Abstract
We report a family with domestic exposure to asbestos and multiple cancers, including eight pleural malignant mesotheliomas and several other lung/pleural tumors. DNA sequence analysis revealed no evidence for an inherited mutation of BAP1. Sequence analysis of other potentially relevant genes, including TP53, CDKN2A and BARD1, also revealed no mutations. DNA microarray analysis of two mesotheliomas revealed multiple genomic imbalances including consistent losses of overlappping segments in 2q, 6q, 9p, 14q, 15q and 22q, but no losses of chromosome 3 harboring the BAP1 locus. However, immunohistochemistry demonstrated loss of nuclear BAP1 staining in 3 of 6 mesotheliomas tested, suggesting that somatic alterations of BAP1 occurred in a subset of tumors from this family. Since mesothelioma could be confirmed in only a single generation, domestic exposure to asbestos may be the predominant cause of mesothelioma in this family. Given the existence of unspecified malignant pleural tumors and lung cancers in a prior generation, the possibility that some other tumor susceptibility or modifier gene(s) may contribute to the high incidence of mesothelioma in this family is discussed. Because the incidence of mesothelioma in this family is higher than expected even in heavily exposed asbestos workers, we conclude that both asbestos and genetic factors have played a role in the high rate of mesothelioma and potentially other pleural/lung cancers seen in this family.
Keywords: malignant mesothelioma, asbestos, familial cancer, cancer predisposition, BAP1
Introduction
Malignant mesothelioma (MM) is an uncommon cancer associated with asbestos exposure. Familial clustering in close relatives has been described in multiple reports and cannot be explained by chance alone (1). In addition to shared asbestos exposures, MM clustering in some families may suggest the contribution of inherited genes (low-penetrance alleles) in the development of this malignancy. A genetic factor predisposing to MM was recently discovered, namely germline mutation of the BAP1 (BRCA1-associated protein 1) gene in two families with a high incidence of MM and only modest exposure to asbestos (2,3). The association of germline BAP1 mutations with familial MM has been confirmed in a series of recent reports (4-7). Germline BAP1 mutations have also been repeatedly associated with other tumor types, including uveal melanomas, cutaneous melanomas, benign melanocytic tumors, kidney cancers, and basal cell carcinomas (3-14). Somatic mutations and/or deletions have been described in MMs of both individuals with germline BAP1 mutations (3) and in sporadic cases lacking a germline mutation (3,15). While Sanger sequencing revealed point mutations in 20-25% of sporadic MMs in these earlier studies (3,15), Yoshikawa et al. found biallelic gene alterations, including homozygous deletions of part or all of the BAP1 gene as well as sequence-level mutations, in 61% of sporadic MMs (16). Subsequent studies with newer next generation and multiplex ligation-dependent probe amplification (MLPA) platforms confirmed a similarly high incidence of BAP1 mutations in this disease (17,18). Based on the reported connection between familial MM and inheritance of a BAP1 mutation, we decided to analyze the BAP1 status in a family with many cases of pleural MM and asbestos exposure.
Materials and methods
Family History and Patient Samples
Over the past 18 years, since the identification of the index patient (III-2) in 1996 and the descripton of four MM cases (19), we have continued to follow this Italian family with multiple cases of MM and other malignancies (Fig. 1). Metaphase-based comparative genomic hybridization (CGH) analysis on tumor samples uncovered DNA losses involving 1p, 6q, 9p, 13q, and 14q (20), each of which is a chromosomal arm that is commonly lost in sporadic MM (21). An update of the history of this extended family was published in 2014 (2). Between 1987 and 2012, there were six women (mean age 62 yrs) and two men (mean age 67 yrs) who developed pleural MM in generation III. In addition to the eight confirmed MMs, two female family members in a prior generation (II-3 and II-8) had pleural cancers (highly suspicious of MM but unconfirmed), without radiological evidence of a primary tumor in the lung or elsewhere. The kindred had exposure to asbestos in the domestic setting; we demonstrated the presence of asbestos in lung tissue from three MM cases, using transmission electron microscopy (III-5: crocidolite, elevated fiber burden, and asbestos bodies) and optical microscopy (III-2 and III-17: asbestos bodies). Furthermore, there was radiological and/or histological evidence of bilateral pleural plaques in six subjects (III-2, III-5, III-6, III-7, III-15 and III-17). For the remaining two MM patients (III-1 and III-4: diagnosed in 1987 and 1988, respectively), X-rays and CT scans were not available for review. Notably, family member II-4 was a brick furnace worker who had significant, lengthy exposure to asbestos occupationally and likely brought asbestos fibers home on his clothes. All seven of his children developed one or more cancer(s) in adulthood, including six with MM. Other family members who developed MM or a pleural cancer of unknown histology, including II-8, III-15 and III-17, were almost daily visitors to the home of II-4.
Figure 1.
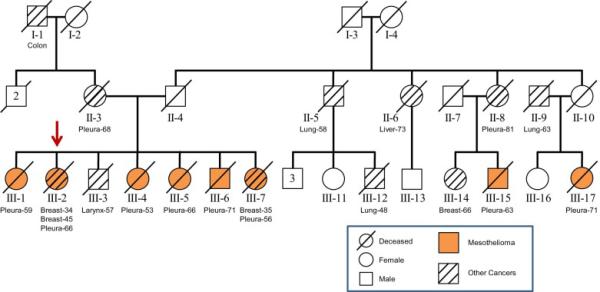
Pedigree showing multiple family members with pleural malignant mesothelioma (MM) in first-degree (6 cases) and third-degree (2 cases) relatives of the proband (arrow) as well as additional cancers in relatives of both paternal and maternal lines. Other malignancies involved the pleura (II-3 and II-8), larynx (III-3), lung (II-5, II-9, and III-12), breast (III-14), liver (II-6), and colon (I-1). Breast cancer also occurred in two MM patients (III-2, III-7) at early ages.
All MM patients in this family are deceased, and only very limited samples were available for mutation and/or immunohistochemical studies. Most specimens were formalin-fixed, paraffin-embedded (FFPE) tissues (from six MM cases). In addition, for one MM case (III-5), a pleural fluid sample was available that lacked any detectable genomic imbalances, based on a chromosome microarray analysis; moreover, ~90% of the cells were cytologically normal as determined by cytopathologic assessment of a corresponding cytospin preparation.
Sequence Analysis
Our main goal was to identify a predisposing (germline) mutation in this family, but unfortunately peripheral blood samples were unavailable; therefore, sequence analysis was performed on three FFPE specimens and one pleural fluid sample from a total of four MM cases. PCR products encompassing all exons and intronic splice regions of BAP1 were amplified for sequencing. Primers consisted of M13-F and M13-R sequences incorporated at the 5’ end to facilitate sequencing. PCR products were gel purified and sequenced using M13-For (GTAAAACGACGGCCAGT) and M13-Rev (CAGGAAACAGCTATGAC) primers. Primers for BAP1 and CDKN2A are shown in Table 1. Due to the inherent DNA fragmentation that can be found when using FFPE samples, primers were chosen to amplify small DNA fragments for Sanger sequencing. Satisfactory sequence data were obtained with all samples tested.
Table 1.
| Bap1 Exons | Forward primer | Reverse Primer |
|---|---|---|
| 1 | GTAAAACGACGGCCAGTGAGCCCAGAGGCGGAGCAG | CAGGAAACAGCTATGACGTCAGGCAGGCGCGTC |
| 2 | GTAAAACGACGGCCAGTGACGCGCCTGCCTGAC | CAGGAAACAGCTATGACCTTGACACCTGCGATGAGGAA |
| 3 | GTAAAACGACGGCCAGTCTCACTCATCAGGGGCTGTC | CAGGAAACAGCTATGACCAGCACTCTGGGTGTAAGGG |
| 4 | GTAAAACGACGGCCAGTAGTGATGACGCAGTGCAAAG | CAGGAAACAGCTATGACCTCCATTTCCACTTCCCAAG |
| 5 | GTAAAACGACGGCCAGTGAGGGGTGCTGTGTATGGG | CAGGAAACAGCTATGACCTGTGAGCCAGGATGAAGGC |
| 6 | GTAAAACGACGGCCAGTTGTGTTCCTTCCGATTCCTGG | CAGGAAACAGCTATGACAAACAGAGTCAGGGCCCAAAA |
| 7 | GTAAAACGACGGCCAGTGGTGGGAGTAGGGGGAGTATC | CAGGAAACAGCTATGACGGTAGGCAGAGACACCCAAC |
| 8 | GTAAAACGACGGCCAGTCAGGGTTTCCTTCTCGCTGA | CAGGAAACAGCTATGACCCCAAAGTAGGTACAGCTCCAG |
| 9 | GTAAAACGACGGCCAGTCCTGCCAGGATATCTGCCTC | CAGGAAACAGCTATGACTCAGAGACAAATGCTGTGGG |
| 10 | GTAAAACGACGGCCAGTAGGTCCTCAGCCCTTAGCTATT | CAGGAAACAGCTATGACTCAGACATTAGCGGGTGGCTC |
| 11 | GTAAAACGACGGCCAGTGGAGGTCCTGCCTGTGTTC | CAGGAAACAGCTATGACTCAAGTAGAGAATCCTGCAAGGG |
| 12 | GTAAAACGACGGCCAGTCCGAGCAGCACTTGTTTGTA | CAGGAAACAGCTATGACGGGATCCGAAGCACCTAGAAC |
| 13a | GTAAAACGACGGCCAGTCGTTCCCTTGCTTCACATCTTCT | CAGGAAACAGCTATGACCCGCTGCTAGTCTTGATGGA |
| 13b | GTAAAACGACGGCCAGTTGGCTGAGAAGCTCAAAGAGTC | CAGGAAACAGCTATGACCGCGTCGGGTTGGCTG |
| 13c | GTAAAACGACGGCCAGTAGTACAGACACGGCCTCTGA | CAGGAAACAGCTATGACGGTTGTAGCGTATGCAGTCAAC |
| 13d | GTAAAACGACGGCCAGTCCCACATCTCCAAGGTGCTT | CAGGAAACAGCTATGACCCTCCTGGGTGCACCAA |
| 14 | GTAAAACGACGGCCAGTAAAGTGTCCTGCACTCTGATGATT | CAGGAAACAGCTATGACGCCTTACCCTCTGCCAGGATTA |
| 15 | GTAAAACGACGGCCAGTGCATGGACTCGCTGCTCATC | CAGGAAACAGCTATGACTGGGTCCTTCTCTGGTCATCAA |
| 16 | GTAAAACGACGGCCAGTTCTGGCAAGATTGGCTCCAG | CAGGAAACAGCTATGACCTCAGCAGGGCATTCCAGTTA |
| 17 | GTAAAACGACGGCCAGTCATGAGAGCCTCAGCTCCT | CAGGAAACAGCTATGACGCAAGAGTGGGCTGCAGAG |
| CDKN2A Exons | Forward primer | Reverse Primer |
|---|---|---|
| 1b | GTAAAACGACGGCCAGTGCGCGCTCAGGGAAGG | CAGGAAACAGCTATGACACAAAACAAGTGCCGAATGC |
| 1a | GTAAAACGACGGCCAGTGAGCGCGGCTGGGAG | CAGGAAACAGCTATGACCAGAGTCGCCCGCCATC |
| 2 | GTAAAACGACGGCCAGTTTAGACACCTGGGGCTTGTG | CAGGAAACAGCTATGACTGGAAGCTCTCAGGGTACAA |
| 3 | GTAAAACGACGGCCAGTTTTCAATGCCGGTAGGGACG | CAGGAAACAGCTATGACAAACGATGCTGTCTTCCATGC |
Immunohistochemistry
Immunohistochemical detection of BAP1 in FFPE tumor tissues was performed using a BAP1 antibody (C4, from Santa Cruz Biotechnology, Santa Cruz, CA), as previously described (15).
Chromosome Microarray Analysis (CMA)
CMA was performed using Affymetrix Oncoscan arrays. Total genomic DNA from each test sample was digested with NspI restriction enzyme and ligated to adapters that recognize cohesive 4-basepair (bp) overhangs. A generic primer that recognizes the adapter sequence was then used to amplify the adapter-ligated DNA fragments. Amplification products were purified using magnetic beads, fragmented, biotin-labeled, and hybridized to arrays according to the manufacturer's recommendations. The hybridized array was then washed and scanned with a GeneChip Scanner 3000 7G. Intensities of probe hybridization were analyzed by using Affymetrix's GeneChip Command Console, and copy number and genotyping analyses were performed using Affymetrix Chromosome Analysis Suite software with default settings.
Multiplex Ligation-Dependent Probe Amplification (MLPA) Analysis
To assess the possibility of a heterozygous or homozygous deletion of all or part of the BAP1 gene, MLPA studies were perfomed on the pleural effusion from subject III-5 by myGenomics (Alpharetta, GA) using a commercially available BAP1 MLPA kit (P417, MRC-Holland, Amsterdam, Netherlands).
Results
To ascertain the potential involvement of a BAP1 mutation, Sanger sequencing was performed on samples from family members III-1, III-2, III-5, and III-7. No point mutations were identified. MLPA analysis of the pleural effusion from case III-5, consisting of mostly normal cells, revealed no deletions in BAP1. We also sequenced the CDKN2A gene in one specimen (III-5) to assess the possibility of a germline point mutation. No mutation was identified, although we did detect a common polymorphism (designated rs11515) in the 3’UTR that does not appear to affect the gene in any obvious way. Additionally, no mutations were identified in TP53 in four tumor tumor tissues from this family.
CMA studies of two MMs revealed multiple genomic imbalances in each tumor. No DNA copy number losses or loss of heterozygosity of chromosome 3, site of the BAP1 locus (3p21), were observed in either tumor. However, overlapping losses of segments in 2q, 6q, 9p, 14q, 15q and 22q were identified. Sequence analysis of the BRCA1-associated ring domain 1 gene, BARD1, located at 2q35 – within the region of overlapping deletions in 2q– revealed no mutation in this gene in pleural fluid cells from case III-05.
In three tumors (cases III-2, III-5, and III-6), BAP1 immunohistochemistry showed nuclear staining (Fig. 2A; III-2 not shown). The other three tumors tested (III-1, III-7 and III-17) showed loss of BAP1 nuclear staining (Fig. 2B; III-7 not shown). Cytoplasmic staining for BAP1 was observed in four tumors, two in association with nuclear positivity (III-5, III-6; Fig. 2A) and two in tumors lacking nuclear staining (III-1, III-7; Fig. 2B; III-7 not shown). Histopathological and genetic findings of six family members with MM for whom detailed imformation was available are shown in Table 2.
Figure 2.
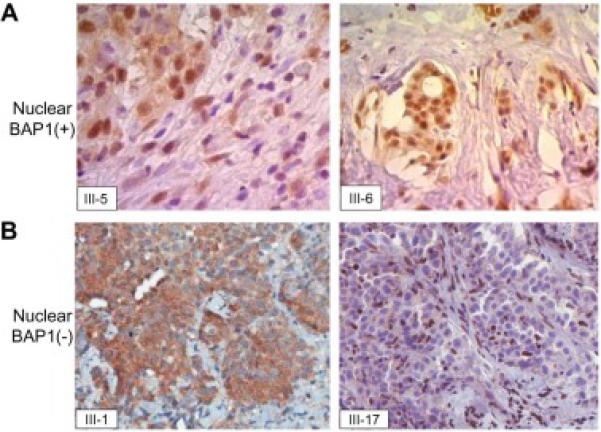
BAP1 immunostaining of MM cells. A) BAP1 nuclear positive staining in cases III-5 and III-6. B) Lack of BAP1 nuclear staining in MM cells from cases III-1 and III-17. Note cytoplasmic immunostaining in MM cells associated either with BAP1 nuclear positivity (III-5 and III-6 in A) or negativity (III-1 in B); note BAP1 nuclear staining in normal stromal cells and lymphocytes (III-17 in B).
Table 2.
Histopathological and genetic findings in six family members with MM.
| Case | Histology | Age at dx. |
Survival (months) |
BAP1 nuclear staining |
BAP1 tumor sequencing status |
CDKN2A sequencing status |
Chromosome Microarray Losses§ |
Chromosome Microarray Gains§ |
|---|---|---|---|---|---|---|---|---|
| III-1 | Epithelial | 59 | 34 | negative | No mutation | Not done | X, 2q, 6q13-qter, 8pter-q12, 9p, 9q, 14, 15q11-q21.1, 22 | 6pter-q13, 8q12-q24, 15q21.1-qter |
| III-2 | Sarcomatoid (desmoplastic) | 66 | 11 | positive | No mutation | Not done | Not done | Not done |
| III-5 | Mixed | 66 | 9 | positive | No mutation | Polymorphism 500C>G in 3′UTR | 1p, 1p, 2q, 4, 6q, 9p, 10q, 11p, 14, 15q11-q21.1, 18p, 20p, 22 | 15q21.1-qter |
| III-6 | Epithelial | 71 | 7 | positive | Not done | Not done | Not done | Not done |
| III-7 | Epithelial | 56 | 50 | negative | No mutation | Not done | Not done | Not done |
| III-17 | Epithelial | 71 | 34 | negative | Not done | Not done | Not done | Not done |
Discussion
Recent studies have revealed germline mutations of BAP1 in familial MM (3-7). In the family presented here, no evidence for an inherited BAP1 mutation was identified, although loss of nuclear BAP1 staining was observed in 3 (III-1, III-7 and III-17) of six MMs tested, suggesting that somatic genetic inactivation of BAP1 occurred in a subset of tumors. Thus, while sequencing of BAP1 in MMs from individuals III-1 and III-7 did not reveal any mutation, loss of nuclear BAP1 staining in these two samples, and in tumor from III-17 – which was not sequenced, may have occurred via somatic deletions of the gene, which would not be identified by Sanger sequencing. Alternatively, somatic epigenetic silencing of BAP1 may have occurred in these cases. A recent immunohistochemistry study demonstrated that sporadic MM patients with loss of expression of BAP1 have a prolonged survival (22). Similarly, our patients with a relatively long survival (III-1, III-7, and III-17; Table 2) showed negative nuclear staining for BAP1.
The high incidence of MM in this family occurred within one generation and did not appear to be transmitted vertically. However, the presence of other cancers in generation II leaves open the possibility of vertical transmission of a cancer susceptibility locus or loci. The family is known to have exposure to asbestos in recent generations, which presumably accounts, at least in part, for the MM clustering among blood-related relatives. By interviewing individual III-17, it emerged that she and her cousin (III-15), both of whom developed MM, had during their childhood spent a great deal of time at the proband's home (III-2), whereas individuals III-14 and III-16 did not. As noted earllier, the proband's father (II-4) had significant, lengthy exposure to asbestos occupationally. The occurrence of two pleural cancer (highly suspicious of MM) in subjects blood-unrelated between them (II-3, proband's mother and II-8, proband's aunt) highlights the possibility of shared domestic exposure, because they frequented the same home. Nevertheless, the aggregation of MM in generation III and the finding of lung cancer (II-5), liver cancer (II-6) and pleural cancer (II-8) in generation II in subjects blood-related to the proband is very striking and suggests the involvement of one or more unknown genetic factors. Since germline BAP1 mutations have been shown to be involved in a tumor syndrome consisting of not just MM but also other tumor types such as cutaneous melanoma, and given that germline mutations of CDKN2A mutations have been identified in some families with a high incidence of cutaneous melanoma (23), sequencing of CDKN2A was performed in one case (III-5) to determine whether a germline CDKN2A mutation might be responsible for the high incidence of MM in this family. The identified polymorphism in CDKN2A is designated by the Single Nucleotide Polymorphism Database (dbSNP) as rs11515. According to information provided in the 1000 Genomes Project, this minor allele occurs with a frequency of 11.5% in the general population. Given the high frequency of this allele, it is unlikely that this variant has a role in MM susceptibility observed in the family presented here. Also, a literature search was performed, using the terms ‘rs11515’ and ‘cancer’ (http://www.ncbi.nlm.nih.gov/pubmed?cmd=search&term=rs11515++cancer), and nearly all the published work indicates that this polymorphism does not increase a person's risk for any type of cancer. Since somatic mutations of TP53 occur in ~15% of MMs (24), and germline mutation of TP53 predisposes to a variety of cancers in patients with Li-Fraumeni Syndrome, we also searched for mutations in this gene in MMs from four members of the family reported here. No point mutations in TP53 were observed.
In summary, an inherited BAP1 mutation does not appear to be involved in the high incidence of MM in the family reported here. Since MM does not appear to be transmitted vertically from one generation to the next, it appears likely that domestic asbestos exposure is the predominant cause of MM in this extended family. However, it is still possible that another susceptibility locus may contribute to the high incidence of MM and other pleural and lung cancers seen in this family. Experimental evidence with knockout mouse models demonstrate that heterozygous germline mutations of tumor suppressor genes such as Cdkn2a and Bap1 are more prone to the development of asbestos-induced MM than genetically normal (wild-type) littermates, but given a sufficient amount of exposure to these carcinogenic fibers, even wild-type mice will develop a significant number of MMs (25, 26). Given that our proband's immediate family experienced certain asbestos exposure in a domestic setting, exposure alone might have been sufficient to cause a high incidence of MM. Notably, however, the proportion of asbestos-exposed individuals who develop MM, even among those who have been heavily exposed occupationally, generally is relatively small. While familial risk for MM has not yet been fully determined to date, an increased risk has been reported among blood relatives in cohorts wherein estimates of asbestos exposure levels for families of workers are known (27). Thus, we speculate that both asbestos and genetic factors have played a role in the high rate of MM and pleural/lung cancers seen in the family presented here.
Acknowledgments
JRT and Fox Chase colleagues are supported by National Cancer Institute grants CA175691 and P30 CA06927, NIEHS grant P42 ES023720 (UPenn Superfund Research and Training Program Center), and a gift from the Local No. 14 Mesothelioma Fund of the International Association of Heat and Frost Insulators & Allied Workers. MC was also supported by a grant from the Mesothelioma Applied Research Foundation. JRT, MC, and JP have a pending patent application on BAP1 mutation testing. JRT has provided consultation regarding genetic aspects of mesothelioma.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosure Statement:
All other authors declare no potential conflict of interest.
References
- 1.Ascoli V, Cavone D, Merler E, et al. Mesothelioma in blood related subjects: report of 11 clusters among 1954 Italy cases and review of the literature. Am J Ind Med. 2007;50:357–69. doi: 10.1002/ajim.20451. [DOI] [PubMed] [Google Scholar]
- 2.Ascoli V, Romeo E, Carnovale Scalzo C, et al. Familial malignant mesothelioma: a population-based study in central Italy (1980-2012). Cancer Epidemiol. 2014;38:273–8. doi: 10.1016/j.canep.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856–9. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiesner T, Fried I, Ulz P, et al. Toward an improved definition of the tumor spectrum associated with BAP1 germline mutations. J Clin Oncol. 2012;30:e337–40. doi: 10.1200/JCO.2011.41.2965. [DOI] [PubMed] [Google Scholar]
- 6.Cheung M, Talarchek J, Schindeler K, et al. Further evidence for germline BAP1 mutations predisposing to melanoma and malignant mesothelioma. Cancer Genet. 2013;206:206–10. doi: 10.1016/j.cancergen.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Betti M, Casalone E, Ferrante D, et al. Inference on germline BAP1 mutations and asbestos exposure from the analysis of familial and sporadic mesothelioma in a high-risk area. Genes Chromosomes Cancer. 2015;54:51–62. doi: 10.1002/gcc.22218. [DOI] [PubMed] [Google Scholar]
- 8.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–21. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Njauw CN, Kim I, Piris A, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PLoS One. 2012;7:e35295. doi: 10.1371/journal.pone.0035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadt K, Choi J, Chung JY, et al. A cryptic BAP1 splice mutation in a family with uveal and cutaneous melanoma, and paraganglioma. Pigment Cell Melanoma Res. 2012;25:815–8. doi: 10.1111/pcmr.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popova T, Hebert L, Jacquemin V, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet. 2013;92:974–80. doi: 10.1016/j.ajhg.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Höiom V, Edsgärd D, Helgadottir H, et al. Hereditary uveal melanoma: a report of a germline mutation in BAP1. Genes Chromosomes Cancer. 2013;52:378–84. doi: 10.1002/gcc.22035. [DOI] [PubMed] [Google Scholar]
- 14.de la Fouchardière A, Cabaret O, Savin L, et al. Germline BAP1 mutations predispose also to multiple basal cell carcinomas. Clin Genet. 2014 doi: 10.1111/cge.12472. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–72. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshikawa Y, Sato A, Tsujimura T, et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci. 2012;103:868–74. doi: 10.1111/j.1349-7006.2012.02223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo Iacono M, Monica V, Righi L, et al. Targeted next-generation sequencing of cancer genes in advanced stage malignant pleural mesothelioma. J Thorac Oncol. 2015;10:492–9. doi: 10.1097/JTO.0000000000000436. [DOI] [PubMed] [Google Scholar]
- 18.Tanji M, Powers A, Luk H, et al. High incidence of somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol. 2015;10:565–76. doi: 10.1097/JTO.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ascoli V, Scalzo CC, Bruno C, et al. Familial pleural malignant mesothelioma: clustering in three sisters and one cousin. Cancer Lett. 1998;130:203–7. doi: 10.1016/s0304-3835(98)00142-6. [DOI] [PubMed] [Google Scholar]
- 20.Ascoli V, Aalto Y, Carnovale-Scalzo C, et al. DNA copy number changes in familial malignant mesothelioma. Cancer Genet Cytogenet. 2001;127:80–2. doi: 10.1016/s0165-4608(00)00420-9. [DOI] [PubMed] [Google Scholar]
- 21.Murthy SS, Testa JR. Asbestos, chromosomal deletions, and tumor suppressor gene alterations in human malignant mesothelioma. J. Cell. Physiol. 1999;180:150–7. doi: 10.1002/(SICI)1097-4652(199908)180:2<150::AID-JCP2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Farzin M, Toon CW, Clarkson A, et al. Loss of expression of BAP1 predicts longer survival in mesothelioma. Pathology. 2015;47:302–7. doi: 10.1097/PAT.0000000000000250. [DOI] [PubMed] [Google Scholar]
- 23.Hansson J. Familial cutaneous melanoma. Adv Exp Med Biol. 2010;685:134–45. doi: 10.1007/978-1-4419-6448-9_13. [DOI] [PubMed] [Google Scholar]
- 24.Altomare DA, Vaslet CA, Skele KL, et al. A mouse model recapitulating molecular features of human mesothelioma. Cancer Res. 2005;65:8090–5. doi: 10.1158/0008-5472.CAN-05-2312. [DOI] [PubMed] [Google Scholar]
- 25.Altomare DA, Menges CW, Xu J, et al. Losses of both products of the Cdkn2a/Arf locus contribute to asbestos-induced mesothelioma development and cooperate to accelerate tumorigenesis. PLoS One. 2011;6:e18828. doi: 10.1371/journal.pone.0018828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Kadariya Y, Cheung M, et al. Germline mutation of Bap1 accelerates development of asbestos-induced malignant mesothelioma. Cancer Res. 2014;74:4388–97. doi: 10.1158/0008-5472.CAN-14-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Klerk N, Alfonso H, Olsen N, et al. Familial aggregation of malignant mesothelioma in former workers and residents of Wittenoom, Western Australia. Int J Cancer. 2013;132:1423–8. doi: 10.1002/ijc.27758. [DOI] [PubMed] [Google Scholar]


