Abstract
The Atypical ChemoKine Receptor 1 (ACKR1) gene, better known as Duffy Antigen Receptor for Chemokines (DARC or Duffy), is responsible for the Duffy Blood Group and plays a major role in regulating the circulating homeostatic levels of pro-inflammatory chemokines. Previous studies have shown that one common variant, the Duffy Null (Fy-) allele that is specific to African Ancestry groups, completely removes expression of the gene on erythrocytes; however, these individuals retain endothelial expression. Additional alleles are associated with a myriad of clinical outcomes related to immune responses and inflammation. In addition to allele variants, there are two distinct transcript isoforms of DARC which are expressed from separate promoters, and very little is known about the distinct transcriptional regulation or the distinct functionality of these protein isoforms. Our objective was to determine if the African specific Fy- allele alters the expression pattern of DARC isoforms and therefore could potentially result in a unique signature of the gene products, commonly referred to as antigens. Our work is the first to establish that there is expression of DARC on lymphoblasts. Our data indicates that people of African ancestry have distinct relative levels of DARC isoforms expressed in these cells. We conclude that the expression of both isoforms in combination with alternate alleles yields multiple Duffy antigens in ancestry groups, depending upon the haplotypes across the gene. Importantly, we hypothesize that DARC isoform expression patterns will translate into ancestry-specific inflammatory responses that are correlated with the axis of pro-inflammatory chemokine levels and distinct isoform-specific interactions with these chemokines. Ultimately, this work will increase knowledge of biological mechanisms underlying disparate clinical outcomes of inflammatory-related diseases among ethnic and geographic ancestry groups.
Introduction
The Duffy Antigen Receptor for Chemokines (DARC), recently renamed Atypical Chemokine Receptor 1 (ACKR1), expresses the red blood cell antigens that define the Duffy Blood Groups for which it was originally discovered [1, 2]. Much of the research involving DARC/ACKR1 deals with its role as the receptor for the malarial parasites Plasmodium vivax and Plasmodium knowlesi [3–5]. It is now known to be a promiscuous atypical chemokine receptor [6], interacting with an array of both classes of chemokines (C-C-L and C-X-C-L); including those involved in inflammation and angiogenesis [3, 7, 8]. DARC/ACKR1 is ubiquitously expressed and highly conserved across placental mammalian species (Boreoeutheria) with over 90% amino acid conservation across primates and over 70% nucleic acid conservation in lower mammals[9]. The main normal function described for DARC/ACKR1 is that it effectively sustains homeostatic levels of circulating chemokines and modulates chemokine gradients between tissues and blood to mediate the influx of neutrophils and monocytes from blood vessels into tissues [10, 11] during immune responses. DARC/ACKR1 has also been implicated to affect cancers as a pro-inflammatory cytokine receptor, specifically in lung cancer etiology, BrCa progression by in vitro studies and allele-specific BrCa patient survival [12–14]. While these studies implicate DARC/ACKR1 in cancer processes, there are still lingering questions concerning how DARC/ACKR1 lends its chemokine binding capacity toward cancer progression. In fact, there is very little investigation of DARC/ACKR1 in regard to the complexity of gene product variants and their distinct role in biological outcomes of immune/inflammatory responses during tumorigenesis.
The DARC/ACKR1 gene products are traditionally referred to as “Duffy blood group antigens” and vary in their expression among different human populations in several ways. There are several antigens described in various clinical reports which presumably represent variants of the gene product [15]. However, the most commonly described variants are those derived from the two major DARC alleles resulting from common Single Nucleotide Polymorphisms (SNPs). One SNP occurs in the gene regulatory region and yields the “Fy- allele” or “Duffy Null” (rs2814778 ), and the other SNP is a missense mutation that yields the “Fy B” and “Fy A” alleles (rs12075). The Duffy Null allele nomenclature refers to a phenotype also known as “Erythrocyte Silent” (Fy es) removing expression of DARC on red blood cells (RBCs) and is usually denoted as “Fy-a-b” to reflect the missing blood group antigens. This allele is rare among individuals of either European or Asian descent but is the most common phenotype in most Africans and African Americans. The Fy- allele has a frequency of nearly 100% in West Africans and greater than 80% of African Americans (Fig 1) [16, 17] as it confers resistance to malaria [18] and became fixed in African populations where malaria is endemic[4, 17]. In addition to these DARC variants, there are 4 other alternate Duffy blood group antigens, usually interpreted as DARC gene allele variants, namely; Fy3, Fy4, Fy5 and Fy6. However, the specific immunogenic domain that defines most of these antigens is not fully elucidated [15, 19]. Of note, one additional important yet rare phenotype, Fy x, expresses the Fy b allele, but is believed to have ‘weak’ expression that is not always detected by the anti-Fyb antibody [6, 20], suggesting the epitope is unique and possibly a yet undescribed additional Duffy antigen. One potential explanation for such complexity among Fy antigens could be the co-expression of alternative DARC gene product isoforms and distinct post-translational modifications (i.e. glycosylation) between the isoforms acting as immunogens. This possibility has not yet been addressed.
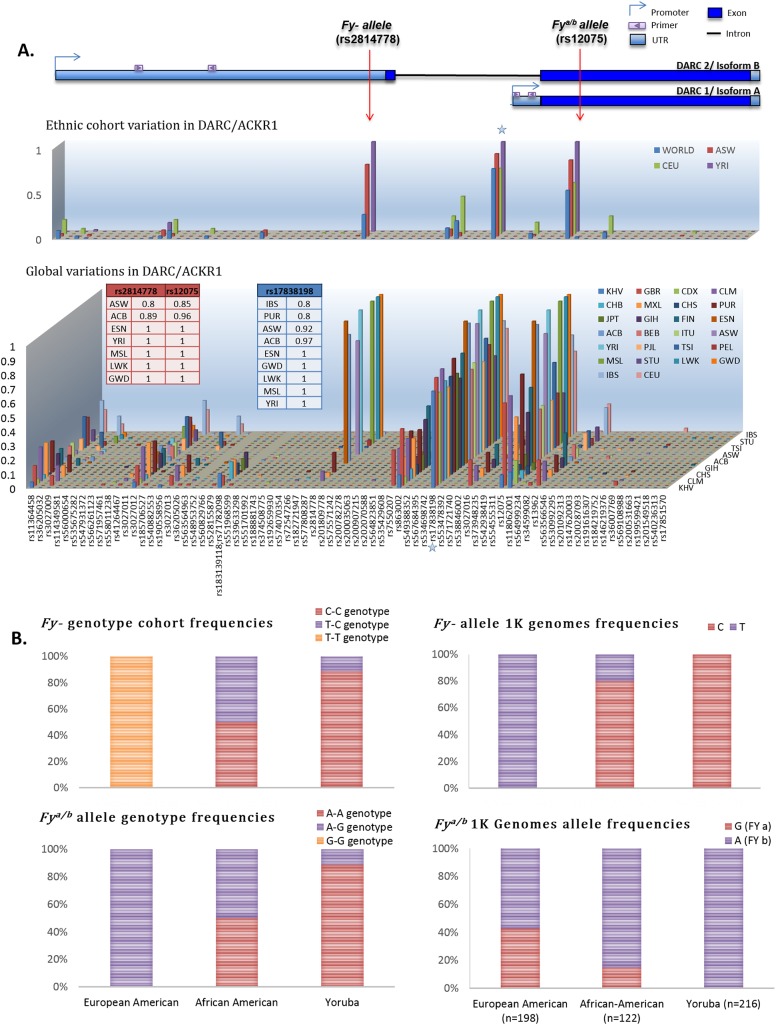
Fig 1. The DARC gene structure and frequencies of the two major alleles; Fy- and Fy a/b in HAPMAP experimental cohort compared to entire 1000 Genomes (1K) populations.
(A). Top, Model of DARC/ACKR1 gene structure indicating the two gene promoter-driven isoforms, DARC1/A and DARC2/B. Primers used for qPCR of transcript variants are indicated as double arrows. Also indicated are the Duffy Null allele (rs2814778) and Fy a/b allele (rs12075). The two transcript products result in unique gene products; Duffy isoform A has 338aa and Duffy isoform B has 336aa. Top graph shows the allele frequencies of all documented polymorphisms in the DARC gene region, specifically in the ethnic cohorts of interest; African American (ASW), West African- Yoruba in Ibadan (YRI) and European American (CEPH). The overall ‘world’ variant frequencies are also indicated, summarizing all sampled populations in the 1000 Genomes. The bottom graph details the individual ancestry groups that encompass the global frequencies across the entire 1000 Genomes dataset for the entire DARC gene region. The X-axis matches the gene model with regard to relative gene structure location. The rs# IDs on the X axis of the bottom graph correspond to those on the top graph, for each SNP. The starred locus indicates a deletion polymorphism that resides in the promoter region of the DARC1/A isoform that has not previously been explicitly reported, described in the text (rs17838198). This locus appears to have been under some selection influence in other populations, similar to the as the malaria selection influence on the Duffy Null allele in African populations, as it is the 3rd most prominent variant in the gene and prevalent in most populations. (B). The genotype frequencies for the Fy- and Fy a/b loci in our cohort are shown in the inset panel on the left. The allele frequencies of Fy- and Fy a/b for the entire corresponding ancestry 1K Genomes panels are shown in the inset panel on the right. The genotype percent distributions of the Fy- genotype across our West African—YRI, African American and European American subpopulations correlate with the high prevalence of the alleles in 1,000 Genome populations. Nearly half of the AA/ASW group is heterozygotes with an additional 40% being CC homozygotes, indicating more than 90% have the Fy- allele, as seen in the adjacent graph. The allele distribution table shows proportion values correlate with previously established numbers and validate our population.
There is long-standing evidence of two separate DARC isoforms [21] that are derived from separate promoters and yield distinct protein products. Specifically, DARC yields two isoform transcripts [17] DARC1/Isoform A (NM001122951.2/UniProt Q16570-2) and DARC2/Isoform B (NM00236.3/UniProt Q16570-1) [22] that create distinct gene products. Each isoform has unique N-terminal amino acids in the extracellular topological domain where chemokine binding occurs. The isoforms contain 17 unique amino acids between them that include distinctive glycosylation and enzymatic cleavage sites. Therefore, there is great potential for the isoforms to diverge in their functionality and lend a level of dynamic complexity to the role of DARC in chemokine regulation. To date, no study has addressed the potential differences in functionality between the isoforms. However, there is clinical evidence of complexities in the Duffy blood group beyond what has been previously described. Specifically, we observe complexity beyond the two major DARC alleles in presentations of varied clinical manifestations linked to DARC including, neutropenia [23, 24], organ system damage in sickle cell disease[25], cancer metastasis regulation[26] as well as hemolytic complications arising from immunological responses among Duffy antigens during blood transfusions [27]. At least in part, these observations could be explained by considering the alternative protein isoforms which would increase the repertoire of antigens 2-fold. In this work, we have shown that both of the DARC isoforms are expressed in lymphoblast lines and at varying levels, relative to the African-specific Fy- allele genotype.
Materials and Methods
Cohort design and 1000 Genomes data
Our study population is a well-defined subset of diverse genetic ancestry groups from genetically described individuals of the International Human Genome Haplotype Mapping Project (HAPMAP)[28–32]. We obtained a subset of HAPMAP lymphoblast cell lines to create a representative panel of genetic ancestry groups, including: Yoruba in Ibadan, Nigeria (abbreviation: YRI); Japanese in Tokyo, Japan (abbreviation: JPT); Han Chinese in Beijing, China (abbreviation: CHB); CEPH (Utah residents with ancestry from northern and western Europe) (abbreviation: CEU). Each of our investigations utilized at least 20 cell lines across the ancestry categories. We obtained access to the 1000 Genomes[33] dataset through the public access portal at www.1000genomes.org.
Genotyping and allele distributions
We conducted independent validation of DARC/ACKR1 allele genotypes in our cohort samples. Duffy Null genotyping was done with PCR amplification and Sanger sequencing. Primers were designed to flank the Fy-. Allele and genotype distributions were then assessed for representative population accuracy in each ancestry group, using GenePop version 4.2 Option 5, to characterize allele frequencies and Fis[34]. Fy a/b allele genotyping was done with the TaqMan Allelic Discrimination protocol. To assess the ancestry distribution in comparison to our results, we utilized the full population genotype set, conducting allele distribution calculations for all SNPs reported in the gene region. We extracted the distributions for the two alleles of interest and they are displayed in Fig 1.
Gene Expression
For in vitro DARC/ACKR1 gene expression evaluations, we used comparative quantitation methods to determine the relative mRNA levels of DARC/ACKR1 isoforms among cell lineages derived from 3 distinct ancestry groups. Specifically, after detection of the transcript with Reverse Transcription PCR, we conducted Real-time PCR using SABiosciences SYBR green protocol on Applied Biosystems 7500 cycler and SDS software version 1.4. The Ct values were transformed to dCT values using RPL13 endogenous control gene, displayed as 1/dCT to preserve directional interpretation of expression levels. We used the manufacturer’s suggested protocols for all assays and primers. Isoform specific primers were designed using the primer-BLAST program through NCBI [35]. Primer sequences were as follows:
DARC1 expression Forward-TCTGGGTATGTCCTCCAGGC
DARC1 expression Reverse-AAGGGCAGTGCAGAGTCATC
DARC2 expression Forward-TCCAATTTCCCAGCACCTCC
DARC2 expression Reverse-GGCTGGTTGGGACTACACTC
For in situ DARC/ACKR1 gene product evaluations in tissues and cells, we used immunohistochemistry to assess the DARC/ACKR1 protein expression. Formalin-fixed cultures were paraffin embedded and sections stained for DARC/ACKR1 using a goat anti-DARC antibody (Novus biologicals: NB100-2421) and detection with biotinylated anti-goat (Biocare), alkaline phosphatase streptavidin (Biogenex), and fast-red chromogen (Biocare).
Results and Discussion
Global and cohort distributions of DARC variants
The global distributions of Duffy Null (Fy-) genotypes have been previously reported in several studies [36–39]. In relation to the gene’s structure, the Fy- SNP involves a T to C substitution in the promoter region of the DARC2 isoform and in the 5’ Untranslated Region (UTR) of the DARC1 isoform (Fig 1A). In our study cohort, this Fy- phenotype is present in over 98% of West African Nigerians in Idaban (YRI) and in 50–80% of African Americans (AA/ASW)[16] and less than 1% of European Americans (CEPH). Intriguingly, we have revealed a previously unexplained variant (rs17838198) that appears to be in Linkage Disequilibrium (LD) with both the Fy- allele and the Fy a/b allele, according to the HapMap data for our ancestry groups of interest. Upon expanding our investigation to include all ancestry groups of the 1,000 Genomes dataset [33] (Fig 1A, S1 Table), we see that rs17838198 is even more prevalent than the Fy a/b allele across most ancestry groups and just as conserved in African populations as the Fy- allele, according to the Minor Allele Frequency data generated by the 1,000 Genomes Project[33]. Of note, rs17838198 is a single nucleotide insertion/deletion adjacent to the DARC1 isoform promoter with unknown effect that could potentially alter DARC gene expression if a transcription factor or polymerase complex binding site is disrupted. We will investigate this possibility further in a later publication.
For this investigation of DARC isoform regulation, we conducted independent genotyping of our HAPMAP [32] sub-population panel to ensure our cohort reflects the expected distributions for each ancestry group. Both the Fy- allele and the Fy a/b allele genotype distributions show the expected pattern of association with ancestry (Fig 1B). All CEPH individuals are TT homozygotes (Fy+), the West Africans-YRI are primarily CC homozygotes (Fy-) with less than 30% being heterozygotes and no TT homozygotes. The African-American group has the highest percentage of heterozygotes and could potentially be uniquely affected by the combination of alleles. The complete lack of TT homozygotes in the African population, with only a single individual with heterozygote status in the entire 1,000 genomes sample set, reaffirms the extent of which natural selection has occurred for the C allele in African regions. The Fya/b locus also shows allele association with ancestry, with the majority of YRI having the A allele, which would confer expression of the Fy b antigen in these individuals. The CEPH population has the highest prevalence of the B allele though all are heterozygotes state. This will result in expression of both the Fy a and Fy b antigen on erythrocytes (Fy a+b+) in this population, as well as on lymphoblasts that we will discuss later. The genotype frequency in AA/ASW individuals appears to be an even mix of both the YRI and CEPH, which is expected given the AA/ASW group is an admixed population, largely made up of both West African and European-American ancestry. Hence, our correlative findings between the 1,000 Genomes and our cohort indicate our representative cohort does reflect the global population of each ancestry group. Interestingly, the YRI individual who is heterozygous for the Duffy Null allele has two copies of the Fy b allele, consistent with prior studies indicating that the Fy(a-b+) phenotype is most prevalent among Blacks who are not Duffy Null, meaning the Fy b allele is segregating with the Duffy Null allele in this population. We can see this most pointedly demonstrated the AA/ASW group, which has the highest percentage (50%) of heterozygotes for the Duffy Null allele. In fact, the AA/ASW population has compound heterozygotes for the Fy- allele and the Fy a/b allele which should have a unique effect on expression of antigens.
DARC protein product is expressed in lymphoblasts and shows dynamic expression levels across Fy- and ancestry groups
In order to investigate ancestry related DARC distinctions, we chose to utilize the lymphoblast lines from the HAPMAP resources. Several genome-wide investigations have indicated DARC/ACKR1 expression in many tissues [40–45], including low level transcripts from microarray data in the cell lineages that were used to derive the HAPMAP lines (S1 Fig). However, there were no explicit reports of these cells expressing the DARC/ACKR1 gene product. Therefore, we conducted IHC staining of our set of HAPMAP lines to measure DARC/ACKR1 protein expression. We observed a large amount of variation of expression across the cohort, documenting the full spectrum of IHC scoring; from 0 (no expression) to 4 (very high expression) (Fig 2A). An analysis of the IHC score distributions among ancestry groups and Fy- genotypes suggests a trend of expression that may be linked to ancestry (Fig 2B). We observed that only the CEPH/CEU ancestry group and the homozygous TT (Fy+) genotype groups have individuals who displayed a score of 0, indicating no expression. Over half of this group only had low to moderate expression (score of 1–2) which correlates with our observed lower transcript levels of the DARC isoforms as well. While this might indicate that some individuals of European descent may not typically express high levels of DARC/ACKR1 in lymphoblasts, almost half of this group also displayed high levels of DARC/ACKR1. This suggests that DARC/ACKR1 expression is highly variable in these cells. In contrast, we observe that the YRI and ASW groups primarily displayed the highest scores (3) indicating very high levels of DARC/ACKR1 product in lymphoblasts from these groups. This suggests that these groups would typically have higher levels of DARC/ACKR1 expression in these cells, with few individuals showing low levels.
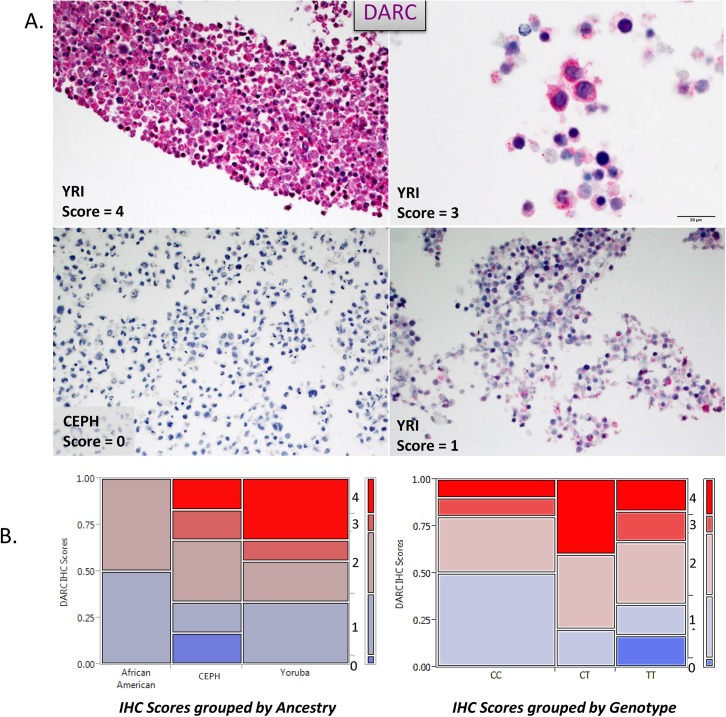
Fig 2. DARC/ACKR1 protein is differentially expressed among lymphoblasts derived from divergent ancestry groups in HAPMAP/ 1,000 (1K) Genomes populations.
(A). Representative IHC images of DARC expression in the lymphoblasts of Africans (YRI) and European Americans (CEPH/CEU). DARC is stained with Vulcan Red chromogen nuclei are stained blue with hematoxylin. (B). Shows a distribution analysis of IHC scores across ancestry groups and Fy- genotypes. The full range of scores (0–4) were observed in our cohort, with clear trends within groups. European Americans were the only ancestry group to have a 0 score, indicating no DARC expression in these cells, correlating with lower transcription of the gene. Similarly, individuals with a homozygous Duffy-positive (TT) genotype were the only to have a score of 0, indicating that lowest levels of DARC expression in lymphoblast are associated with the European lineage and the TT genotype, in contrast to higher DARC expression in the lymphocytes of African lineages. Accordingly, the majority of high lymphoblast IHC scores (3 or 4) were in the African lineage. All African Americans only showed moderate expression levels in lymphoblasts.
All in all, published reports and data sources that indicate little to no expression in these cells are likely biased with over-representation of samples and data from people of European ancestry, with very few samples from people of African ancestry. Such bias leads to incorrect conclusions concerning the spatial expression pattern of DARC/ACKR1 (S1 Fig), and specifically now in lymphoblasts. While our small sample size would not avail statistical significance in differences among the CEPH/CEU, YRI and AA/ASW groups, we have clearly demonstrated that DARC/ACKR1 is expressed in these cells, at varying levels among individuals; including, extremely high protein product levels among the CEPH and YRI individuals (Fig 2A). These findings indicate that our HAPMAP lines are ideal for investigations of differential DARC expression and regulation.
Both DARC1 and DARC2 isoforms are expressed in lymphoblasts and have distinct expression levels
We next investigated whether the DARC protein products expressed by lymphoblasts could be the result of distinct DARC isoform expression. Because the ancestry-specific Fy- allele results in a cell-type-specific loss of DARC expression, it was plausible that there is dynamic regulation of the gene in certain contexts, given that individuals who lack erythrocyte DARC/ACKR1 expression still express the gene in other tissues, such as endothelial tissues [7, 46–48]. In addition, previous reports indicated that the erythrocyte silencing of DARC/ACKR1 results from loss of a single transcription factor binding site. Therefore, given the isoforms are expressed from distinct promoters, we hypothesized that there may be significant variation of isoform expression throughout the global population associated with the Fy- allele.
Using the well-defined genetic ancestry HAPMAP cohort, we investigated whether the Fy- genotype was associated with differential expression of DARC isoforms. We anticipated that the Fy- allele may facilitate removal of DARC/ACKR1expression from lymphocytes if the hematopoietic transcription factor (GATA-3), reported to be the regulator in erythrocytes [49], was also the regulator in lymphoblasts. Therefore, we decided to determine if this would also be true of lymphocyte lineages. However, we found the opposite to be true. Using RT-PCR we were able to detect mRNA expression of both DARC/ACKR1 isoforms, (DARC1/A and DARC2/B) in all individuals, including those with Fy- genotypes. However, the levels of DARC isoform transcripts were very dynamic among the cohort. Therefore, we quantified the differential levels of isoform-specific transcripts using qPCR (Fig 3A). An ANOVA across the entire group indicated the overall isoform transcript levels were significantly variant (p = 0.0007), suggesting the transcripts have distinct expression among our lymphoblast lines, derived from divergent ancestries (Fig 3A). The African and African American groups have a higher level of expression in these lines relative to CEPH-European Americans. The lower level of DARC/ACKR1 isoform transcripts also correlates with the lower IHC scores in this population (Fig 2B). These findings illustrate the Fy- allele may be linked to altered expression of the gene in different cell types or tissues and not just removal of its expression in erythrocytes.
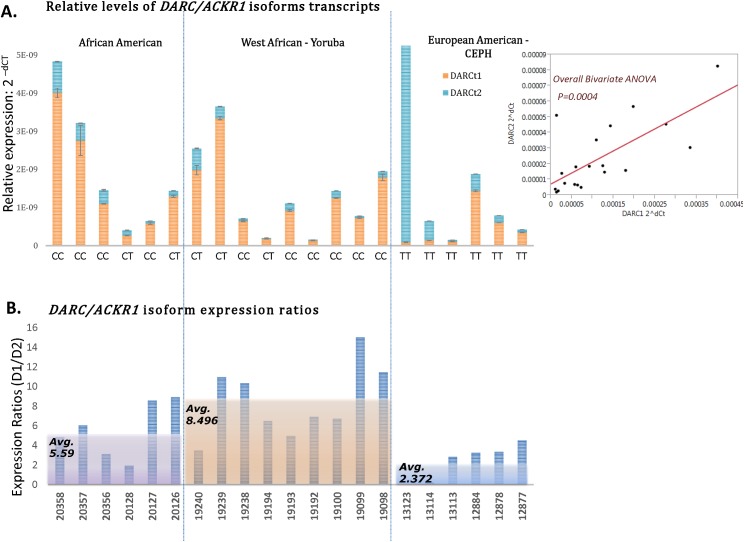
Fig 3. Differential expression of DARC isoforms in lymphoblasts derived from 1K Genomes populations.
(A) Shows the relative expression levels of DARC 1 and DARC 2 transcripts among our HAPMAP cohort panel. The Fy- genotypes are indicated in the X axis of the top graph (A) and correlated HAPMAP cell line IDs are indicated in the X axis of the bottom graph (B). The “C” allele indicates the Duffy Null mutation. There is a clear trend of higher expression in the African and African Americans, with DARC 1 showing prominent expression. (B) Shows the relative ratios of DARC1/DARC2 isoforms in our cohort. The average ratio values for each ancestry category are shown as overlapping shaded box insets to display the values of each ancestry group. The highest ratio in the African category indicates the greatest difference between isoform expression values. C. ANOVA statistics indicate there are significant differences in DARC isoform transcripts across the cohort groups. The overall fitness across the entire group is mainly due to correlated expression of the isoforms for individuals with the C allele. There is clear trend of higher expression of the DARC 1 isoform in African and African American lineages, relative to European Americans. These data show a trend for lower expression in the European American (CEPH) categories and TT homozygotes for both DARC isoforms. These data indicate differences in isoform regulation, associated with the Fy- “Duffy Null” allele.
To visualize the relationship of DARC/ACKR1 isoforms relative to each other within individuals, we calculated the relative expression ratios of [DARC1/DARC2] (Fig 3B). This perspective reveals a significant difference in isoform transcript levels among the ancestry groups. The West African group shows the highest average ratio, indicating the largest difference between isoform regulation. Additionally, the CEPH group shows the lowest average ratio, indicating that while the levels of DARC/ACKR1 are lower in this population, the relative levels between isoforms is more correlated and may be suggestive of distinct promoter regulation differences. Accordingly, the African American ratios are averaged between the YRI and CEPH groups, similar to the genotype distributions.
Differential DARC isoform expression is associated with genetic ancestry and Fy-genotypes
To thoroughly interrogate the differential expression of DARC/ACKR1 isoforms, we conducted a series of statistical tests to determine associations with either ancestry or the Fy- genotype. First, to determine if the specific isoform levels are associated with ethnicity, we conducted an ANOVA for each isoform across ethnicities. While we detected a trend for relatively lower and more tightly regulated levels in the EA group for both isoforms (Fig 4); however, this finding isn’t statistically significant (p = 0.482) possibly due to our small sample size. Similarly, when an ANOVA was conducted for each isoform across Fy- genotype groups, we found trends of expression levels with higher levels of both isoforms in individuals with the Fy- alleles.
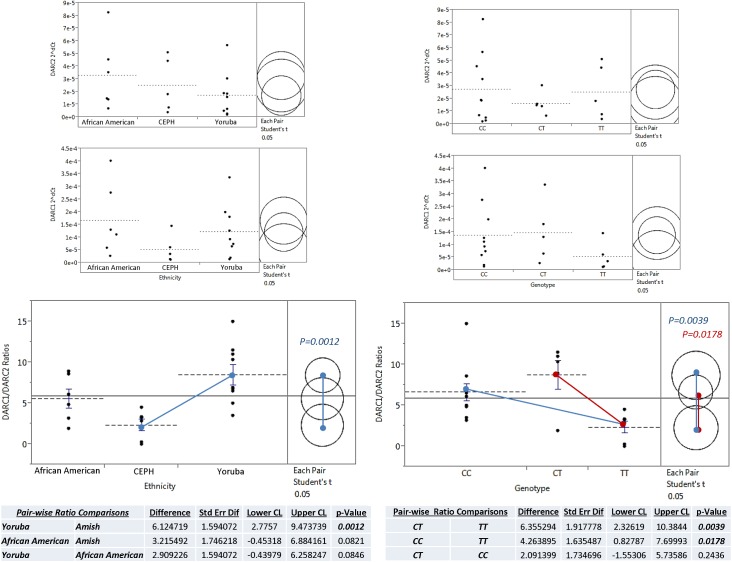
Fig 4. Statistical comparisons of relative levels of DARC isoform transcripts and ratios among ancestry and Fy- genotype groups.
Statistical analyses across ancestry groups are on the left and genotype groups are on the right. ANOVA graphs indicate the individual group means (dotted blue line) of DARC isoform expression (top) and ratios (bottom) for stated categories. No statistically significant difference in individual transcript isoforms was identified across groups; however, significant differences were measured among ratios of DARC1/DARC2 isoforms. The means connection lines (solid blue and red) indicate the statistically significant comparisons. The significant p-values for pairwise T-test analyses of means is shown in the colors corresponding with each pair. The only statistically significant comparison across ancestry groups were between Amish and Yoruba groups. Across genotypes, there was a significant difference in DARC isoform ratios between TT genotype and both CT and CC genotypes. Significant differences between the isoform ratios indicate the isoforms are differentially regulated and the control of regulation is altered in isoform promoters with the C allele (Fy-).
Given that the ratio of DARC1/DARC2 revealed the relationship of relative isoform expression, we conducted pair-wise comparisons on these values to determine if relative levels of DARC1/DARC2 isoforms was associated with ancestry or Fy- genotypes. The results were significant. Across ancestry groups, the largest and only significant difference in relative isoform expression was between the CEPH and YRI categories (Pvalue = 0.0012). The African American group was an equal medium between the YRI and CEPH group and not significantly different from either. This observation also reflects the genotype distributions previously described. Across Fy- genotypes there was a significant difference between the categories with the “C” allele compared to the homozygous T allele. This finding indicates that the Duffy Null Fy- genotype results in significantly altered regulation and expression of the DARC isoforms and this is associated with African Ancestry.
Conclusions
Impact of altered DARC gene isoform levels among ancestry groups
IHC analysis indicates varying levels of DARC/ACKR1 gene product among our cohort ancestry groups with the consistently the higher levels of in the African group lymphoblasts. This is in contrast to the Duffy Null phenotype status of these individuals, where of no DARC/ACKR1 is expressed on erythrocytes. Intriguingly, the transcript variant reported to be impacted by the Fy- allele (DARC/ACKR1-B) is not the transcript affected in lymphoblast cells. Our results indicate that the levels of the DARC/ACKR1-A/Isoform A transcript show a significantly higher expression level among Fy- genotypes relative to DARC2 isoforms levels. The implications of this altered isoform regulation could have a huge impact on our understanding of DARC/ACKR1 functionality. For instance, to date the repertoire of Duffy antigens includes only 2 distinct protein products, the result of the Fy a/b allele variation in the coding region. However, we have shown that the predominant transcript variant, in lymphoblasts, is the DARC/ACKR1-A isoform, which corresponds to the 338aa product variant. All studies to date have only addressed the 336aa product variant. This leaves a huge void of information concerning the distinct functions of the isoforms and especially in the context of the role DARC/ACKR1 plays in immunobiology; specifically, the impact of immunogenic potential for the 17 distinct amino acids between the isoforms, the potential of interactions between isoforms, the potential for isoform-specific affinity to certain chemokines as well as the tissue specificity or isoform spatial expression that relates to each of these characteristics. Summarily, we can conclude that each isoform expressed in an individual will also harbor the Fy a or Fy b allele, yielding 4 distinct DARC/ACKR1 protein products for individuals who are heterozygous at the Fya/b locus. Of note, given our findings, we can confidently predict that certain individuals are likely expressing up to 4 different protein variants of DARC/ACKR1 in circulating cells (Table 1) given the expression levels of transcripts (Fig 1) and levels of DARC/ACKR1 protein products (Fig 2). The combinations of these Duffy antigens could impact hemolytic interactions as well as regulation of chemokine levels in circulation. Indeed, perhaps the unexplained complexity with regard to alternate Duffy antigens (Fy5, Fy 6, etc.) could be due to the previously ignored isoform variants. In fact, DARC/ACKR1 is a glycoprotein with unique prediction sites for extracellular, isoform-specific N-terminal glycosylation and therefore the potential for distinct isoform-specific interactions with chemokines is highly likely.
Table 1. Summary of DARC/ACKR1 genotyping for Duffy null and A/B alleles, DARC/ACKR1 protein isoform expression and predicted Duffy antigen phenotypes.
| Ancestry | Cell Line | DARC isoform 1 mRNA | DARC isoform 2 mRNA | rs2814778 Fyes- allele | rs12075 Fya/b allele | Expected Erythroid Phenotype | Anticipated Epithelial Phenotype | Potential # of antigens on RBC | Potential # of antigens on WBC |
|---|---|---|---|---|---|---|---|---|---|
| Amish | GM12877 | + | + | TT | AG | Fy (a+b+) | Fy (1a+1b+2a+2b+) | 2–4 | 4 |
| GM12878 | + | + | TT | AG | Fy (a+b+) | ||||
| GM12884 | + | + | TT | AG | Fy (a+b+) | ||||
| GM13113 | + | + | TT | AG | Fy (a+b+) | ||||
| GM13114 | + | + | TT | AG | Fy (a+b+) | ||||
| GM13123 | + | + | TT | AG | Fy (a+b+) | ||||
| West African—Yoruba | GM19098 | + | + | TC | AA | Fy (a-b+) | Fy (1a- ,1b+ ,2a- ,2b+) | 1–2 | 2 |
| GM19099 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| GM19100 | + | + | CC | AG | Fy (a-b-) | Fy (1a+1b+2a+2b+) | 0 | 4 | |
| GM19192 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| GM19193 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| GM19194 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| GM19238 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| GM19239 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| GM19240 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| African American | GM20126 | + | + | TC | AG | Fy (a-b+) | Fy (1a +/-, 1b +, 2a +/- 2b + ) | 1–2 | 2–4 |
| GM20127 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| GM20128 | + | + | TC | AG | Fy (a-b+) | Fy (1a +/- ,1b + 2a + /-2b + ) | 1–2 | 2–4 | |
| GM20356 | + | + | TC | AA | Fy (a-b+) | Fy (1a- ,1b+ ,2a- ,2b+) | 1–2 | 2 | |
| GM20357 | + | + | CC | AA | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 2 | |
| GM20358 | + | + | CC | AG | Fy (a-b-) | Fy (1a- ,1b+ ,2a- ,2b+) | 0 | 4 |
We genotyped the Duffy null and Fy A/B alleles in our HapMap Cohort to validate the genotypes of the 1000 genomes populations of Amish, Yoruba and African Americans with expected results. The distribution of the genotypes for both the Fy- and Fya/b alleles show correlation with ancestry groups. We have detected the expression of DARC/ACKR1 isoforms in all lines and based on this have predicted the Duffy antigens anticipated to be expressed in these cells as well as other epithelial cells. Strikingly, we find that individuals who are erythrocyte-silent (es) null (alias-Duffy null) and express no Duffy antigens on red blood cells may express up to four different Duffy antigens on lymphoblasts (italics). Additionally, only two antigens are traditionally noted for individuals with Fy+ genotypes (Fya or Fyb); however, we have detected the expression of both isoform transcripts in these individuals which suggests there could be as many as 4 antigens expressed, even in erythrocytes (bold italics). Lastly, African Americans have a high frequency of compound heterozygosity, yielding a unique range of potential Duffy antigens on both the erythrocytes and lymphoblasts which may also translate to epithelial cells. Because the Duffy null and A/B alleles tend to be linked, and the Duffy positive allele tends to segregate with the Fyb allele, this suggests most of the African American compound heterozygotes will express the B antigen on erythrocytes. In addition, because both transcript isoforms are expressed off the same gene (harboring the Fyb allele) this will potentially result in expression of two separate antigens on erythrocytes and potentially up to 4 in lymphoblasts or other epithelial cells.
Of utmost importance, with regard to ancestry and Fy- individuals, we have now begun to uncover variation in DARC/ACKR1 isoform levels in distinct tissues which may translate into variation of tissue-specific inflammatory responses, including those observed in the tumor microenvironment. With this study, we have begun to investigate distinct isoform-specific functions by characterizing these isoform differences among ancestry groups. More studies are now required to investigate how these isoforms function, which may completely transform our understanding of inflammatory actions within the vast array of chemokines, even with regard to known DARC/ACKR1—chemokine interactions among different classes of chemokines—none of which have been defined with respect to the isoforms. The physiologic consequences of differences in DARC/ACKR1 isoform levels in Fy+ and Fy- individuals is completely unexplored and we hypothesize that distinct DARC isoform expression patterns will define a unique inflammatory status among Fy- individuals which may systemically influence disease susceptibility and clinical outcome, overall an array of diseases in which DARC has already been implicated to influence.
Supporting Information
Systemic expression levels of DARC in humans. Data from indicated microarray expression datasets were used to generate an eFP Browser image that depicts the unbiquitous expression of DARC/ACKR1 in the skeletal, digestive and immune systems. Adapted from the eFP by R.Patel. Images by E.T. Hamanishi. Data from GSE1133, E-GEOD-7307, GSE3526, GSE2361, GSE19650, E-GEOD-6257. Data normalized by MAS 5.0 method TGT value 100
(TIFF)
List of abbreviations for the 1,000 Genomes populations mentioned in Fig 1.
(PNG)
Acknowledgments
We would like to thank undergraduate researchers, Casey Garrett, Ashley Chukwu, Briana Bennett and Matthew Johnson for technical support. We thank Dr. Matthew Boegehold for critical review of our work and formative feedback.
Data Availability
Data are available from the 1,000 Genomes website and FTP servers (http://browser.1000genomes.org/index.html).
Funding Statement
The authors received no specific funding for this work outside of internal seed/start-up funding from the institution.
References
- 1. Vescio LA, Farina D, Rogido M, Sola A. Hemolytic disease of the newborn caused by anti-Fyb. Transfusion. 1987;27(4):366 Epub 1987/07/01. . [DOI] [PubMed] [Google Scholar]
- 2. Weinstein L, Taylor ES. Hemolytic disease of the neonate secondary to anti-Fya. American journal of obstetrics and gynecology. 1975;121(5):643–5. Epub 1975/03/01. . [DOI] [PubMed] [Google Scholar]
- 3. Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, et al. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261(5125):1182–4. Epub 1993/08/27. . [DOI] [PubMed] [Google Scholar]
- 4. Livingstone FB. The Duffy blood groups, vivax malaria, and malaria selection in human populations: a review. Human biology. 1984;56(3):413–25. Epub 1984/09/01. . [PubMed] [Google Scholar]
- 5. VanBuskirk KM, Sevova E, Adams JH. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(44):15754–9. Epub 2004/10/23. 10.1073/pnas.0405421101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pogo AO, Chaudhuri A. The Duffy protein: a malarial and chemokine receptor. Seminars in hematology. 2000;37(2):122–9. Epub 2000/05/03. . [DOI] [PubMed] [Google Scholar]
- 7. Pruenster M, Rot A. Throwing light on DARC. Biochemical Society transactions. 2006;34(Pt 6):1005–8. Epub 2006/11/01. 10.1042/BST0341005 . [DOI] [PubMed] [Google Scholar]
- 8. Szabo MC, Soo KS, Zlotnik A, Schall TJ. Chemokine class differences in binding to the Duffy antigen-erythrocyte chemokine receptor. The Journal of biological chemistry. 1995;270(43):25348–51. Epub 1995/10/27. . [DOI] [PubMed] [Google Scholar]
- 9. Tournamille C, Blancher A, Le Van Kim C, Gane P, Apoil PA, Nakamoto W, et al. Sequence, evolution and ligand binding properties of mammalian Duffy antigen/receptor for chemokines. Immunogenetics. 2004;55(10):682–94. Epub 2004/01/09. 10.1007/s00251-003-0633-2 . [DOI] [PubMed] [Google Scholar]
- 10. Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS genetics. 2009;5(1):e1000360 Epub 2009/01/31. 10.1371/journal.pgen.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mayr FB, Spiel AO, Leitner JM, Firbas C, Kliegel T, Jilma-Stohlawetz P, et al. Duffy antigen modifies the chemokine response in human endotoxemia. Critical care medicine. 2008;36(1):159–65. Epub 2007/11/17. 10.1097/01.CCM.0000297875.55969.DB . [DOI] [PubMed] [Google Scholar]
- 12. Wang J, Ou ZL, Hou YF, Luo JM, Shen ZZ, Ding J, et al. Enhanced expression of Duffy antigen receptor for chemokines by breast cancer cells attenuates growth and metastasis potential. Oncogene. 2006;25(54):7201–11. Epub 2006/06/21. 10.1038/sj.onc.1209703 . [DOI] [PubMed] [Google Scholar]
- 13. Addison CL, Belperio JA, Burdick MD, Strieter RM. Overexpression of the duffy antigen receptor for chemokines (DARC) by NSCLC tumor cells results in increased tumor necrosis. BMC cancer. 2004;4:28 Epub 2004/06/25. 10.1186/1471-2407-4-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu XF, Li LF, Ou ZL, Shen R, Shao ZM. Correlation between Duffy blood group phenotype and breast cancer incidence. BMC cancer. 2012;12:374 Epub 2012/08/30. 10.1186/1471-2407-12-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Talano JA, Hillery CA, Gottschall JL, Baylerian DM, Scott JP. Delayed hemolytic transfusion reaction/hyperhemolysis syndrome in children with sickle cell disease. Pediatrics. 2003;111(6 Pt 1):e661–5. Epub 2003/06/05. . [DOI] [PubMed] [Google Scholar]
- 16. Schmid P, Ravenell KR, Sheldon SL, Flegel WA. DARC alleles and Duffy phenotypes in African Americans. Transfusion. 2012;52(6):1260–7. Epub 2011/11/16. 10.1111/j.1537-2995.2011.03431.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, et al. The global distribution of the Duffy blood group. Nature communications. 2011;2:266 Epub 2011/04/07. 10.1038/ncomms1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oliveira TY, Harris EE, Meyer D, Jue CK, Silva WA Jr. Molecular evolution of a malaria resistance gene (DARC) in primates. Immunogenetics. 2012;64(7):497–505. Epub 2012/03/08. 10.1007/s00251-012-0608-2 . [DOI] [PubMed] [Google Scholar]
- 19. Le Pennec PY, Rouger P, Klein MT, Robert N, Salmon C. Study of anti-Fya in five black Fy(a-b-) patients. Vox sanguinis. 1987;52(3):246–9. Epub 1987/01/01. . [DOI] [PubMed] [Google Scholar]
- 20. Tournamille C, Le Van Kim C, Gane P, Le Pennec PY, Roubinet F, Babinet J, et al. Arg89Cys substitution results in very low membrane expression of the Duffy antigen/receptor for chemokines in Fy(x) individuals. Blood. 1998;92(6):2147–56. Epub 1998/09/10. . [PubMed] [Google Scholar]
- 21. Iwamoto S, Li J, Omi T, Ikemoto S, Kajii E. Identification of a novel exon and spliced form of Duffy mRNA that is the predominant transcript in both erythroid and postcapillary venule endothelium. Blood. 1996;87(1):378–85. Epub 1996/01/01. . [PubMed] [Google Scholar]
- 22. Carvalho TL, Ribolla PE, Curi RA, Mota LS. Characterization and transcriptional analysis of the promoter region of the Duffy blood group, chemokine receptor (DARC) gene in cattle. Veterinary immunology and immunopathology. 2009;132(2–4):153–9. Epub 2009/06/30. 10.1016/j.vetimm.2009.05.016 . [DOI] [PubMed] [Google Scholar]
- 23. Reiner AP, Lettre G, Nalls MA, Ganesh SK, Mathias R, Austin MA, et al. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT). PLoS genetics. 2011;7(6):e1002108 Epub 2011/07/09. 10.1371/journal.pgen.1002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thobakgale CF, Ndung'u T. Neutrophil counts in persons of African origin. Current opinion in hematology. 2014;21(1):50–7. Epub 2013/11/22. 10.1097/MOH.0000000000000007 . [DOI] [PubMed] [Google Scholar]
- 25. Afenyi-Annan A, Kail M, Combs MR, Orringer EP, Ashley-Koch A, Telen MJ. Lack of Duffy antigen expression is associated with organ damage in patients with sickle cell disease. Transfusion. 2008;48(5):917–24. Epub 2008/02/06. 10.1111/j.1537-2995.2007.01622.x . [DOI] [PubMed] [Google Scholar]
- 26. Shen H, Schuster R, Stringer KF, Waltz SE, Lentsch AB. The Duffy antigen/receptor for chemokines (DARC) regulates prostate tumor growth. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2006;20(1):59–64. Epub 2006/01/06. 10.1096/fj.05-4764com . [DOI] [PubMed] [Google Scholar]
- 27. Chan-Shu SA. The second example of anti-Duffy5. Transfusion. 1980;20(3):358–60. Epub 1980/05/01. . [DOI] [PubMed] [Google Scholar]
- 28. International HapMap C. The International HapMap Project. Nature. 2003;426(6968):789–96. 10.1038/nature02168 . [DOI] [PubMed] [Google Scholar]
- 29. International HapMap C, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buchanan CC, Torstenson ES, Bush WS, Ritchie MD. A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data. Journal of the American Medical Informatics Association: JAMIA. 2012;19(2):289–94. Epub 2012/02/10. 10.1136/amiajnl-2011-000652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. A haplotype map of the human genome. Nature. 2005;437(7063):1299–320. Epub 2005/10/29. 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. The International HapMap Project. Nature. 2003;426(6968):789–96. Epub 2003/12/20. 10.1038/nature02168 . [DOI] [PubMed] [Google Scholar]
- 33. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. Epub 2012/11/07. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Molecular ecology resources. 2008;8(1):103–6. Epub 2008/01/01. 10.1111/j.1471-8286.2007.01931.x . [DOI] [PubMed] [Google Scholar]
- 35. Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC bioinformatics. 2012;13:134 Epub 2012/06/20. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meny GM. The Duffy blood group system: a review. Immunohematology / American Red Cross. 2010;26(2):51–6. Epub 2010/10/12. . [PubMed] [Google Scholar]
- 37. Nemesure B, Wu SY, Hennis A, Leske MC. Distribution of Duffy Antigen Receptor for Chemokines (DARC) and Risk of Prostate Cancer in Barbados, West Indies. Journal of immigrant and minority health / Center for Minority Public Health. 2014. Epub 2014/01/09. 10.1007/s10903-013-9970-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nickel RG, Willadsen SA, Freidhoff LR, Huang SK, Caraballo L, Naidu RP, et al. Determination of Duffy genotypes in three populations of African descent using PCR and sequence-specific oligonucleotides. Human immunology. 1999;60(8):738–42. Epub 1999/08/10. . [DOI] [PubMed] [Google Scholar]
- 39. Parasol N, Cohen N, Zemishlany Z, Lerer B, Kosower NS. Duffy antigen/receptor for chemokines (DARC): genotypes in Ashkenazi and non-Ashkenazi Jews in Israel. Human biology. 2001;73(2):307–13. Epub 2001/07/12. . [DOI] [PubMed] [Google Scholar]
- 40. Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(16):6062–7. Epub 2004/04/13. 10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7(2):67–80. Epub 2006/03/31. 10.1007/s10048-006-0032-6 . [DOI] [PubMed] [Google Scholar]
- 42. Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM, et al. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86(2):127–41. Epub 2005/06/14. 10.1016/j.ygeno.2005.04.008 . [DOI] [PubMed] [Google Scholar]
- 43. Hiraoka N, Yamazaki-Itoh R, Ino Y, Mizuguchi Y, Yamada T, Hirohashi S, et al. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology. 2011;140(1):310–21. Epub 2010/10/20. 10.1053/j.gastro.2010.10.009 . [DOI] [PubMed] [Google Scholar]
- 44. Johnson LA, Clasper S, Holt AP, Lalor PF, Baban D, Jackson DG. An inflammation-induced mechanism for leukocyte transmigration across lymphatic vessel endothelium. The Journal of experimental medicine. 2006;203(12):2763–77. Epub 2006/11/23. 10.1084/jem.20051759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kalogeropoulos M, Varanasi SS, Olstad OK, Sanderson P, Gautvik VT, Reppe S, et al. Zic1 transcription factor in bone: neural developmental protein regulates mechanotransduction in osteocytes. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24(8):2893–903. Epub 2010/04/01. 10.1096/fj.09-148908 . [DOI] [PubMed] [Google Scholar]
- 46. Patterson AM, Siddall H, Chamberlain G, Gardner L, Middleton J. Expression of the duffy antigen/receptor for chemokines (DARC) by the inflamed synovial endothelium. The Journal of pathology. 2002;197(1):108–16. Epub 2002/06/26. 10.1002/path.1100 . [DOI] [PubMed] [Google Scholar]
- 47. Segerer S, Regele H, Mac KM, Kain R, Cartron JP, Colin Y, et al. The Duffy antigen receptor for chemokines is up-regulated during acute renal transplant rejection and crescentic glomerulonephritis. Kidney international. 2000;58(4):1546–56. Epub 2000/09/30. 10.1046/j.1523-1755.2000.00316.x . [DOI] [PubMed] [Google Scholar]
- 48. Peiper SC, Wang ZX, Neote K, Martin AW, Showell HJ, Conklyn MJ, et al. The Duffy antigen/receptor for chemokines (DARC) is expressed in endothelial cells of Duffy negative individuals who lack the erythrocyte receptor. The Journal of experimental medicine. 1995;181(4):1311–7. Epub 1995/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nature genetics. 1995;10(2):224–8. Epub 1995/06/01. 10.1038/ng0695-224 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Systemic expression levels of DARC in humans. Data from indicated microarray expression datasets were used to generate an eFP Browser image that depicts the unbiquitous expression of DARC/ACKR1 in the skeletal, digestive and immune systems. Adapted from the eFP by R.Patel. Images by E.T. Hamanishi. Data from GSE1133, E-GEOD-7307, GSE3526, GSE2361, GSE19650, E-GEOD-6257. Data normalized by MAS 5.0 method TGT value 100
(TIFF)
List of abbreviations for the 1,000 Genomes populations mentioned in Fig 1.
(PNG)
Data Availability Statement
Data are available from the 1,000 Genomes website and FTP servers (http://browser.1000genomes.org/index.html).