Abstract
The associations between CYP1A1 polymorphisms and risk of leukemia have been studied extensively, but the results have been inconsistent. Therefore, in this study, we performed a meta-analysis to clarify associations of three CYP1A1 polymorphisms (T3801C, A2455G, and C4887A) with the risks of acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and chronic myeloid leukemia (CML). Medline, EMBASE, and China National Knowledge Infrastructure databases were searched to collect relevant studies published up to April 20, 2015. The extracted data were analyzed statistically, and pooled odds ratios with 95% confidence intervals were calculated to quantify the associations. Overall, 26 publications were included. Finally, T3801C was associated with an increased risk of AML in Asians under the dominant model. For A2455G, the risk of ALL was increased among Caucasians in the recessive model and the allele-contrast model; A2455G was also associated with an increased risk of CML among Caucasians under the recessive model, dominant model, and allele-contrast model. For C4887A, few of the included studies produced data. In conclusion, the results suggest that Asians carrying the T3801C C allele might have an increased risk of AML and that Caucasians with the A2455G GG genotype might have an increased risk of ALL. Further investigations are needed to confirm these associations.
Keywords: CYP1A1, ALL, AML, CML, polymorphism
Background
Leukemia is a malignant tumor of the hematopoietic system.1 There are four common types of leukemia: acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphoblastic leukemia, and chronic myeloid leukemia (CML). Environmental exposures and genetic susceptibility play roles in the etiology of leukemia.2 For environmental exposures, benzene, ionizing radiation, and cytotoxic therapy are some of the proposed causes of acute leukemia; for genetic susceptibility, the most common type is the single nucleotide polymorphism (SNP) that consists of a variation at a single base pair. Depending on where it is located, SNPs can interfere with a gene’s function, affecting metabolic pathways and further affecting the course of the disease and its progress. SNPs in xenobiotic system, DNA repair system, and cell regulation have been identified as risk factors in childhood leukemia.3,4
CYP1A1 and leukemia
Cytochrome P450 (CYP) enzyme catalyzes the phase I metabolism reaction. The CYP1A1 gene (CYP1A1) is a member of the CYP family that participates in the metabolism of xenobiotics and endogenous compounds, particularly polycyclic aromatic hydrocarbons such as benzo[a]pyrene.5 Genetic polymorphisms have been reported for CYP1A1 that may alter the function of enzyme and thus influence the ability of enzymes to metabolize the chemical carcinogens and mutagens,6,7 which may influence how susceptible individuals are to contracting leukemia.8
SNPs in CYP1A1
The most commonly studied SNP in CYP1A1 is the T3801C polymorphism (also referred to as CYP1A1*2A, m1, or rs4646903). This SNP was localized to chromosome 15q229 and is characterized by a T to C mutation at nucleotide 3,801 in the 3′-flanking region of CYP1A1. Some studies have indicated that the T3801C polymorphism can alter the level of gene expression or the stability of messenger RNA (mRNA), resulting in the enzyme exhibiting highly inducible activity.10 Another common genetic mutant site is A2455G (also referred to as CYP1A1*2C, m2 allele, or rs1048943), which has been widely examined with regard to cancer susceptibility.11–13 This SNP is characterized by an A to G mutation at nucleotide 2,455 in the region of CYP1A1.14 The C4887A mutation (CYP1A1*4 allele) is also an important mutant site, which results in the replacement of Thr by Asn in codon 461, near the site of the A2455G mutation.15 Previous research has shown that this polymorphism can alter the level of gene expression or the stability of mRNA, resulting in the enzyme exhibiting highly inducible activity. In other words, certain variant genotypes of CYP1A1 that change the enzymatic activity appear to crucially influence the risk of leukemia.16
Aim of this study
Previous studies of the relationships between CYP1A1 polymorphisms and risk of leukemia have generated controversial results, and so whether CYP1A1 polymorphisms are a risk factor for leukemia remains uncertain. In this study, we performed a quantitative meta-analysis with the aim of generating more reliable results by applying an analysis with greater statistical power.
Materials and methods
Search strategy
We searched Medline (PubMed), EMBASE, and CNKI (China National Knowledge Infrastructure) databases for publications written in English or Chinese up to April 20, 2015. The search was based on the following Boolean combination of keywords: “CYP1A1” (“Cytochrome P450 1A1”) AND [“ALL” (“acute lymphocytic leukemia” OR “acute lymphoblastic leukemia”) OR “AML” (“acute myeloid leukemia” OR “acute myeloblastic leukemia” OR “acute myelocytic leukemia”) OR “CML” (“chronic myeloid leukemia” OR “chronic myelocytic leukemia” OR “chronic myeloblastic leukemia”) OR “leukemia” (“leucocythemia”)]. All searched studies were retrieved, and their bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were searched manually to find additional eligible studies.
Inclusion criteria
Published studies were included if they fulfilled the following criteria:
Use of a case-control design.
Providing sufficient data on the distribution of CYP1A1 polymorphisms in leukemia and in controls or sufficient information for such data to be calculated.
Focusing on the associations between CYP1A1 polymorphisms and the risk of AML, ALL, and/or CML.
Data extraction
Two investigators independently carried out an eligibility evaluation and data abstraction for each potentially eligible study. Any disagreements were further discussed and resolved by consensus. The following data were abstracted from each of the included studies: name of the first author, year of publication, race distribution of the study population, genotype distributions of the cases and controls, and numbers of cases and controls. We also checked that the distribution of the genotypes in controls conformed with Hardy–Weinberg equilibrium (HWE).
Statistical analysis
The meta-analysis examined the overall associations under the recessive model (CC versus TC + TT for T3801C, GG versus AG + AA for A2455G, and CC versus AC + AA for C4887A), the dominant model (CC + TC versus TT for T3801C, GG + AG versus AA for A2455G, and CA + AA versus CC for C4887A), and the allele-contrast model (C allele versus T allele for T3801C, G allele versus A allele for A2455G, and A allele versus C for C4887A).
The raw data for genotype distribution were used to calculate the study-specific estimates of odds ratio (OR) and 95% confidence interval (CI). Review Manager (version 5.3) software (The Nordic Cochrane Centre, Copenhagen, Denmark) was used to implement the meta-analysis. The presence of heterogeneity was assessed using Cochran’s Q statistic and quantified using the I2 statistic, where a value above 50% indicates the presence of a very high degree of heterogeneity.17
ORs and 95% CIs were calculated using the Z-test to assess the associations between the CYP1A1 SNPs and risk of leukemia. The threshold for statistical significance was set at P<0.05. The fixed effects model was used when no significant clinical or statistical heterogeneity was present (I2≤50%),18 whereas the random effects model was used when there was substantial heterogeneity (I2>50%).
When substantial heterogeneity was present, sensitivity analysis was performed by excluding individual studies; outlying studies were identified and excluded, and the I2 estimates for these different sets of studies were examined.
Potential publication bias was estimated by constructing funnel plots. If most of the data appeared at the top of a funnel plot and were distributed approximately symmetrically, this was considered to indicate the absence of obvious publication bias and vice versa.19 Funnel plots were not constructed when there were too few analyzed studies.
Results
Overview of the characteristics of the included studies
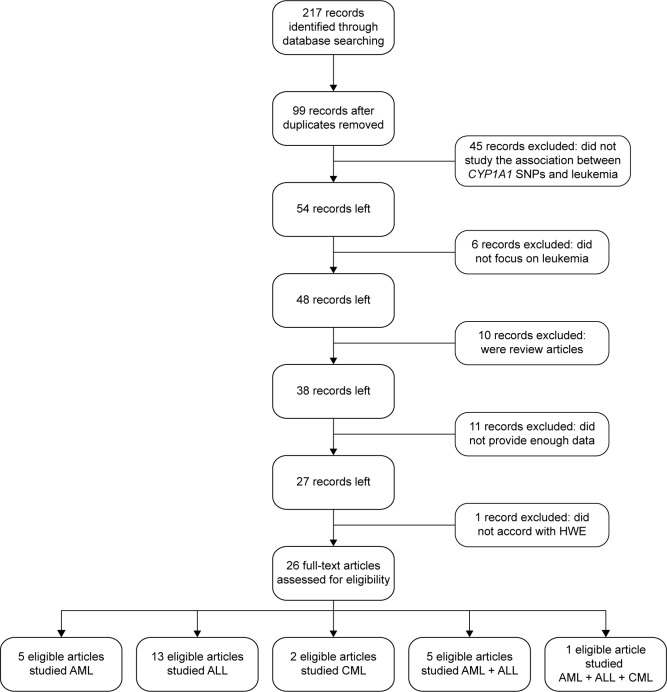
The flow chart for the study selection process is depicted in Figure 1. We selected 217 articles identified by the Boolean combination of the 5 keywords listed in the “Search strategy” section. Checking for duplicates resulted in the removal of 118 articles. Of the remaining 99 articles, 45 did not report studies of the associations between CYP1A1 SNPs and leukemia, 6 did not focus on leukemia, 10 were review articles, and 11 did not provide enough data, and so all of these articles were also discarded. Moreover, the genotypes in controls did not conform with HWE in the study of Chen et al.20 Therefore, ultimately only 26 studies related to the relationships between CYP1A1 SNPs and the risk of leukemia remained in this meta-analysis, among which 13, 5, and 2 related to ALL, AML, and CML, respectively, 5 articles related to both ALL and AML, and 1 article related to all three disease types.
Figure 1.
Flow chart of study selection.
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; HWE, Hardy–Weinberg equilibrium; SNP, single nucleotide polymorphism.
Study characteristics
The basic data of the 26 articles included in the meta-analysis were extracted and are listed in Table 1. In the study of Majumdar et al21 the distributions of the A2455G and C4887A genotypes in controls did not conform with HWE (although the T3801C genotypes did), and so this study was excluded when we analyzed the roles of the CYP1A1 A2455G and C4887A polymorphisms. Overall, the meta-analysis included the following studies:
Three studies involving 272 cases and 402 controls related to the role of the A2455G polymorphism in the risk of CML.
Four studies involving 846 cases and 2,201 controls related to the role of the A2455G polymorphism in the risk of AML.
Ten studies involving 1,330 cases and 3,688 controls related to the role of the T3801C polymorphism in the risk of AML.
One study involving 193 cases and 273 controls related to the role of the C4887A polymorphism in the risk of AML.
Eight studies involving 1,734 cases and 2,194 controls related to the role of the A2455G polymorphism in the risk of ALL.
Sixteen studies involving 2,299 cases and 3,209 controls related to the role of the T3801C polymorphism in the risk of ALL.
One study involving 177 cases and 295 controls related to the role of the C4887A polymorphism in the risk of ALL.
Table 1.
Study characteristics
| Study | Ethnicity | Cases | SNP | Number of cases | Number of controls | PHWE in controls |
|---|---|---|---|---|---|---|
| Agha et al29 | Caucasian | ALL | T3801C, C4887A | 186 | 200 | 0.0577 |
| Aydin-Sayitoglu et al30 | Caucasian | ALL | T3801C | 249 | 140 | 0.3057 |
| AML | T3801C | 249 | 140 | 0.3057 | ||
| Bolufer et al31 | Caucasian | ALL | T3801C | 443 | 454 | 0.151 |
| AML | T3801C | 443 | 454 | 0.151 | ||
| Bonaventure et al32 | Caucasian | ALL | T3801C | 493 | 549 | 0.2309 |
| AML | T3801C | 493 | 549 | 0.2309 | ||
| Krajinovic et al27 | Caucasian | ALL | T3801C | 177 | 304 | 0.3941 |
| C4887A | 0.1355 | |||||
| Ouerhani et al33 | Caucasian | ALL | T3801C | 100 | 106 | 0.5731 |
| Swinney et al12 | Caucasian | ALL | T3801C, A2455G | 258 | 646 | 0.1272 |
| Yamaguti et al13 | Caucasian | ALL | T3801C | 99 | 99 | 0.1701 |
| A2455G | 0.9485 | |||||
| Selvin49 | Caucasian | ALL | A2455G | 175 | 175 | 0.9347 |
| Gao et al34 | Asian | ALL | T3801C | 78 | 112 | 0.1723 |
| A2455G | 0.1357 | |||||
| Joseph et al35 | Asian | ALL | T3801C | 118 | 118 | 0.5939 |
| A2455G | 0.141 | |||||
| Lee et al36 | Asian | ALL | T3801C | 164 | 164 | 0.4015 |
| A2455G | 0.4514 | |||||
| Suneetha et al37 | Asian | ALL | T3801C | 92 | 150 | 0.806 |
| Balta et al38 | Mixed | ALL | T3801C | 144 | 185 | 0.0907 |
| AML | T3801C | 144 | 185 | 0.0907 | ||
| Canalle et al39 | Mixed | ALL | T3801C | 113 | 221 | 0.083 |
| Clavel et al40 | Mixed | ALL | T3801C | 219 | 105 | 0.1861 |
| AML | T3801C | 219 | 105 | 0.1861 | ||
| Gallegos-Arreola et al41 | Mixed | ALL | T3801C | 210 | 228 | 0.7728 |
| Razmkhah et al43 | Mixed | ALL | A2455G | 85 | 94 | 0.4353 |
| AML | A2455G | 85 | 94 | 0.9543 | ||
| CML | A2455G | 85 | 94 | 0.9543 | ||
| Gallegos-Arreola et al48 | Mixed | ALL | A2455G | 136 | 136 | 0.315 |
| D’Alo et al44 | Caucasian | AML | T3801C | 193 | 273 | 0.1604 |
| A2455G | 0.4279 | |||||
| C4887A | 0.7257 | |||||
| Jiang et al45 | Asian | AML | T3801C | 98 | 120 | 0.1062 |
| Kim et al46 | Asian | AML | T3801C | 415 | 1,700 | 0.6805 |
| A2455G | 0.0688 | |||||
| Majumdar et al21 | Asian | AML | T3801C | 110 | 126 | 0.8415 |
| Yamaguti et al47 | Mixed | AML | T3801C | 133 | 133 | 0.1381 |
| A2455G | 0.3451 | |||||
| Al-Achkar et al42 | Caucasian | CML | A2455G | 126 | 172 | 0.3253 |
| Taspinar et al22 | Caucasian | CML | A2455G | 107 | 135 | 0.5889 |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; PHWE, probability of adherence to the Hardy–Weinberg equilibrium; SNP, single nucleotide polymorphism.
Meta-analysis results
The main results of the meta-analysis are listed in Table 2.
Table 2.
ORs and heterogeneity results for CYP1A1 meta-analysis
| SNP | Cancer | Ethnicity | n | Genetic model | I2 | Contrast model | P, ORs (95% CIs) |
|---|---|---|---|---|---|---|---|
| T3801C | ALL | Caucasian | 8 | Recessive | 26% | Fixed effects | 0.28, 1.31 (0.81, 2.12) |
| Dominant | 61% | Random effects | 0.10, 1.30 (0.95, 1.79) | ||||
| Allele contrast | 32% | Fixed effects | 0.05, 1.18 (1.00, 1.39) | ||||
| Asian | 4 | Recessive | 63% | Random effects | 0.73, 1.14 (0.54, 2.39) | ||
| Dominant | 68% | Random effects | 0.06, 1.64 (0.98, 2.73) | ||||
| Allele contrast | 80% | Random effects | 0.19, 1.35 (0.86, 2.14) | ||||
| Mix | 4 | Recessive | 76% | Random effects | 0.21, 2.68 (0.57, 12.62) | ||
| Dominant | 82% | Random effects | 0.32, 1.34 (0.75, 2.39) | ||||
| Allele contrast | 92% | Random effects | 0.34, 1.42 (0.69, 2.91) | ||||
| A2455G | ALL | Caucasian | 4 | Recessive | 0% | Fixed effects | 0.002, 2.23 (1.36, 3.68) |
| Dominant | 39% | Fixed effects | 0.12, 1.22 (0.95, 1.58) | ||||
| Allele contrast | 41% | Fixed effects | 0.010, 1.31 (1.07, 1.61) | ||||
| Asian | 3 | Recessive | 11% | Fixed effects | 0.14, 1.57 (0.87, 2.83) | ||
| Dominant | 72% | Random effects | 0.29, 1.40 (0.76, 2.59) | ||||
| Allele contrast | 0% | Fixed effects | 0.89, 1.02 (0.80, 1.29) | ||||
| Mix | 1 | Recessive | Fixed effects | ||||
| Dominant | Fixed effects | 0.94, 1.03 (0.45, 2.34) | |||||
| Allele contrast | Fixed effects | 0.94, 1.03 (0.47, 2.26) | |||||
| C4887A | ALL | Caucasian | 1 | Recessive | Fixed effects | 0.61, 1.67 (0.23, 11.99) | |
| Dominant | Fixed effects | 0.14, 0.57 (0.27, 1.21) | |||||
| Allele contrast | Fixed effects | 0.22, 0.65 (0.33, 1.30) | |||||
| T3801C | AML | Caucasian | 4 | Recessive | 0% | Fixed effects | 0.26, 1.74 (0.66, 4.55) |
| Dominant | 47% | Fixed effects | 0.16, 0.82 (0.62, 1.08) | ||||
| Allele contrast | 59% | Random effects | 0.63, 0.90 (0.59, 1.37) | ||||
| Asian | 3 | Recessive | 0% | Fixed effects | 0.60, 0.93 (0.71, 1.22) | ||
| Dominant | 0% | Fixed effects | 0.03, 1.26 (1.02, 1.55) | ||||
| Allele contrast | 0% | Fixed effects | 0.23, 1.09 (0.95, 1.26) | ||||
| Mix | 3 | Recessive | 0% | Fixed effects | 0.72, 1.19 (0.46, 3.05) | ||
| Dominant | 74% | Random effects | 0.65, 1.24 (0.48, 3.22) | ||||
| Allele contrast | 72% | Random effects | 0.81, 1.11 (0.48, 2.55) | ||||
| A2455G | AML | Caucasian | 1 | Recessive | |||
| Dominant | Fixed effects | 0.47, 0.78 (0.39, 1.53) | |||||
| Allele contrast | Fixed effects | 0.48, 0.78 (0.40, 1.53) | |||||
| Asian | 1 | Recessive | Fixed effects | 0.39, 0.79 (0.47, 1.35) | |||
| Dominant | Fixed effects | 0.42, 1.09 (0.88, 1.36) | |||||
| Allele contrast | Fixed effects | 0.72, 1.03 (0.87, 1.23) | |||||
| Mix | 2 | Recessive | 0% | Fixed effects | 0.50, 1.64 (0.39, 6.92) | ||
| Dominant | 0% | Fixed effects | 0.0009, 1.96 (1.32, 2.90) | ||||
| Allele contrast | 0% | Fixed effects | 0.002, 1.72 (1.22, 2.43) | ||||
| C4887A | AML | Caucasian | 1 | Recessive | Fixed effects | 0.39, 2.85 (0.26, 31.63) | |
| Dominant | Fixed effects | 0.005, 2.16 (1.27, 3.69) | |||||
| Allele contrast | Fixed effects | 0.004, 2.08 (1.26, 3.44) | |||||
| A2455G | CML | Caucasian | 2 | Recessive | 17% | Fixed effects | 0.004, 19.61 (2.62, 146.76) |
| Dominant | 69% | Random effects | 0.0010, 3.93 (1.74, 8.87) | ||||
| Allele contrast | 1% | Fixed effects | P,0.00001, 3.92 (2.61, 5.90) | ||||
| Mix | 1 | Recessive | Fixed effects | 0.89, 0.80 (0.03, 20.00) | |||
| Dominant | Random effects | 0.79, 0.88 (0.34, 2.29) | |||||
| Allele contrast | Fixed effects | 0.70, 0.84 (0.34, 2.07) |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CI, confidence interval; CML, chronic myeloid leukemia; OR, odds ratio; SNP, single nucleotide polymorphism.
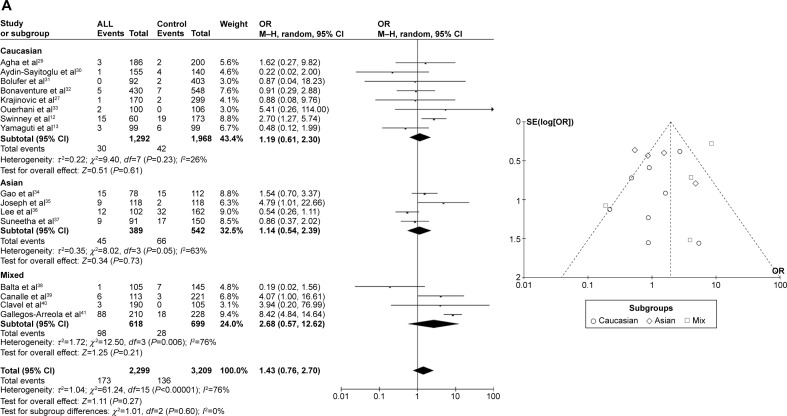
CYP1A1 T3801C was not associated with ALL risk in Caucasians or Asians
In the meta-analysis of 16 studies that involved data from 2,299 cases and 3,209 controls and investigated the effect of the T3801C polymorphism on the risk of ALL, a high degree of heterogeneity was observed under the recessive model (I2=76%, P<0.00001), dominant model (I2=68%, P<0.00001), and allele-contrast model (I2=80%, P<0.00001). A sensitivity analysis was conducted since I2 exceeded 50% for all three models. However, removing any particular article from the analysis did not reduce the degree of heterogeneity, and so the random effects model was used. This identified an association between the T3801C SNP and the risk of ALL under the dominant model (P=0.006, OR =1.39, 95% CI =1.10–1.76) and the allele-contrast model (P=0.03, OR =1.31, 95% CI =1.02–1.67), but not under the recessive model (P=0.27, OR =1.43, 95% CI =0.76–2.70). Moreover, the funnel plot did not reveal any obvious publication bias.
Different results can be obtained when including different studies due to population differences in the T3801C mutation frequency or linkage disequilibrium block. Subgroup analysis according to race was conducted, which revealed no marked associations in the three genetic comparisons among Asians (recessive model: P=0.73, OR =1.14, 95% CI =0.54–2.39; dominant model P=0.06, OR =1.64, 95% CI =0.98–2.73; and allele-contrast model: P=0.19, OR =1.35, 95% CI =0.86–2.14) and Caucasians (recessive model: P=0.28, OR =1.31, 95% CI =0.81–2.12; dominant model: P=0.10, OR =1.30, 95% CI =0.95–1.79; and allele-contrast model: P=0.05, OR =1.18, 95% CI =1.00–1.39) (Figure 2).
Figure 2.
Meta-analysis of the association between CYP1A1 T3801C gene polymorphism and ALL risk under three models: (A) recessive, (B) dominant, and (C) allele contrast.
Abbreviations: ALL, acute lymphoblastic leukemia; CI, confidence interval; M–H, Mantel–Haenszel type; OR, odds ratio; SE, standard error.
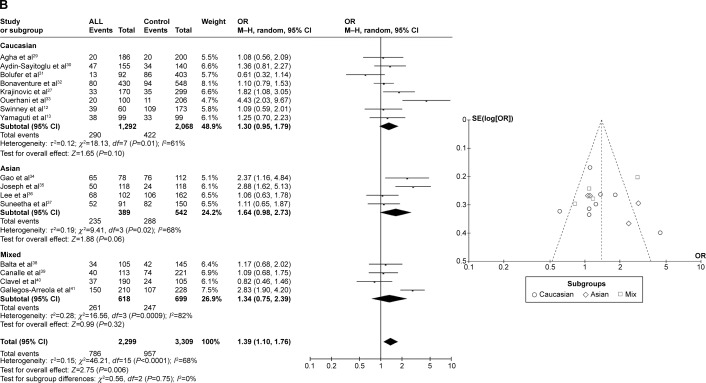
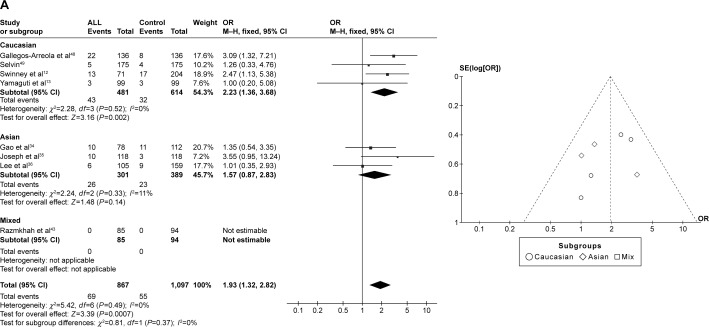
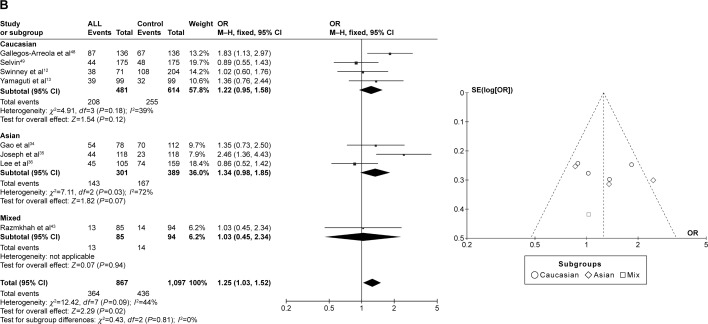
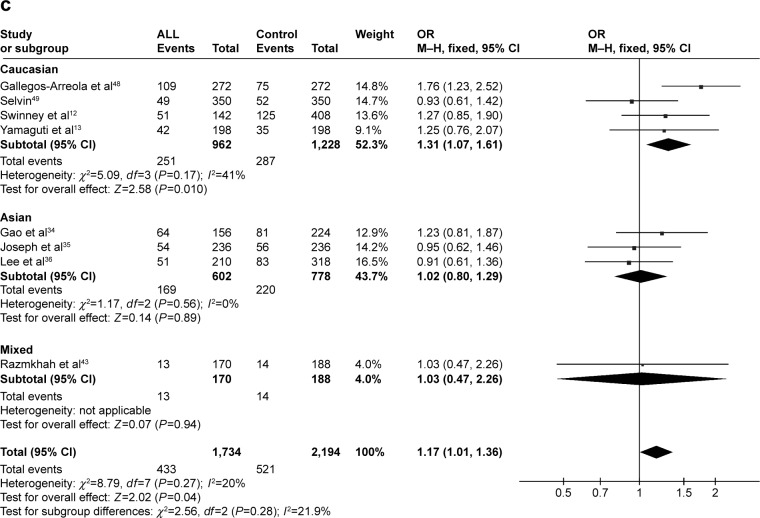
CYP1A1 A2455G was associated with ALL risk in Caucasians
In the meta-analysis of eight studies that involved the data from 1,734 patients and 2,194 controls and investigated the effect of the A2455G polymorphism on the risk of ALL, the forest plot revealed no heterogeneity under the recessive model (I2=0%, P=0.49), dominant model (I2=44%, P=0.09), and allele-contrast model (I2=20%, P=0.27). The fixed effects model was therefore used for these three contrast models, which revealed an association between the A2455G polymorphism and the risk of ALL under the recessive model (P=0.0007, OR =1.93, 95% CI =1.32–2.82), the dominant model (P=0.02, OR =1.25, 95% CI =1.03–1.52), and the allele-contrast model (P=0.04, OR =1.17, 95% CI =1.01–1.36). The funnel plot did not reveal any obvious publication bias.
In subgroup analysis according to race, the risk of leukemia was higher in Caucasians under the recessive model (P=0.002, OR =2.23, 95% CI =1.36–3.68) and the allele-contrast model (P=0.010, OR =1.31, 95% CI =1.07–1.61), but not under the dominant model (P=0.12, OR =1.22, 95% CI =0.95–1.58) (Figure 3).
Figure 3.
Meta-analysis of the association between CYP1A1 A2455G gene polymorphism and ALL risk under three models: (A) recessive, (B) dominant, and (C) allele contrast.
Abbreviations: CML, chronic myeloid leukemia; CI, confidence interval; M–H, Mantel–Haenszel type; OR, odds ratio; ALL, acute lymphoblastic leukemia.
CYP1A1 C4887A was not associated with ALL risk in Caucasians
In the meta-analysis of the single study that involved 177 cases and 295 controls (all of whom were Caucasians) and investigated the effect of the C4887A polymorphism on the risk of ALL, no association was found between the CYP1A1 C4887A SNP and the risk of ALL under any contrast model when using the fixed effects model.
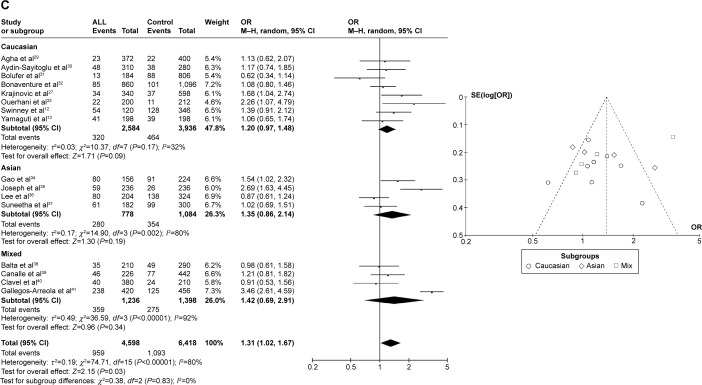
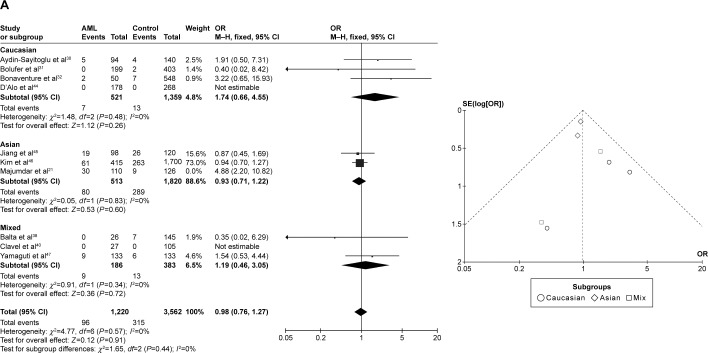
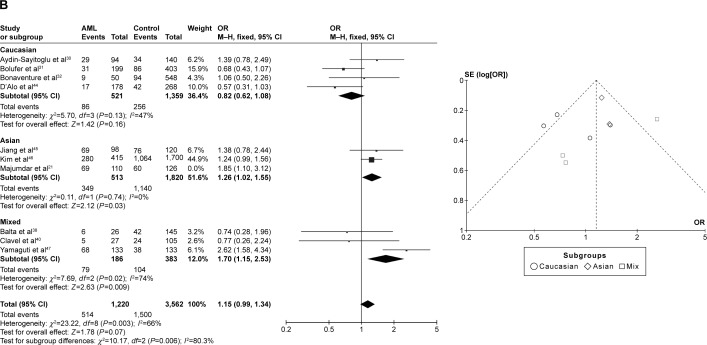
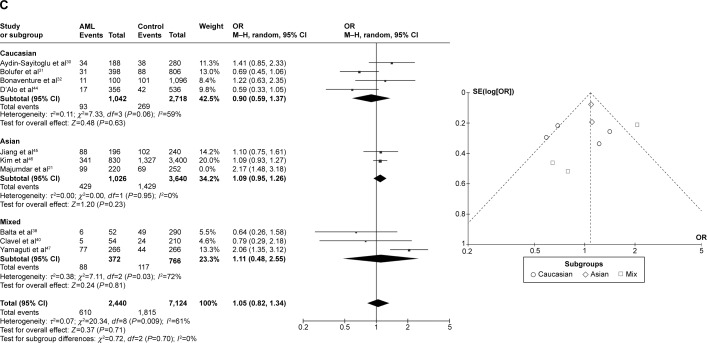
CYP1A1 T3801C was associated with AML risk in Asians
In the meta-analysis of ten studies that involved 1,330 cases and 3,688 controls and investigated the effect of the T3801C polymorphism on the risk of AML, a high degree of heterogeneity was observed under the recessive model (I2=62%, P=0.010), dominant model (I2=65%, P=0.002), and allele-contrast model (I2=72%, P=0.0002). A sensitivity analysis was therefore conducted, which revealed that the study of Majumdar et al21 was an outlier. Removing their data from the analysis reduced the degree of heterogeneity. Under the allele-contrast model, the degree of heterogeneity was still high, and so a random effects model was applied. No association was found between the T3801C SNP and the risk of AML under the recessive model (P=0.91, OR =0.98, 95% CI =0.76–1.27), the dominant model (P=0.07, OR =1.15, 95% CI =0.99–1.34), or the allele-contrast model (P=0.71, OR =1.05, 95% CI =0.82–1.34). The funnel plot did not reveal any obvious publication bias.
However, in subgroup analysis according to race, the risk was increased among Asians under the dominant model (P=0.03, OR =1.26, 95% CI =1.02–1.55), but not under the recessive model (P=0.60, OR =0.93, 95% CI =0.71–1.22) or the allele-contrast model (P=0.23, OR =1.09, 95% CI =0.95–1.26) (Figure 4).
Figure 4.
Meta-analysis of the association between CYP1A1 T3801C gene polymorphism and AML risk under three models: (A) recessive, (B) dominant, and (C) allele contrast.
Abbreviations: AML, acute myeloid leukemia; CI, confidence interval; M–H, Mantel–Haenszel type; OR, odds ratio; SE, standard error.
CYP1A1 A2455G was not associated with AML risk in Caucasians or Asians
The meta-analysis of four studies that involved 846 patients and 2,201 controls – one study involved Caucasians, one study involved Asians, and two studies involved mixed races – found no association between the A2455G SNP and the risk of AML under any contrast model.
CYP1A1 C4887A was associated with AML risk in Caucasians
In the meta-analysis of the single study that involved 193 cases and 273 controls (all of whom were Caucasians), when using the fixed effects model there was a significant association between the C4887A SNP and the risk of AML under the dominant model (P=0.005, OR =2.16, 95% CI =1.27–3.69) and the allele-contrast model (P=0.004, OR =2.08, 95% CI =1.26–3.44). However, this relationship needs to be confirmed in further studies.
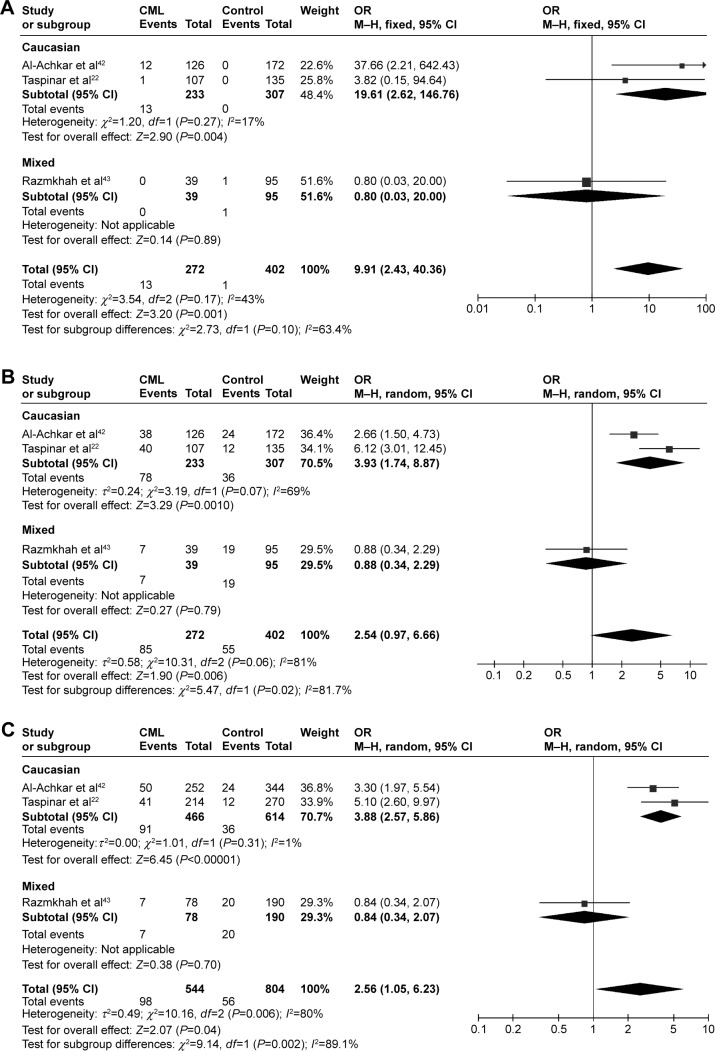
CYP1A1 A2455G was associated with CML risk in Caucasians
In the meta-analysis of the three studies that involved 272 patients and 402 controls and investigated the effect of the A2455G polymorphism on the risk of CML, the forest plot revealed no heterogeneity under the recessive model (I2=43%, P=0.17), and so the fixed effects model was applied. There was a high degree of heterogeneity under the dominant model (I2=81%, P=0.006) and the allele-contrast model (I2=80%, P=0.006), and so a sensitivity analysis was conducted. However, removing any particular study from the analysis did not reduce the degree of heterogeneity, and so the random effects model was used for these two contrast models. An association was found between the A2455G SNP and the risk of CML under the recessive model (P=0.001, OR =9.91, 95% CI =2.43–40.36) and the allele-contrast model (P=0.04, OR =2.56, 95% CI =1.05–6.23).
In subgroup analysis according to race, the risk of leukemia was increased in Caucasians under the recessive model (P=0.004, OR =19.61, 95% CI =2.62–146.76), dominant model (P=0.0010, OR =3.93, 95% CI =1.74–8.87), and allele-contrast model (P<0.00001, OR =3.92, 95% CI =2.61–5.90) (Figure 5).
Figure 5.
Meta-analysis of the association between CYP1A1 A2455G gene polymorphism and CML risk under three models: (A) recessive, (B) dominant, and (C) allele contrast.
Abbreviations: ALL, acute lymphoblastic leukemia; CI, confidence interval; M–H, Mantel–Haenszel type; OR, odds ratio; SE, standard error; CML, chronic myeloid leukemia.
Discussion
This study identified 26 articles describing research into the associations between the CYP1A1 T3801C, CYP1A1 A2455G, and CYP1A1 C4887A polymorphisms and the risks of ALL, AML, and CML. The conclusions drawn using the data obtained in these studies were not consistent. This situation prompted the use of meta-analysis in this study – which is a common method applied in systematic reviews – to integrate these contradictions, and to obtain more accurate results. All of the finally included studies were of high quality and had a rigorous scientific design, accurate data, and clear results. The main conclusion is that the presence of the CYP1A1 T3801C polymorphism increased the risks of AML among Asians, while the CYP1A1 A2455G polymorphism increased the risks of ALL among Caucasians.
Few studies have investigated the CYP1A1 polymorphism as a possible risk factor for CML.22 The present meta-analysis found an association between the A2455G SNP and the risk of CML among Caucasians. However, this result was based on a relatively small number of cases where the statistical power was weak, and so, this conclusion should be treated with caution. The accumulation of disadvantageous environmental factors might be required to develop chronic disease, and so, investigating environment – genetic interactions – would be meaningful for this kind of chronic disease. Although, in this study, we endeavored to extract relevant information regarding smoking and alcohol consumption from the primary literature, insufficient data were obtained. Further investigations concerning the interactions between smoking, alcohol consumption, and gene variations with CML are required.
This meta-analysis included studying the associations between three SNPs (T3801C, A2455G, and C4887A) and the risk of AML. The results showed that the CYP1A1 T3801C polymorphism might not be significantly correlated with the risk of AML. Possible racial variations in presentation, treatment patterns, and survival with respect to AML might exist,23 which made subgroup analysis according to race necessary. We found a significant increased risk among Asians under the dominant model, but not under the recessive or allele-contrast model. Compared with the meta-analysis of Zhuo et al24 who also researched the association between T3801C and the risk of AML, the dominant model for Asians utilized a fixed effects in the present meta-analysis, which makes our conclusions more convincing. For A2455G, the subgroup analyses found no association among either Asians or Caucasians with the risk of AML. The meta-analysis of Zhuo et al25 also investigated the association between A2455G and the risk of AML, but our conclusions are more convincing due to the inclusion of a larger number of articles. In addition, we believe that the article of Zhuo et al contained some data errors.25 The research data of Majumdar et al21 defined the allele *2B as an m1 (T3801C) and m2 (A2455G) combined locus, and so the data for *2B cannot represent the data for A2455G. Moreover, the raw data for A2455G genotypes in controls did not conform with HWE, and so our meta-analysis excluded that group of data. Zhuo et al also included the combined mutation data, which undermines the credibility of their meta-analysis.25 Our meta-analysis included only one study of the C4887A SNP and revealed an association between this SNP and the risk of AML under the dominant and allele-contrast models. However, the sample was too small to reliably characterize the intrinsic link between the CYP1A1 C4887A polymorphism and risk of AML. Further investigations are needed to confirm this conclusion.
Our meta-analysis also studied the associations between three SNPs (T3801C, A2455G, and C4887A) and the risk of ALL. The results showed that the CYP1A1 T3801C polymorphism might have a strong correlation with an increased risk of ALL. However, in the subgroup analyses stratified by race, the data did not reveal such an association among either Asians or Caucasians when they were analyzed separately. Our analysis included more studies than that of Zhuo et al26 which also researched the association between T3801C and the risk of ALL, and so our conclusions are more plausible. The results for the CYP1A1 A2455G polymorphism indicated that this is significantly correlated with the risk of ALL among Caucasians. Again, our analysis included more studies than that of Zhuo et al25 which also researched the association between A2455G and ALL. In addition, we believe that the article of Zhuo et al contained some data errors.25 Krajinovic et al27 found that CYP1A1*2B represents both m1 (T3801C) and m2 (A2455G), and so, the data for *2B cannot represent the data for A2455G alone. Moreover, Zhuo et al included combined mutation data, which would produce incorrect results.25 More importantly, our study only extracted data for a single race from the study of Swinney et al12 which makes our race subgroup analyses more credible. Only one study related to the CYP1A1 C4887A polymorphism was included in our meta-analysis. The results showed that this polymorphism might not be correlated with the risk of ALL. Also, the subgroup analyses found no association among either Asians or Caucasians when they were analyzed separately. The sample was too small to reliably characterize the intrinsic link between the CYP1A1 C4887A polymorphism and the risk of ALL, and so further studies are needed to clarify this.
There are two implications of this study that are particularly significant: 1) the CYP1A1 A2455G polymorphism can be used as a screening index for the risk of ALL and CML, while the CYP1A1 T3801C polymorphism can be used as a screening index for the risk of AML; and 2) genetic screening can be used to identify individuals with the susceptible genotype who could take early defense measures to reduce their risk of leukemia.
Three limitations of this meta-analysis should be considered when interpreting its findings. First, our findings only related to the relationships between specific genes and diseases, whereas leukemia is influenced by multiple factors, and so, future studies need to consider age, race, family history, and environmental and lifestyle factors when attempting to draw more accurate conclusions. Second, a relatively small number of eligible studies were included in this meta-analysis, which could limit the generalizability of the results. This also restricted the statistical power of the subgroup analyses. As is well known, the inclusion of studies with very small samples may lead to overestimations of the associations.28 Third, only published studies were included in the present meta-analysis. A certain degree of publication bias is always present due to the possibility of nonsignificant or negative findings not being published.
In conclusion, this meta-analysis suggests that Asians who carry the T3801C C allele might have an increased risk of AML, while Caucasians with the A2455G GG genotype might have an increased risk of ALL. In addition, we found an association between the CYP1A1 A2455G polymorphism and risk of CML among Caucasians. Future large studies of these sites are recommended in order to confirm the findings of this study and also to eventually identify the underlying mechanisms.
Acknowledgments
This work was supported by the Natural Science Foundation of Shaanxi Province (number 2015JM8415) and the Fundamental Research Funds for the Central Universities of China (number 2011jdhz55).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Huang Y, Xie D, Tang N, et al. XRCC1 Arg399Gln variation and leukemia susceptibility: evidence from 2,647 cases and 5,518 controls. Tumour Biol. 2014;35(1):799–808. doi: 10.1007/s13277-013-1110-7. [DOI] [PubMed] [Google Scholar]
- 2.Infante-Rivard C, Labuda D, Krajinovic M, Sinnett D. Risk of childhood leukemia associated with exposure to pesticides and with gene polymorphisms. Epidemiology. 1999;10(5):481–487. [PubMed] [Google Scholar]
- 3.Perera FP, Mooney LA, Dickey CP, et al. Molecular epidemiology in environmental carcinogenesis. Environ Health Perspect. 1996;104(Suppl 3):441–443. doi: 10.1289/ehp.96104s3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brisson GD, Alves LR, Pombo-de-Oliveira MS. Genetic susceptibility in childhood acute leukaemias: a systematic review. Ecancermedicalscience. 2015;9:539. doi: 10.3332/ecancer.2015.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guengerich FP, Shimada T. Activation of procarcinogens by human cytochrome P450 enzymes. Mutat Res. 1998;400(1–2):201–213. doi: 10.1016/s0027-5107(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 6.Dufour C, Svahn J, Bacigalupo A, et al. Genetic polymorphisms of CYP3A4, GSTT1, GSTM1, GSTP1 and NQO1 and the risk of acquired idiopathic aplastic anemia in Caucasian patients. Haematologica. 2005;90(8):1027–1031. [PubMed] [Google Scholar]
- 7.Hung RJ, Boffetta P, Brockmoller J, et al. CYP1A1 and GSTM1 genetic polymorphisms and lung cancer risk in Caucasian non-smokers: a pooled analysis. Carcinogenesis. 2003;24(5):875–882. doi: 10.1093/carcin/bgg026. [DOI] [PubMed] [Google Scholar]
- 8.Takanashi M, Morimoto A, Yagi T, et al. Impact of glutathione S-transferase gene deletion on early relapse in childhood B-precursor acute lymphoblastic leukemia. Haematologica. 2003;88(11):1238–1244. [PubMed] [Google Scholar]
- 9.Crofts F, Cosma GN, Currie D, Taioli E, Toniolo P, Garte SJ. A novel CYP1A1 gene polymorphism in African-Americans. Carcinogenesis. 1993;14(9):1729–1731. doi: 10.1093/carcin/14.9.1729. [DOI] [PubMed] [Google Scholar]
- 10.Shah PP, Saurabh K, Pant MC, Mathur N, Parmar D. Evidence for increased cytochrome P450 1A1 expression in blood lymphocytes of lung cancer patients. Mutat Res. 2009;670(1–2):74–78. doi: 10.1016/j.mrfmmm.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YD, Tan LN, Zhang XL, Wei HY, Xiong H, Hu Q. Meta-analysis of cytochrome P4501A1 MspI gene polymorphism and childhood acute leukemia. Biomed Environ Sci. 2011;24(6):683–687. doi: 10.3967/0895-3988.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Swinney RM, Beuten J, Collier AR, et al. Polymorphisms in CYP1A1 and ethnic-specific susceptibility to acute lymphoblastic leukemia in children. Cancer Epidemiol Biomarkers Prev. 2011;20(7):1537–1542. doi: 10.1158/1055-9965.EPI-10-1265. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguti GG, Lourenco GJ, Silveira VS, Tone LG, Lopes LF, Lima CS. Increased risk for acute lymphoblastic leukemia in children with cytochrome P450A1 (CYP1A1)- and NAD(P)H:quinone oxidoreductase 1 (NQO1)-inherited gene variants. Acta Haematol. 2010;124(3):182–184. doi: 10.1159/000320275. [DOI] [PubMed] [Google Scholar]
- 14.Kawajiri K, Nakachi K, Imai K, Yoshii A, Shinoda N, Watanabe J. Identification of genetically high risk individuals to lung cancer by DNA polymorphisms of the cytochrome P450IA1 gene. FEBS Lett. 1990;263(1):131–133. doi: 10.1016/0014-5793(90)80721-t. [DOI] [PubMed] [Google Scholar]
- 15.Cascorbi I, Brockmoller J, Roots I. A C4887A polymorphism in exon 7 of human CYP1A1: population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res. 1996;56(21):4965–4969. [PubMed] [Google Scholar]
- 16.Rojas M, Cascorbi I, Alexandrov K, et al. Modulation of benzo[a] pyrene diolepoxide-DNA adduct levels in human white blood cells by CYP1A1, GSTM1 and GSTT1 polymorphism. Carcinogenesis. 2000;21(1):35–41. doi: 10.1093/carcin/21.1.35. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Lopez E, Martin-Guerrero I, Ballesteros J, Garcia-Orad A. A systematic review and meta-analysis of MTHFR polymorphisms in methotrexate toxicity prediction in pediatric acute lymphoblastic leukemia. Pharmacogenomics J. 2013;13(6):498–506. doi: 10.1038/tpj.2012.44. [DOI] [PubMed] [Google Scholar]
- 18.Gurion R, Gafter-Gvili A, Vidal L, et al. Has the time for first-line treatment with second generation tyrosine kinase inhibitors in patients with chronic myelogenous leukemia already come? Systematic review and meta-analysis. Haematologica. 2013;98(1):95–102. doi: 10.3324/haematol.2012.063172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen HC, Hu WX, Liu QX, et al. Genetic polymorphisms of metabolic enzymes CYP1A1, CYP2D6, GSTM1 and GSTT1 and leukemia susceptibility. Eur J Cancer Prev. 2008;17(3):251–258. doi: 10.1097/CEJ.0b013e3282b72093. [DOI] [PubMed] [Google Scholar]
- 21.Majumdar S, Mondal BC, Ghosh M, et al. Association of cytochrome P450, glutathione S-transferase and N-acetyl transferase 2 gene polymorphisms with incidence of acute myeloid leukemia. Eur J Cancer Prev. 2008;17(2):125–132. doi: 10.1097/CEJ.0b013e3282b6fd68. [DOI] [PubMed] [Google Scholar]
- 22.Taspinar M, Aydos SE, Comez O, Elhan AH, Karabulut HG, Sunguroglu A. CYP1A1, GST gene polymorphisms and risk of chronic myeloid leukemia. Swiss Med Wkly. 2008;138(1–2):12–17. doi: 10.4414/smw.2008.12036. [DOI] [PubMed] [Google Scholar]
- 23.Bierenbaum J, Davidoff AJ, Ning Y, Tidwell ML, Gojo I, Baer MR. Racial differences in presentation, referral and treatment patterns and survival in adult patients with acute myeloid leukemia: a single-institution experience. Leuk Res. 2012;36(2):140–145. doi: 10.1016/j.leukres.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuo W, Zhang L, Wang Y, Zhu B, Chen Z. CYP1A1 MspI polymorphism and acute myeloid leukemia risk: meta-analyses based on 5,018 subjects. J Exp Clin Cancer Res. 2012;31:62. doi: 10.1186/1756-9966-31-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuo W, Zhang L, Zhu B, Qiu Z, Chen Z. Association between CYP1A1 Ile462Val variation and acute leukemia risk: meta-analyses including 2,164 cases and 4,160 controls. PLoS One. 2012;7(10):e46974. doi: 10.1371/journal.pone.0046974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuo W, Zhang L, Qiu Z, Zhu B, Chen Z. Does cytochrome P450 1A1 MspI polymorphism increase acute lymphoblastic leukemia risk? Evidence from 2,013 cases and 2,903 controls. Gene. 2012;510(1):14–21. doi: 10.1016/j.gene.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 27.Krajinovic M, Labuda D, Richer C, Karimi S, Sinnett D. Susceptibility to childhood acute lymphoblastic leukemia: influence of CYP1A1, CYP2D6, GSTM1, and GSTT1 genetic polymorphisms. Blood. 1999;93(5):1496–1501. [PubMed] [Google Scholar]
- 28.Das P, Shaik AP, Bammidi VK. Meta-analysis study of glutathione-S-transferases (GSTM1, GSTP1, and GSTT1) gene polymorphisms and risk of acute myeloid leukemia. Leuk Lymphoma. 2009;50(8):1345–1351. doi: 10.1080/10428190903003236. [DOI] [PubMed] [Google Scholar]
- 29.Agha A, Shabaan H, Abdel-Gawad E, El-Ghannam D. Polymorphism of CYP1A1 gene and susceptibility to childhood acute lymphoblastic leukemia in Egypt. Leuk Lymphoma. 2014;55(3):618–623. doi: 10.3109/10428194.2013.809527. [DOI] [PubMed] [Google Scholar]
- 30.Aydin-Sayitoglu M, Hatirnaz O, Erensoy N, Ozbek U. Role of CYP2D6, CYP1A1, CYP2E1, GSTT1, and GSTM1 genes in the susceptibility to acute leukemias. Am J Hematol. 2006;81(3):162–170. doi: 10.1002/ajh.20434. [DOI] [PubMed] [Google Scholar]
- 31.Bolufer P, Collado M, Barragan E, et al. The potential effect of gender in combination with common genetic polymorphisms of drug-metabolizing enzymes on the risk of developing acute leukemia. Haematologica. 2007;92(3):308–314. doi: 10.3324/haematol.10752. [DOI] [PubMed] [Google Scholar]
- 32.Bonaventure A, Goujon-Bellec S, Rudant J, et al. Maternal smoking during pregnancy, genetic polymorphisms of metabolic enzymes, and childhood acute leukemia: the ESCALE study (SFCE) Cancer Causes Control. 2012;23(2):329–345. doi: 10.1007/s10552-011-9882-9. [DOI] [PubMed] [Google Scholar]
- 33.Ouerhani S, Cherif N, Bahri I, Safra I, Menif S, Abbes S. Genetic polymorphisms of NQO1, CYP1A1 and TPMT and susceptibility to acute lymphoblastic leukemia in a Tunisian population. Mol Biol Rep. 2013;40(2):1307–1314. doi: 10.1007/s11033-012-2174-y. [DOI] [PubMed] [Google Scholar]
- 34.Gao J, Huang Y, Liang J, Fang X. Relationship between genetic polymorphisms of CYP1A1 and ALL. Chin J Birth Health Heredity. 2003;11:21–24. [Google Scholar]
- 35.Joseph T, Kusumakumary P, Chacko P, Abraham A, Radhakrishna PM. Genetic polymorphism of CYP1A1, CYP2D6, GSTM1 and GSTT1 and susceptibility to acute lymphoblastic leukaemia in Indian children. Pediatr Blood Cancer. 2004;43(5):560–567. doi: 10.1002/pbc.20074. [DOI] [PubMed] [Google Scholar]
- 36.Lee KM, Ward MH, Han S, et al. Paternal smoking, genetic polymorphisms in CYP1A1 and childhood leukemia risk. Leuk Res. 2009;33(2):250–258. doi: 10.1016/j.leukres.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suneetha KJ, Nancy KN, Rajalekshmy KR, Rama R, Sagar TG, Rajkumar T. Role of glutathione-s-transferase and CYP1A1*2A polymorphisms in the therapy outcome of south Indian acute lymphoblastic leukemia patients. Indian J Med Paediatr Oncol. 2011;32(1):25–29. doi: 10.4103/0971-5851.81886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balta G, Yuksek N, Ozyurek E, et al. Characterization of MTHFR, GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes in childhood acute leukemia. Am J Hematol. 2003;73(3):154–160. doi: 10.1002/ajh.10339. [DOI] [PubMed] [Google Scholar]
- 39.Canalle R, Burim RV, Tone LG, Takahashi CS. Genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukemia. Environ Mol Mutagen. 2004;43(2):100–109. doi: 10.1002/em.20003. [DOI] [PubMed] [Google Scholar]
- 40.Clavel J, Bellec S, Rebouissou S, et al. Childhood leukaemia, polymorphisms of metabolism enzyme genes, and interactions with maternal tobacco, coffee and alcohol consumption during pregnancy. Eur J Cancer Prev. 2005;14(6):531–540. doi: 10.1097/00008469-200512000-00007. [DOI] [PubMed] [Google Scholar]
- 41.Gallegos-Arreola MP, Gonzalez-Garcia JR, Figuera LE, Puebla-Perez AM, Delgado-Lamas JL, Zuniga-Gonzalez GM. Distribution of CYP1A1*2A polymorphism in adult patients with acute lymphoblastic leukemia in a Mexican population. Blood Cells Mol Dis. 2008;41(1):91–94. doi: 10.1016/j.bcmd.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Al-Achkar W, Azeiz G, Moassass F, Wafa A. Influence of CYP1A1, GST polymorphisms and susceptibility risk of chronic myeloid leukemia in Syrian population. Med Oncol. 2014;31(5):889. doi: 10.1007/s12032-014-0889-4. [DOI] [PubMed] [Google Scholar]
- 43.Razmkhah F, Pazhakh V, Zaker F, Atashrazm F, Sheikhi M. Frequency of CYP1A1*2C polymorphism in patients with leukemia in the Iranian population. Lab Med. 2011;42(4):220–223. [Google Scholar]
- 44.D’Alo F, Voso MT, Guidi F, et al. Polymorphisms of CYP1A1 and glutathione S-transferase and susceptibility to adult acute myeloid leukemia. Haematologica. 2004;89(6):664–670. [PubMed] [Google Scholar]
- 45.Jiang L, Chen M, Qin G. Association between the polymorphisms of cytochrome P4501A1 and glutathione S-transferase M1, T1 Genes and acute myeloid leukemia in Guangxi. Guangxi Med J. 2008;80:464–466. [Google Scholar]
- 46.Kim HN, Kim NY, Yu L, et al. Association of GSTT1 polymorphism with acute myeloid leukemia risk is dependent on smoking status. Leuk Lymphoma. 2012;53(4):681–687. doi: 10.3109/10428194.2011.625576. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguti GG, Lourenco GJ, Costa FF, Lima CS. High risk of ‘de novo’ acute myeloid leukaemia in individuals with cytochrome P450 A1 (CYP1A1) and NAD(P)H:quinone oxidoreductase 1 (NQO1) gene defects. Eur J Haematol. 2009;83(3):270–272. doi: 10.1111/j.1600-0609.2009.01272.x. [DOI] [PubMed] [Google Scholar]
- 48.Gallegos-Arreola MP, Batista-Gonzalez CM, Delgado-Lamas JL, et al. Cytochrome P4501A1 polymorphism is associated with susceptibility to acute lymphoblastic leukemia in adult Mexican patients. Blood Cells Mol Dis. 2004;33(3):326–329. doi: 10.1016/j.bcmd.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Selvin S. Cytochrome P450 1A1 polymorphism and childhood leukemia: an analysis of matched pairs case-control genotype data. Cancer Epidemiol Biomarkers Prev. 2004;13(8):1371–1374. [PubMed] [Google Scholar]