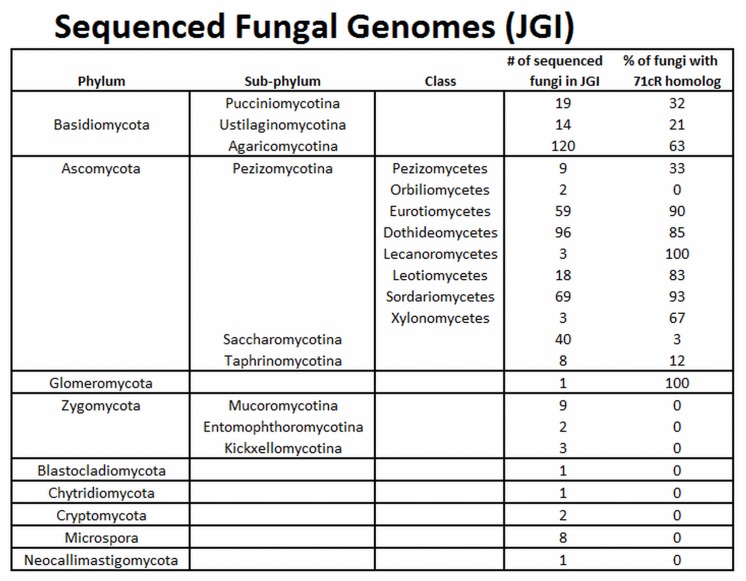
To analyze the distribution of 71cR among fungal species, orthologs of the 71cR protein sequence were identified from sequenced fungal genomes in the JGI MycoCosm portal [33,34]. With one exception, 71cR orthologs were found in fungal genomes in all classes within the Dikarya (Basidiomycota and Ascomycota), including both mycelial and yeast species (Fig 7). Distribution was not ubiquitous, however, ranging from less than 5% to 100% of sequenced genomes within a taxonomic group. 71cR orthologs were especially common within the Ascomycota sub-phylum Pezizomycotina and the Basidiomycota sub-phylum Agaricomycotina. Cercospora spp. are in class Dothideomycetes within sub-phylum Pezizomycotina, and 85% of the 96 sequenced species within this class contained 71cR orthologs. While the Cercospora canescens genome contained a close ortholog, we were unable to identify an ortholog within the Cercospora zeae-maydis genome sequence. Distribution was less or lacking outside of the Dikarya. The one sequenced genome from the Glomeromycota (Rhizophagus irregularis) also had a 71cR ortholog, but 71cR orthologs were not found from other clades traditionally grouped into Zygomycota (such as Mucoromycotina and Kickxellomycotina) or from basal fungal lineages such as Neocallimastigomycota, Chytridiomycota, and Microsporidia (Fig 7). Due to the low number of sequenced genomes from these basal fungal lineages, however, it is not possible to definitively determine whether 71cR orthologs are completely absent within these taxa.
Fig 7. Distribution of 71cR orthologs among sequenced fungal species.
For each genome sequence available in the JGI MycoCosm portal, tblastn was used to search the filtered model transcripts for 71cR orthologs. To the right of each taxon, the number of analyzed genome sequences is shown, along with the percent of genomes in the taxon for which a 71cR ortholog was identified.
Because all orthologs of 71cR are hypothetical proteins, we searched for conserved domains in the protein sequence. A search on the Conserved Domain Database in the NCBI website [39] showed that a 131 amino acid portion of the 71cR protein aligns with a Nuclear Transport Factor 2 (NTF2-like) super-family domain. This result was confirmed using Pfam software [41] (version 2014_07), and the sequence was further identified as being in the DUF (domain of unknown function) 1348 family, one of the 24 families in the NTF2-like super-family. Of the 24 families in the NTF2-like super-family, 16 have known functions, and the remaining eight families, including DUF1348, are not characterized [41]. The DUF1348 family domain is present in 613 different UniProtKB proteins from 562 different organisms, 79 of which are in Eukarya and 534 of which are in Bacteria [41].
As the DUF1348 domain has not been characterized, relationships between the 71cR protein and proteins with known functions in the NTF2-like super-family were analyzed in an attempt to identify a putative function for 71cR. Maximum likelihood analysis was done for amino acid sequences of three DUF1348 domains including 71cR, as well as 54 proteins from different organisms representing each of the 16 characterized domains within the NTF2-like superfamily (S2 Fig). Some of the NTF2 families formed monophyletic clades. For example, the four scytalone dehydratase (SDH) sequences formed a monophyletic clade with a bootstrap value of 53%. However, sequences for most of the NTF2 families were polyphyletic, with little or no bootstrap support to cluster families within clades (S2 Fig). 71cR formed a clade with two other DUF1348 proteins from Talaromyces and Pseudomonas (S2 Fig) with a bootstrap value of 68%. The next closest domain was from a SnoaL-like polyketide cyclase from Methylomicrobium album from the NTF2 SnoaL1 domain family. Bootstrap support for the association with the SnoaL-like polyketide cyclase was very low (21%), however, thus no firm conclusions can be drawn about function from this analysis.
Secondary and tertiary structure prediction of 71cR
As the phylogenetic analyses did not enable us to make conclusions about function, we analyzed secondary and tertiary structure of the 71cR amino acid sequence as these structures are important for their biological functions. The secondary and tertiary structure of the 71cR amino acid sequence was analyzed using Raptor software [35]. The secondary structure of 71cR is predicted to have 7 beta sheets and 4 alpha helices (Fig 8A). For the tertiary structure predictions, Raptor software predicted the best template to be Protein PFL_3262 from Pseudomonas fluorescens (2imj:A). The predicted model has a p-value of 2.96e-11. The predicted structure of 71cR protein is a half barrel structure with 7 beta sheets enclosing the alpha helix (Fig 8B). The two other alpha helices are positioned near beta sheets #4, 5, and 6 (Fig 8B). The predicted model also states that this protein forms a dimer. The Raptor server predicted that 71cR binds steroids, and identified 12 ligand binding sites for steroid binding (A19, W23, N43, K61, Y68, F86, Y88, Y90, C102, E106, R119, M121). The pocket multiplicity value predicted for steroid binding is 74, and we compared it to the pocket multiplicity of a known steroid delta-isomerase (Accession number: P07445); its pocket multiplicity was also 74, providing high support for 71cR binding to steroids. To determine whether the predicted ligand binding sites are conserved in 71cR, 130 of its closest orthologs (S1 Table) were aligned to identify conserved residues within the sequence (Fig 9). The 71cR sequence was mostly conserved throughout its length. All of the ligand binding sites found in the Raptor analysis except Y90 and C102 were shown to be conserved.
Fig 8. 71cR secondary and tertiary structure predictions from Raptor software.
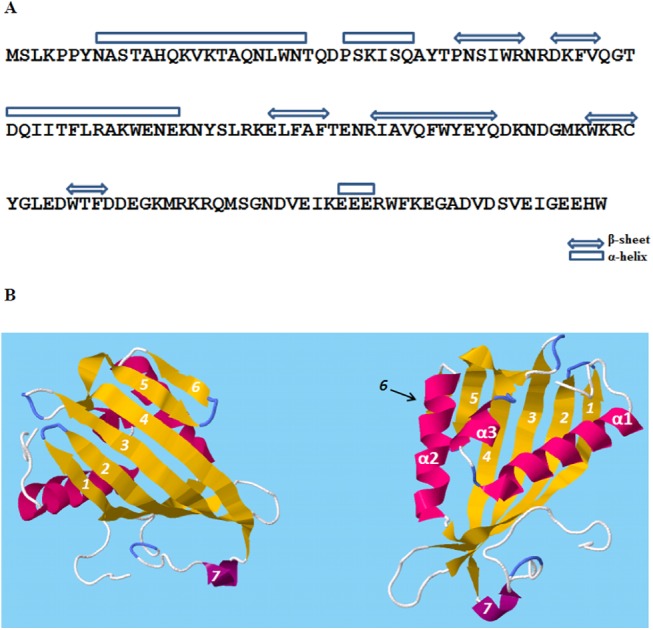
A. Secondary structure showing 4 alpha helixes and 7 beta sheets. B. Predicted tertiary structure; the two figures represent the same structure from different angles. The beta sheets are numbered from 1–7, and alpha helixes are numbered from α1-α3. 71cR is predicted to have a half-barrel structure with 5 beta sheets enclosing the α1 helix. This protein is predicted to form a dimer to complete the barrel structure.
Fig 9. Alignment of 71cR amino acid sequence with the 130 amino acid sequences in S1 Table.
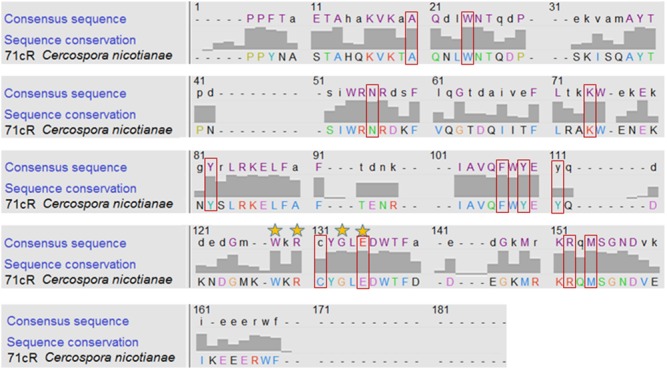
Chimera 1.10.1 was used to display the degree of conservation for each residue within the alignment of the conserved domain [38]. 71cR C. nicotianae sequence is shown along with the consensus sequence from the whole dataset. The numbers above the amino acid residues indicate the positions within the sequence alignment. Color of each amino acid was determined according to the ClustalX [42] color scheme. The bars show the conservation between 71cR and all of the 130 sequences in S1 Table. Stars on top of the amino acids indicate the complete conservation of that sequence except the outgroup (E on position 135 is completely conserved throughout every single one including the outgroup). The red boxes around the letters show the ligand binding sites predicted by Raptor software.
We then used the Dali server [36] which aligns tertiary structures of proteins. The crystal structure of the 71cR is not available. Thus we aligned the crystal structure of the closest ortholog with a crystal structure, protein PFL_3262 from Pseudomonas fluorescens (PDB ID: 2IMJ). This analysis predicts that the PFL_3262 tertiary structure aligns most closely with steroid delta-isomerase proteins, with z values of 14.1–15.4. This finding suggests that 71cR orthologs are most similar to steroid delta-isomerase proteins, a finding consistent with the Raptor software’s prediction of steroid binding. To further investigate this association, we re-ran the analysis with a known steroid delta-isomerase protein (Accession number: 3t8n). Z-scores between the steroid delta-isomerase protein (3t8n) and other steroid delta-isomerase proteins were higher, in the range of 20.4–25.7. We conclude that 71cR is similar to steroid delta-isomerase proteins, but is not this precise enzyme. In agreement with the similarity to steroid delta-isomerase proteins, which are localized in the cytosol, the Predict Protein server [37] identified no transmembrane helices or disulphide bridges in 71cR and predicted localization in the cytosol.
Analysis of 71cR effect on cercosporin
Steroid delta-isomerase proteins catalyze isomerization reactions of steroids. We thus decided to test if 71cR may be catalyzing a reaction to degrade or modify the structure of cercosporin. We tested possible degradation by treating cultures of resistant N. crassa 71cR transformants (#379 and 380) and wt with cercosporin, harvesting the mycelium, and extracting and quantifying cercosporin from the mycelium and from the medium. We found no detectable differences in the amount of cercosporin recovered from the resistant transformants as compared to wt (data not shown), suggesting no degradation of cercosporin. In addition, we used confocal microscopy to generate λ-stacks to compare fluorescence emission spectra of the treated hyphae. We were unable to detect a difference between wild-type and resistant N. crassa 71cR transformants (data not shown). Thus we do not have evidence for degradation or modification of cercosporin by the 71cR transformants.
Discussion
The 185 EST sequences recovered from the subtractive library provided a large set of candidate gene products that may have a role in cercosporin resistance [7]. Among this list are three transporters, ATR1, ATR2 and CFP, that have been shown to have a role in resistance [10,12,13]. In this paper, we characterized genes in the library encoding hypothetical proteins. These were of particular interest to us, as resistance to 1O2 is poorly understood. To understand putative functions of the proteins encoded by the genes, the EST sequences were blasted against the NCBI protein database. Most of the ESTs aligned with orthologs with high similarity, however all of the orthologs identified encode proteins of unknown function. We then used the full length sequences of the closest orthologs to identify functional domains (Table 3). The results revealed diverse functional domains, most of which do not correspond to known mechanisms of cercosporin or 1O2 resistance.
To test the possible role of these hypothetical proteins in cercosporin resistance, we assayed for changes in gene expression when the C. nicotianae cercosporin-sensitive atr1 transporter mutant was exposed to cercosporin toxicity. This mutant is sensitive due to a mutation in the ATR1 transporter involved in transport of cercosporin out of the cell [12], and has been used previously to identify genes that are upregulated under conditions of cercosporin toxicity and impart cercosporin resistance when expressed in sensitive fungi [13]. Only two of the 20 hypothetical protein-encoding genes, 24cF and 71cR, were significantly up-regulated. These results led us to further characterize them for a possible role in resistance.
To test the ability of these genes to increase cercosporin resistance, the cercosporin-sensitive fungus N. crassa was transformed with the complete gene sequences of 71cR and 24cF under the control of a constitutive promoter, and the resulting transformants were screened for increases in cercosporin resistance, an assay previously utilized to identify other cercosporin resistance genes [13,17]. Under the conditions of our cercosporin-sensitivity assay, growth of wt N. crassa is inhibited by about 70% whereas growth of the resistant C. nicotianae is inhibited by about 40%. No significant increase in resistance was seen in any of the 24cF transformants tested, however significant increases in cercosporin resistance were found in some 71cR transformants, with inhibition as low as 20%. Resistance of the 71cR transformants varied, with some showing no increase. This variation might be explained by the random integration of the insert and different levels of transgene transcripts [43]. To confirm that the resistance seen in 71cR transformants was due to the transgene, expression of the 71cR transgene was tested from randomly picked transformants, two that showed significant resistance and two that did not. The results showed that transformants that showed increased resistance had high expression of the gene compared to the expression of the gene in the transformants that did not have increased resistance. Thus for 71cR transformants, expression of cercosporin resistance correlated with 71cR gene expression. For 24cF, each of the transformants assayed had high levels of expression without imparting increases in resistance. Thus we concluded that 24cF alone cannot impart significant levels of cercosporin resistance. Given that 24cF is upregulated under conditions of cercosporin toxicity, it is possible that it can play a role in resistance in combination with other genes. Our domain analysis (Table 3) showed that the protein has domains similar to those of phospholipid methyltransferases in the PEMT superfamily, but the precise function of this protein is unknown and further characterization will be needed to explore how it might contribute to resistance.
To further characterize the role of 71cR in cercosporin resistance, 71cR was disrupted in wt C. nicotianae, and resistance to cercosporin was assayed. The resistance of the disruptants to cercosporin was not significantly different than the wt C. nicotianae. The cercosporin production in 71cR disruptants was also compared to wt, and no difference in cercosporin production was observed. We hypothesized that the disruption of 71cR may be compensated for by induction of other resistance genes. We thus assayed two of the 71cR disruptants for expression of known resistance genes and for genes in the cercosporin biosynthetic pathway, when the disruptants were treated with cercosporin. Three genes, CFP, CTB5 and CTB7, were found to be induced in the 71cR disruptant mutants when they were exposed to cercosporin toxicity. CFP is an MFS transporter previously characterized to have a role in cercosporin resistance [10]. We have also shown that CFP is strongly induced by cercosporin toxicity in cercosporin-sensitive atr1 and atr2 mutants of C. nicotianae [13], thus CFP induction may be a general response of C. nicotianae against cercosporin toxicity, perhaps by facilitating export of the toxin out of the cells. The two CTB genes encode oxidoreductases found in the cercosporin biosynthetic cluster [2,44]. Oxidoreductase activity has been hypothesized to be involved in cercosporin resistance through reduction of cercosporin [2] and these genes are also induced in the cercosporin-sensitive atr2 background [13]. Mutants for CTB5 and CTB7 have not been shown to be altered in resistance [40,44]; however, possible compensation by upregulation of other resistance genes in ctb5 and ctb7 mutants has not been investigated. Further studies with ctb5 and ctb7 disruption mutants need to be done to conclusively characterize a possible role in cercosporin resistance.
The ability of 71cR to impart cercosporin resistance in N. crassa led us to further characterize it. To identify the closest orthologs of 71cR, tblastn and blastp searches were done against the NCBI database, and 71cR was found to have high homology (up to 84% amino acid similarity) to hypothetical proteins from diverse fungi including members of both Ascomycota and Basidiomycota. To better understand species distribution within the fungal kingdom, we searched for orthologs of 71cR in each sequenced fungal genome from JGI using tblastn. This analysis showed that 71cR orthologs are widely distributed in the Basidiomycota and Ascomycota but that distribution is not ubiquitous within species in these groups. In the Ascomycota, orthologs were common in the Pezizomycotina, especially in the classes Dothidiomycetes (containing Cercospora), Eurotiomycetes, and Sordariomycetes. Orthologs of 71cR are also present in most fungal genomes in other subphyla in Ascomycota and Basidiomycota. Rhizophagus irregularis, the only sequenced genome from Glomeromycota, also had an ortholog of 71cR. However, 71cR orthologs were absent from sequenced genomes from other clades traditionally included in Zygomycota, as well as basal fungal lineages such as Chytridiomycota and Microsporidia (Fig 7). Although there are limited genomes available in JGI from these more basal lineages, the absence of this ortholog could indicate that 71cR evolved later in the evolutionary history of fungi.
As the most conserved orthologs to 71cR were not characterized for function, we used the Conserved Domain Database in the NCBI website [39] and Pfam [41] to identify domains. 71cR was shown to have an uncharacterized domain in the DUF1348 family in the NTF2 superfamily. The NTF2-like superfamily contains families with a common fold that results in a cone-like shape with a cavity inside. Although sharing a common fold, the proteins have very poor sequence similarity [45], thus explaining the lack of bootstrap support to cluster families within clades in our phylogenetic analysis (S2 Fig). The NTF2-like superfamily contains both enzymatically active and non-enzymatically active proteins. SnoaL polyketide cyclase, scytalone dehydratase, limonene-1,2-epoxide hydrolase and δ5-3-ketosteroid isomerase are examples of the enzymatically active group, which are typically intracellular [45]. Non-enzymatically active NTF2-like protein domains are mainly extracellular. The examples of proteins with non-enzymatically active NTF2-like domains are the C-terminus of calcium/calmodulin-dependent protein kinase II [46], Mba1, a putative ribosome-binding receptor [47], proteins involved in DNA transfer during bacterial conjugation [48], and immunity proteins in the bacterial polymorphic toxin systems [49]. Our phylogenetic analysis confirmed that 71cR formed a clade with other proteins containing a DUF1348 domain. The next closest characterized ortholog was a SnoaL-like polyketide cyclase from Methylomicrobium album, however, the bootstrap value for this relationship was extremely low (21%). Therefore, we were unable to draw any conclusions about the possible function of 71cR from the phylogenetic analysis.
Since we could not predict a function of 71cR based on sequence homology, we turned to characterization of the 71cR secondary and tertiary structures. Raptor software [35] predicted that the 71cR protein has antiparallel beta strands forming a half barrel structure with one of the main alpha helix structures buried inside this half barrel. The protein is predicted to form a dimer. Solvent Accessibility prediction [35] shows several ligand binding regions and predicted localization in the cytosol. With this information, the 71cR protein was predicted to have an enzymatically active group as it is intracellular. Furthermore, the Raptor server predicted that 71cR binds steroids (pocket multiplicity of 74), and predicted amino acid residues that would be important for ligand binding. We confirmed that most of the predicted ligand binding sites are conserved throughout the close orthologs of 71cR.
Further analysis of tertiary structure was done using the Dali server [36]. Using Dali, the 71cR protein aligned closely with steroid delta-isomerase proteins. Steroid delta-isomerase proteins are common in diverse organisms from bacteria to higher eukaryotes. They catalyze the isomerization of steroids by transferring protons in a two-step reaction [50], and are involved in both steroid biosynthesis [51] and steroid degradation pathways [52]. Although this was the closest association, z values were not high enough to conclude with confidence that 71cR is a steroid delta-isomerase, and it may catalyze a different reaction. We hypothesized that 71cR may impart resistance by degrading or altering the structure of the polyketide cercosporin molecule, but were unable to obtain evidence to support this hypothesis. It is interesting that so many fungi have orthologs to 71cR as most of these are not known to produce photoactivated perylenequinones or to be resistant to such compounds. It is possible that the enzymes, although all having DUF1348 domains, are very different. Alternatively, it is possible that they carry out similar reactions, but that the substrates are different in different species. Further research will be required to characterize the specific function of the 71cR protein as well as orthologs in other fungal species.
In summary, we have shown that the 71cR gene, encoding a hypothetical protein, is upregulated in C. nicotianae in response to cercosporin toxicity, and that expression of this gene in the cercosporin-sensitive fungus N. crassa can impart cercosporin resistance. C. nicotianae disruption mutants are not more sensitive to cercosporin than wt, but other genes previously documented to play a role in resistance are up-regulated in these mutants upon exposure to cercosporin toxicity, suggesting that the fungus compensates for loss of resistance genes by up-regulation of other resistance mechanisms. Analysis of 71cR sequence, phylogeny, conserved domains, and secondary and tertiary structure identify the protein as having an NTF2-like superfamily domain with unknown function, to be intracellular, localized in the cytosol, to have enzymatic activity, and to have similarities to steroid delta-isomerase proteins. Current research is focused on expression of 71cR in plants to determine if it has utility for engineering Cercospora- and cercosporin-resistant plants.
Supporting Information
Cercosporin resistance assay of N. crassa transformed with genes encoding hypothetical proteins from Cercospora nicotianae. Cultures grown under continuous light on Vogel’s medium supplemented with 10 μM cercosporin (right) and with 0.5% acetone (left) that was used to solubilize cercosporin. Top: N. crassa transformed with 71cR showing resistance to cercosporin. Middle: N. crassa transformant lacking resistance. Bottom: N. crassa wild type with colony margins at 21 hours (time used in resistance assay) marked on plate.
(TIF)
RAxML was used for maximum likelihood analysis of the sequences, which include the 71cR amino acid sequence, a hypothetical protein from Talaromyces stipitatus (XP_002479782.1) (a close ortholog of 71cR), and a Pseudomonas fluorescens protein (Protein Pfl_3262) in the same domain family (DUF1348), along with conserved domains of characterized proteins within the NTF2-like super family. Families within the NTF2-like superfamily are indicated in bold, followed by the name of the species from which the sequence was found. Domain abbreviations are: DUF1348, domain of unknown function 1348; RHBS, ring hydroxylating beta subunit; SnoaL, polyketide cyclase; LEH, limonene epoxide hydrolase; PHEN, phenazine biosynthesis protein; NTF2, nuclear transport factor 2; VirB8, type IV secretion assembly factor; WI12, wound induced protein; MTR2, nuclear pore RNA shuttling protein; Tim44, mitochondrial import protein; CDPK, Ca2+/calmodulin-dependent protein kinase; LBD, lumazine-binding domain; TRPEP, transpeptidase; MBA1, mitochondrial import protein; SDH, scytalone dehydratase. The red box indicates the clade containing the DUF1348 sequences. The 71cR protein from C. nicotianae is shown in red text. Bootstrap values for each relationship are presented on the tree, and the scale bar represents substitutions per site. The accession numbers with the species names where these proteins were found are included in S2 Table.
(TIF)
This dataset was used in phylogenetics analysis of closely related sequences and analysis of conserved amino acid residues (Fig 9). Sequences are listed in order of similarity to the 71cR protein sequence.
(PDF)
These protein sequences were used for phylogenetics analysis with conserved domains to identify the most closely related characterized ortholog of 71cR.
(PDF)
Acknowledgments
We thank The Cellular and Molecular Imaging Facility at North Carolina State University for assistance with the usage of their microscope. We also thank Dr. Marc Cubeta of the Department of Plant Pathology at NC State for the help with the phylogenetic analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding was provided by US-Egypt Science and Technology Joint Fund through the USDA cooperative agreement# 58-3148-1-161, http://sites.nationalacademies.org/pga/dsc/egypt/index.htm. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Daub ME, Ehrenshaft M (2000) The photoactivated Cercospora toxin cercosporin: Contributions to plant disease and fundamental biology. Ann Rev Phytopathol 38: 461–490. [DOI] [PubMed] [Google Scholar]
- 2. Daub ME, Herrero S, Chung KR (2013) Reactive oxygen species in plant pathogenesis: the role of perylenequinone photosensitizers. Antiox Redox Signal 19: 970–989. [DOI] [PubMed] [Google Scholar]
- 3. Daub ME, Herrero S, Taylor TV (2010) Strategies for the development of resistance to cercosporin, a toxin produced by Cercospora species In: Lartey RT, Weiland JJ, Panella L, Crous PW, Windels CE, editors. Cercospora Leaf Spot of Sugar Beet and Related Species. St. Paul, MN: APS Press; pp. 157–172. [Google Scholar]
- 4. Jenns AE, Daub ME (1995) Characterization of mutants of Cercospora nicotianae sensitive to the toxin cercosporin. Phytopathology 85: 906–912. [Google Scholar]
- 5. Jenns AE, Scott DL, Bowden EF, Daub ME (1995) Isolation of mutants of the fungus Cercospora nicotianae altered in their response to singlet-oxygen-generating photosensitizers. Photochem Photobiol 61: 488–493. [Google Scholar]
- 6. Chung KR, Daub ME, Kuchler K, Schuller C (2003) The CRG1 gene required for resistance to the singlet oxygen-generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem Biophys Res Commun 302: 302–310. [DOI] [PubMed] [Google Scholar]
- 7. Herrero S, Amnuaykanjanasin A, Daub ME (2007) Identification of genes differentially expressed in the phytopathogenic fungus Cercospora nicotianae between cercosporin toxin-resistant and -susceptible strains. FEMS Microbiol Lett 275: 326–337. [DOI] [PubMed] [Google Scholar]
- 8. Daub ME, Leisman GB, Clark RA, Bowden EF (1992) Reductive detoxification as a mechanism of fungal resistance to singlet-oxygen-generating photosensitizers. Proc Natl Acad Sci USA 89: 9588–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ehrenshaft M, Bilski P, Li M, Chignell CF, Daub ME (1999) A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA 96: 9374–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Callahan T, Rose M, Meade M, Ehrenshaft M, Upchurch R (1999) CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Mol Plant-Microbe Interact 12: 901–910. [DOI] [PubMed] [Google Scholar]
- 11. Choquer M, Lee MH, Bau HJ, Chung KR (2007) Deletion of a MFS transporter-like gene in Cercospora nicotianae reduces cercosporin toxin accumulation and fungal virulence. FEBS Lett 581: 489–494. [DOI] [PubMed] [Google Scholar]
- 12. Amnuaykanjanasin A, Daub ME (2009) The ABC transporter ATR1 is necessary for efflux of the toxin cercosporin in the fungus Cercospora nicotianae . Fung Genet Biol 46: 146–158. [DOI] [PubMed] [Google Scholar]
- 13. Beseli A, Amnuaykanjanasin A, Herrero S, Thomas E, Daub ME (2015) Membrane transporters in self resistance of Cercospora nicotianae to the photoactivated toxin cercosporin. Curr Genet 10.1007/s00294-015-0486-x [DOI] [PubMed] [Google Scholar]
- 14. Upchurch RG, Rose MS, Eweida M, Zuo W (2005) Expression of the cercosporin transporter, CFP, in tobacco reduces frog-eye lesion size. Biotechnol Lett 27: 1543–1550. [DOI] [PubMed] [Google Scholar]
- 15. Bilski P, Li MY, Ehrenshaft M, Daub ME, Chignell CF (2000) Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem Photobiol 71: 129–134. [DOI] [PubMed] [Google Scholar]
- 16. Herrero S, Daub ME (2007) Genetic manipulation of vitamin B-6 biosynthesis in tobacco and fungi uncovers limitations to up-regulation of the pathway. Plant Sci 172: 609–620. [Google Scholar]
- 17. Beseli A, Goulart da Silva M, Daub M (2015) The role of Cercospora zeae-maydis homologs of Rhodobacter sphaeroides 1O2-resistance genes in resistance to the photoactivated toxin cercosporin. FEMS Microbiol Lett 362:1–7. [DOI] [PubMed] [Google Scholar]
- 18. Vogel HJ (1956) A convenient growth medium for Neurospora (Medium N). Microbial Genet Bull 13: 42–43. [Google Scholar]
- 19. Deininger P (1990) Molecular cloning: a laboratory manual. Anal Biochem 186: 182–183. [Google Scholar]
- 20. Sweigard J, Chumley F, Carroll A, Farrall L, Valent B (1997) A series of vectors for fungal transformation. Fung Genet Newslett 44: 52–53. [Google Scholar]
- 21. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 22. Winer J (1999) Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro . Anal Biochem 270: 41–49. [DOI] [PubMed] [Google Scholar]
- 23. Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW (2000) Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and Real-Time methods. Anal Biochem 285: 194–204. [DOI] [PubMed] [Google Scholar]
- 24. Wang H, Meiqing Q, Cutler AJ (1993) A simple method of preparing plant samples for PCR. Nucleic Acids Res 21: 4153–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. You BJ, Lee MH, Chung KR (2009) Gene-specific disruption in the filamentous fungus Cercospora nicotianae using a split-marker approach. Arch Microbiol 191: 615–622. 10.1007/s00203-009-0489-4 [DOI] [PubMed] [Google Scholar]
- 26. Jenns AE, Daub ME, Upchurch RG (1989) Regulation of cercosporin accumulation in culture by medium and temperature manipulation. Phytopathology 79: 213–219. [Google Scholar]
- 27. Edgar R (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5): 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddison W, Maddison D (2015) Mesquite: a modular system for evolutionary analysis. Version 3.04 http://mesquiteprojectorg.
- 29. Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18 (5): 691–699. [DOI] [PubMed] [Google Scholar]
- 30. Keane T, Creevey C, Pentony M, Naughton T, Mclnerney J (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evolutionary Biology 6: 29 10.1186/1471-2148-6-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silvestro D, Michalak I (2012) RaxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12:333–337. [Google Scholar]
- 32. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grigoriev I, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, et al. (2014) MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42(1): D699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP, et al. (2011) Fueling the future with fungal genomics. Mycology 2(3):192–209. [Google Scholar]
- 35. Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, et al. (2012) Template-based protein structure modeling using the RaptorX web server. Nature Protocols 7: 1511–1522. 10.1038/nprot.2012.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–W549. 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, et al. (2014) PredictProtein—an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 10.1093/nar/gku36638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng ED, et al. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 13: 1605–1612. [DOI] [PubMed] [Google Scholar]
- 39. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Geer LY, et al. (2011) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41: D384–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen HQ, Lee MH, Chung KR (2007) Functional characterization of three genes encoding putative oxidoreductases required for cercosporin toxin biosynthesis in the fungus Cercospora nicotianae . Microbiology 153: 2781–2790. [DOI] [PubMed] [Google Scholar]
- 41. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. (2014) The Pfam protein families database. Nucleic Acids Res 42: D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith KM, Kothe GO, Matsen CB, Khlafallah TK, Adhvaryu KK, Hemphill M, et al. (2008) The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenet Chromatin 1:5 10.1186/1756-8935-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen H, Lee MH, Daub ME, Chung KR (2007) Molecular analysis of the cercosporin biosynthetic gene cluster in Cercospora nicotianae . Mol Microbiol 64: 755–770. [DOI] [PubMed] [Google Scholar]
- 45. Eberhardt RY, Chang Y, Bateman A, Murzin AG, Axelrod HL, Hwang WC, et al. (2013) Filling out the structural map of the NTF2-like superfamily. BMC Bioinformatics 14: 327 10.1186/1471-2105-14-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Griffith LC, Lu CS, Sun XX (2003) CaMKII, an enzyme on the move: regulation of temporospatial localization. Mol Interv 3(7): 386–403. [DOI] [PubMed] [Google Scholar]
- 47. Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM (2006) Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J 25(8): 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goessweiner-Mohr N, Grumet L, Arends K, Pavkov-Keller T, Gruber CC, Gruber K, et al. (2013) The 2.5 A structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J Biol Chem 288(3): 2018–2028. 10.1074/jbc.M112.428847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang D, De Souza R, Anantharaman V, Iyer L, Aravind L (2012) Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7: 18 10.1186/1745-6150-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chakravorty D, Hammes-Schiffer S (2010) Impact of mutation on proton transfer reactions in ketosteroid isomerase: insights from molecular dynamics simulations. J Am Chem Soc 132(21): 7549–7555. 10.1021/ja102714u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Talalay P; Benson AM (1972) Δ5-3-ketosteroid isomerase In: Boyer PD. The Enzymes 6 (3rd ed.). Academic Press; pp. 591–618. [Google Scholar]
- 52. Horinouchi M, Kurita T, Hayashi T, Kudo T (2010) Steroid degradation genes in Comamonas testosteroni TA441: Isolation of genes encoding a Δ4(5)-isomerase and 3α- and 3β-dehydrogenases and evidence for a 100 kb steroid degradation gene hot spot. J Steroid Biochem Mol Biol 122(4): 253–263. 10.1016/j.jsbmb.2010.06.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cercosporin resistance assay of N. crassa transformed with genes encoding hypothetical proteins from Cercospora nicotianae. Cultures grown under continuous light on Vogel’s medium supplemented with 10 μM cercosporin (right) and with 0.5% acetone (left) that was used to solubilize cercosporin. Top: N. crassa transformed with 71cR showing resistance to cercosporin. Middle: N. crassa transformant lacking resistance. Bottom: N. crassa wild type with colony margins at 21 hours (time used in resistance assay) marked on plate.
(TIF)
RAxML was used for maximum likelihood analysis of the sequences, which include the 71cR amino acid sequence, a hypothetical protein from Talaromyces stipitatus (XP_002479782.1) (a close ortholog of 71cR), and a Pseudomonas fluorescens protein (Protein Pfl_3262) in the same domain family (DUF1348), along with conserved domains of characterized proteins within the NTF2-like super family. Families within the NTF2-like superfamily are indicated in bold, followed by the name of the species from which the sequence was found. Domain abbreviations are: DUF1348, domain of unknown function 1348; RHBS, ring hydroxylating beta subunit; SnoaL, polyketide cyclase; LEH, limonene epoxide hydrolase; PHEN, phenazine biosynthesis protein; NTF2, nuclear transport factor 2; VirB8, type IV secretion assembly factor; WI12, wound induced protein; MTR2, nuclear pore RNA shuttling protein; Tim44, mitochondrial import protein; CDPK, Ca2+/calmodulin-dependent protein kinase; LBD, lumazine-binding domain; TRPEP, transpeptidase; MBA1, mitochondrial import protein; SDH, scytalone dehydratase. The red box indicates the clade containing the DUF1348 sequences. The 71cR protein from C. nicotianae is shown in red text. Bootstrap values for each relationship are presented on the tree, and the scale bar represents substitutions per site. The accession numbers with the species names where these proteins were found are included in S2 Table.
(TIF)
This dataset was used in phylogenetics analysis of closely related sequences and analysis of conserved amino acid residues (Fig 9). Sequences are listed in order of similarity to the 71cR protein sequence.
(PDF)
These protein sequences were used for phylogenetics analysis with conserved domains to identify the most closely related characterized ortholog of 71cR.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.