Abstract
Vinculin is filamentous (F)-actin-binding protein enriched in integrin-based adhesions to the extracellular matrix (ECM). Whereas studies in 2-dimensional (2D) tissue culture models have suggested that vinculin negatively regulates cell migration by promoting cytoskeleton–ECM coupling to strengthen and stabilize adhesions, its role in regulating cell migration in more physiologic, 3-dimensional (3D) environments is unclear. To address the role of vinculin in 3D cell migration, we analyzed the morphodynamics, migration, and ECM remodeling of primary murine embryonic fibroblasts (MEFs) with cre/loxP-mediated vinculin gene disruption in 3D collagen I cultures. We found that vinculin promoted 3D cell migration by increasing directional persistence. Vinculin was necessary for persistent cell protrusion, cell elongation, and stable cell orientation in 3D collagen, but was dispensable for lamellipodia formation, suggesting that vinculin-mediated cell adhesion to the ECM is needed to convert actin-based cell protrusion into persistent cell shape change and migration. Consistent with this finding, vinculin was necessary for efficient traction force generation in 3D collagen without affecting myosin II activity and promoted 3D collagen fiber alignment and macroscopical gel contraction. Our results suggest that vinculin promotes directionally persistent cell migration and tension-dependent ECM remodeling in complex 3D environments by increasing cell–ECM adhesion and traction force generation.—Thievessen, I., Fakhri, N., Steinwachs, J., Kraus, V., McIsaac, R. S., Gao, L., Chen, B.-C., Baird, M. A., Davidson, M. W., Betzig, E., Oldenbourg, R., Waterman, C., M., Fabry, B. Vinculin is required for cell polarization, migration, and extracellular matrix remodeling in 3D collagen.
Keywords: 3D cell migration, cell morphodynamics, traction force generation, integrin
Cell migration is critical for embryonic development and tissue regeneration and can have a major impact on diseases such as cancer metastasis or autoimmunity. During tissue morphogenesis and neoplastic processes, many cells migrate in a mesenchymal mode, which is characterized by a highly polarized cell shape with actin-based protrusions and adhesions to the extracellular matrix (ECM). Mesenchymal cell migration requires the integration of actin cytoskeleton-based force generation and cell adhesion to the ECM through integrin adhesion receptors. How force generation and ECM adhesion affect cell motility has been studied mainly by using 2-dimensional (2D) cell culture substrates (1), which cannot replicate the mechanics of cell migration in vivo, where cells have to overcome the steric hindrance of the ECM to move forward. How traction force generation and ECM adhesion are converted into cell motion in 3-dimensional (3D) environments remains unclear (2–5). As 3D matrices of collagen, fibrin, or cell-derived matrices are being used more frequently (6), answering these questions becomes increasingly important.
The coordinated migration of cells in vivo often requires a remodeling of the ECM, which modulates haptic and mechanical guidance cues for the cells (7–10). In turn, excessive ECM remodeling may promote cell motility in pathologic conditions such as cancer metastasis (11, 12). Cell motion and ECM remodeling both depend on cellular traction forces that enable cells to move forward and apply tension, thus remodeling the ECM. It remains unknown, however, which adhesion and cytoskeleton proteins are necessary for traction force generation and tension-dependent remodeling of complex 3D matrices.
The adhesion protein vinculin is present in cell–cell junctions and in focal adhesions (FAs), where it links filamentous (F)-actin to the integrin-binding protein talin, to transmit forces between the actin cytoskeleton and ECM (13–16). Vinculin restrains cell migration velocity on 2D substrates (13, 16, 17). Consistent with this effect, vinculin expression is reduced in several metastatic human tumors, and cancer cells overexpressing vinculin show reduced metastatic capacity when injected into nude mice (18, 19), suggesting that vinculin also inhibits cell migration in vivo. However, as tumor metastasis involves other vinculin-dependent processes, such as cell survival, apoptosis, and cell proliferation, it is unclear whether vinculin decreases tumor metastasis in the mouse model by restraining cell migration or by other mechanisms. Indeed, vinculin seems to affect cell migration in 3D environments fundamentally differently than 2D cell migration. It promotes, rather than impairs, the invasion of fibroblasts into 3D collagen gels and the invasion of cancer cells in vitro and in vivo in an ECM-dependent manner (16, 20). The collagen invasiveness of different cancer cells correlates with the magnitude and anisotropy of contractile forces generated by the cells and with the anisotropy of cell shape (21), suggesting that vinculin also promotes 3D collagen invasion by increasing contractile force generation and cell shape anisotropy. Indeed, on 2D substrates, vinculin affects both cellular force generation and morphology (13). However, how vinculin influences the morphology and contractile forces of cells in 3D matrices is unknown.
We report the effects of vinculin gene disruption on the migration, morphodynamics, force generation, and ECM remodeling of primary murine embryonic fibroblasts (MEFs) in a 3D collagen type I matrix. Vinculin promoted persistent protrusion, traction force generation, stable polarization, and directionally persistent migration in 3D collagen. Moreover, vinculin was necessary for efficient fiber alignment and contraction of 3D collagen. Our results suggest a critical role for vinculin-dependent traction force generation in coordinating cell motility and ECM remodeling in complex 3D environments.
MATERIALS AND METHODS
Isolation of primary MEFs and vinculin gene disruption
Primary vinculin-deficient MEFs and parental control MEFs were generated as described elsewhere (13). Animals were maintained according to guidelines approved by the National Heart, Lung, and Blood Institute Animal Care and Use Committee [National Institutes of Health, Bethesda, MD, USA].
Cloning and construction of fluorescent protein conjugates
The tandem tomato (tdTomato)-farnesyl and –F-tractin fusion constructs were generated by using C1 or N1 vector backbones (Clontech-style). The tdTomato fluorescent protein cDNA was amplified with a 5′ primer encoding an AgeI site and a 3′ primer encoding a BspEI site (C1) or a NotI site (N1) for insertion into C1 or N1 vector backbones (Clontech-style). PCR products were purified, digested, and ligated into similarly treated enhanced green fluorescent protein (EGFP)-C1 and EGFP-N1 vector backbones, yielding tdTomato-C1 and tdTomato-N1 cloning vectors. To generate the F-tractin fusion, residues 9–40 (MARPRGAGPCSPGLERAPRRSVGELRLLFEA) of the F-tractin–binding domain of rat ITPKA (NM_031045.2) were synthesized (Integrated DNA Technologies, Coralville, IA, USA) and annealed. The resulting product and tdTomato-N1 were sequentially digested with HindIII and BamHI and ligated to yield tdTomato–F-tractin. tdTomato-farnesyl was constructed by using the 20-amino-acid (KLNPPDESGPGCMSCKCVLS) farnesylation signal from c-Ha-Ras (NM_001130442.1). To produce the farnesyl fusion, pEGFP-farnesyl (Clontech Laboratories, Inc., Mountain View, CA, USA) was sequentially digested with AgeI and BspEI. The purified product was ligated to a similarly treated tdTomato-C1 cloning vector, to yield tdTomato-farnesyl. Plasmid DNA was purified with the Plasmid Maxi kit (Qiagen, Valencia, CA, USA) and sequence verified (Bioanalytical and Molecular Cloning DNA Sequencing Laboratory, Florida State University, Tallahassee, FL, USA). Proper localization of fusion proteins was tested by transfection in HeLa cells (CCL2 line; American Type Culture Collection, Manassas, VA,) using Effectene (Qiagen) and ∼1 μg vector.
Transfection and collagen gel preparations
MEFs were either nucleofected (solution MEF2, program T20; Lonza, Basel Switzerland) or transfected (Lipofectamine 2000, Life Technologies, Carlsbad, CA, USA) with the appropriate constructs, according to manufacturer’s instructions and subsequently cultured for 15–18 h. Collagen I gel preparations for cell morphodynamics and 3D traction force measurements were prepared as described previously (22). In brief, a 2.4 mg/ml collagen I solution containing 0.8 mg/ml collagen R (rat tail collagen I; Serva, Heidelberg, Germany), 1.6 mg/ml collagen G (bovine collagen I; Biochrom, Cambridge, United Kingdom), and 25 mM NaHCO3 in DMEM was prepared under sterile conditions on ice. Collagen polymerization was induced by adjusting the pH to ∼10 through the addition of NaOH to a final concentration of 15 mM. Then, 20,000 cells/ml collagen mixture were immediately resuspended, and 4 ml cell suspensions were plated in 6 cm dishes. For Bessel beam and liquid crystal polarizing (LCPol)Scope microscopy, collagen matrix contraction, 3D cell migration, and cell morphology measurements, a 2.4 mg/ml collagen I solution containing 2.4 mg/ml collagen I (rat tail collagen I; BD Biosciences, Franklin Lakes, NJ, USA) and 25 mM NaHCO3 in DMEM was prepared under sterile conditions on ice. Collagen polymerization was induced by adding NaOH to a final concentration of 15 mM, 20,000 cells/ml collagen mixture were immediately resuspended, and the cell suspension was pipetted, either into 24-well plates (collagen matrix contraction; 0.2 ml/well) or on 22 × 22 mm glutaraldehyde-activated coverslips in 3.5 cm dishes (Bessel beam and LCPolScope microscopy, 3D cell migration, morphology; 85 μl/coverslip). The gels were allowed to polymerize for 2 h at 37°C and 5% CO2 in a humidified incubator, before medium (DMEM, 20% FBS, penicillin, and streptomycin) was added. 3D cultures were imaged 15–18 h after gelation, unless otherwise indicated.
Western blot analysis
Cells were scraped for 1 min in ice-cold Laemmli buffer containing protease inhibitors (Roche Diagnostics, Indianapolis, IN, USA) and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO, USA), triturated 8–10 times through a 25-gauge needle, boiled for 5 min, and frozen in aliquots. Proteins (20 μg/lane) were separated by SDS-PAGE and electroblotted to PVDF-membrane (Millipore, Billerica, MA, USA). The membranes were blocked [5% bovine serum albumin (BSA) in Tris-buffered saline/0.1% Tween20], immunoprobed with primary and horseradish peroxidase (HRP)–conjugated secondary antibodies and developed with ECL substrate (Thermo Scientific-Pierce Biotechnology, Rockford, IL, USA). Antibodies included monoclonal anti-vinculin (1:1000; Sigma-Aldrich), rabbit polyclonal anti-pS19myosin light chain (MLC)-2 (1:1000; Cell Signaling Technology, Danvers, MA, USA) and anti-MLC-2 (1:1000; Cell Signaling Technology), and anti-mouse and -rabbit HRP conjugates (1:2.000; Jackson ImmunoResearch Laboratory, West Grove, PA, USA).
Collagen matrix contraction assay
A 200 μl cell suspension (4 × 105 cells/ml collagen I; 2.4 mg/ml) per well was plated into BSA (3%/PBS)-coated 24-well plates and kept for 2 h at 37°C for polymerization. Then, 2 ml medium was added on top of the gels. Gels containing control and vinculin-knockout (Vcl-KO) MEFs as well as gels without cells as control were cultured for 24 h before imaging.
Fibronectin-assembly assay
Cells (3 × 105) were seeded on uncoated coverslips placed in 3.5 cm dishes and incubated in DMEM/20% FCS for 4 h. Fibronectin (FN) was then added to the medium to a final concentration of 10 μg/ml. Samples and negative controls without added FN were cultured for another 16 h, washed 2× with PBS, fixed in 4% paraformaldehyde/PBS (20 min, room temperature), and blocked (2% BSA/PBS) for 1 h at room temperature. The cells were incubated overnight at 4°C with mouse monoclonal anti-FN (1:200; Sigma-Aldrich), washed 3 times in PBS, incubated with Alexa488-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch Laboratory) for 1 h at room temperature, washed 3 times in PBS, and mounted in fluorescent mounting medium (Ibidi, Martinsried, Germany), before laser scanning confocal imaging of FN.
Microscopy
Phase-contrast imaging of MEFs migrating in 3D collagen I ECM was performed on an inverted microscope system (TE300; Nikon, Melville NY, USA), with a ×20/0.75 Plan Apo objective and 0.52 NA condenser. Images were taken in 1 plane every 10 min for 12 h. Spinning-disk confocal (SDC) imaging of tdTomato-farnesyl labeled cells for aspect ratio and quantification of lamellipodial protrusion was performed on an inverted microscope system (TE2000E2; Nikon) (23), with a ×60/1.2 NA Plan Apo violet-corrected water-immersion objective. For aspect ratio measurements, image z-stacks over the cell depth were taken at 1 μm z-section distance. Lamellipodial protrusion dynamics of F-tractin–labeled cells were analyzed on single-plane images series of horizontally protruding regions (cell edge in focus throughout the duration of the time lapse) at 5 s intervals. Laser scanning confocal imaging of cells expressing tdTomato-farnesyl for quantification of morphodynamics was performed on an upright SP5X laser scanning confocal microscope (Leica, Wetzlar, Germany), with a ×20/1.0 NA dip-in water-immersion objective. Image z-stacks were taken over the cell depth at 0.5–1 μm z-section distance at 5 min intervals. The same system and objective lens were used for collagen reflection-based imaging of 3D traction forces. Image z-stacks of individual cells in 400 × 400 × 370 μm volumes were taken at a 0.74 μm z-section distance and 488 nm wavelength, before and after cell relaxation in 6.25 μg/ml cytochalasin D for 45 min at room temperature. The SP5X system was also used for laser scanning confocal imaging of FN assembled by MEFs. 3D image stacks of 234 × 234 × 10 μm were acquired at 0.2 μm z-section distance with a ×63/1.4 NA Plan Apo oil objective lens. Collagen matrix contraction was imaged on a Nikon SMZ15000 binocular microscope. 4D Bessel beam imaging of tdTomato–F-tractin–labeled MEFs was performed on a custom built Bessel beam plane illumination microscope (24). Image stacks of a 65 × 65 × 50 μm image volume were taken every 30 s, with a ∼500 nm thick scanning 2-photon Bessel beam and a 0.8 NA objective lens. For polarization microscopy, the alignment of collagen fibers in the ECM surrounding Vcl-KO and control cells was analyzed with the LCPol microscope (25). The LC-PolScope was implemented on an inverted microscope stand (200M; Zeiss, Jenna, Germany) with a ×20/0.5 NA objective lens, 0.55 NA long-working distance (LWD) condenser, a halogen lamp with 546/30 nm interference filter, and polarization analysis hardware and software (www.OpenPolScope.org).
Image analysis
3D cell migration
The x/y-displacement of nuclei was manually tracked (MetaMorph; Molecular Dynamics, Sunnyvale, CA, USA). Instantaneous velocities (displacement per time between consecutive frames) were determined, and averaged for each cell.
Cell polarization
Image stacks were maximum z-projected (MetaMorph), and the aspect ratio of the cell was determined.
Reconstruction of Bessel beam images
3D image stacks taken by 2-photon Bessel beam plane illumination microscopy were deconvolved by Amira software (FEI, Hillsboro, OR, USA) by using the measured point-spread function.
3D lamellipodium protrusion velocity
Kymographs were placed through lamellipodial protrusions, and lamellipodium protrusion velocity was determined.
Cell protrusion persistence
Confocal image stacks were maximum projected, thresholded, and binarized. Custom-written MatLab software (MathWorks, Natick, MA, USA) was used to position a minimum bounding circle around cell segments at each time point, to define the outermost cell borders. The peripheral segmented cell area within distances of 2–20 μm from the minimum bounding circle toward the circle center was recorded at each time point. The spatial overlap of these subsegments between multiple combinations of 2 time points was calculated for increasing time intervals over the course of a movie. The decay of protrusion overlap over time was fitted to a stretched exponential function, to derive the time constant τ as an indicator for the protrusion remodeling time.
Cell reorientation dynamics
The main axis orientation of segmented cells was recorded by using the MatLab Regionprops function, and the angle change in axis orientation was calculated at 5 min intervals.
Protrusion range and cell body shape
Minimum area outer bonding ellipses and maximum area inner bounding ellipses were manually fitted (ImageJ; National Institutes of Health, Bethesda, MD, USA) to the cells on maximum projections of confocal image stacks. Aspect ratios were calculated as the ratio of the major to minor axes of the respective ellipse. The protrusion range was calculated as the area of the minimum outer bounding ellipse.
3D traction force reconstruction and analysis
Custom-written C++ software was used to calculate 3D collagen displacement fields and to reconstruct force fields (to be published separately). In brief, 3D particle velocimetry was used to calculate collagen displacement fields. To reconstruct 3D force fields, artificial displacement fields were generated based on a nonlinear material model, to account for collagen strain stiffening and fiber buckling, and fitted to the measured displacement fields, by using the minimal amount of total force necessary to achieve a good fit.
3D collagen matrix contraction
Collagen plugs were manually segmented, and the plug area was quantified (MetaMorph). Gel contraction was determined as the ratio of collagen plug size after culture time and initial plug size.
Quantification of collagen fiber alignment
Collagen birefringence retardance and slow axis orientation were calculated as described previously (25).
FN fibril assembly
Individual z-sections of an image stack were corrected for extracellular background, and fluorescence intensities for all voxels of an entire image stack were integrated to quantify total FN amounts assembled or distributed according to their gray values.
Statistical analysis
Normally distributed data were analyzed with the 2-tailed Student’s t test for unequal variance, with a significance value specific for each analysis (0.05, if not mentioned). Nonnormally distributed data were analyzed with the Mann-Whitney U test, yielding a significance value specific for each analysis (0.05, if not mentioned). The angle distribution of cell axis reorientation was analyzed with the Kolmogorov-Smirnov test.
RESULTS
Vinculin promotes cell elongation, stable orientation, and directionally persistent migration in 3D collagen ECM
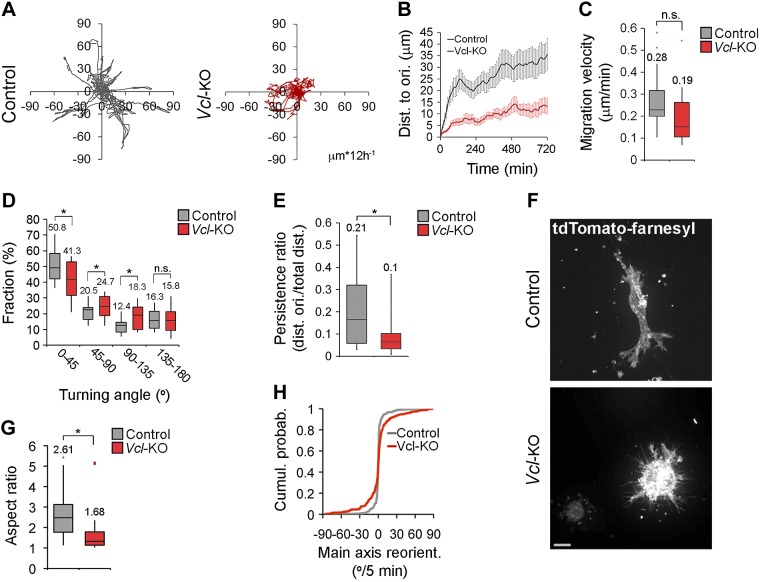
To investigate how vinculin facilitates 3D collagen invasion (16), we characterized its effects on 3D migration at the level of single cells. To exclude clonal variations and long-term adaptation of our cells to vinculin loss and cell culture conditions, we used vinculin-deficient primary MEFs (Vcl-KO MEF), generated by gene excision induced in cells derived from E13.5 Vclflox/flox embryos (13). We embedded control and Vcl-KO MEFs in collagen I gels (2.4 mg/ml), recorded a phase-contrast time-lapse image series of individual cells over 12 h, and tracked the displacement of their nuclei in the x/y direction. This method revealed an impaired migration of Vcl-KO MEFs, which reached less than half the distance to the origin compared with the control MEFs (Fig. 1A, B; Supplemental Movie 1). Whereas the migration velocity of Vcl-KO MEFs was only slightly (nonsignificantly; P = 0.062) reduced vs. control MEFs (Fig. 1C), the distribution of the turning angle between consecutive frames (10 min) showed a significant shift toward wider angles in Vcl-KO vs. control MEFs (Fig. 1D). Consistent with this, the persistence ratio (distance to origin/total distance) was significantly lower for Vcl-KO vs. control MEFs (Fig. 1E). These results demonstrate that vinculin promotes the migration of primary MEFs in 3D collagen by increasing the directional persistence.
Figure 1.
Vinculin is necessary for the persistent migration and normal polarization of primary MEFs in 3D collagen. A) Tracks of 17 control and 16 Vcl-KO MEFs, migrating in 2.4 mg/ml collagen I gels, monitored for 12 h at a 10 min frame rate. Tracks were recorded in the x/y dimensions. B) Quantification of distance from the origin over time. n = 17 (control) and 16 (Vcl-KO) MEFs; error bars, sem. C) Box-and-whisker plot of cell migration velocity. n = 17 (control) and 16 (Vcl-KO) MEFs. Numbers indicate means; P = 0.062, Student’s t test. D) Plots of turning angle distribution. n = 17 (control) and n = 16 (Vcl-KO) MEFs; data as in (C). *P < 0.05, Student’s t test. E) Plot of persistence ratio (distance to the origin:total distance) of control and Vcl-KO MEFs after 12 h migration. n = 17 (control) and 16 (Vcl-KO) MEFs; data as in (C). *P < 0.05, Student’s t test. F) Maximum projections of confocal stacks of live control and Vcl-KO MEFs expressing tdTomato-farnesyl. Scale bar, 10 μm. G) Plot of aspect ratio of live control and Vcl-KO MEFs in 3D collagen. n = 27 (control) and 33 (Vcl-KO) MEFs; data as in (C). *P < 0.005, Student’s t test. H) Cumulative probability distribution of the reorientation angle of the cell main axis of tdTomato-farnesyl labeled control and Vcl-KO MEFs at 5 min frame rate (Fig. 3A; Supplemental Movie 3). n = 336 frames from 12 control MEFs and 280 frames from 10 Vcl-KO MEFs, Kolmogorov-Smirnov test.
To investigate how vinculin promotes directional persistence in collagen, we first characterized the morphology of control and Vcl-KO MEFs in 3D collagen. We embedded control and Vcl-KO MEFs, expressing tdTomato-farnesyl to label cell membranes in collagen I and imaged the cells with SDC microscopy 24 h after embedding. Whereas control MEFs displayed an elongated morphology typical of fibroblasts in 3D ECM, Vcl-KO MEFs appeared rounded (Fig. 1F). Quantification of the aspect ratio revealed that Vcl-KO MEFs were significantly less elongated than control MEFs (Fig. 1G). To test whether the rounder morphology of Vcl-KO MEFs was related to their decreased directional persistence, we analyzed whether loss of vinculin affected the orientation of the cell’s main axis over time. We recorded a confocal time-lapse image series of control and Vcl-KO MEFs expressing tdTomato-farnesyl in 3D collagen (Supplemental Movie 3), segmented the cells over time, and analyzed the reorientation angles of the cell main axis at 5 min intervals. While control MEFs showed a stable orientation of the cell main axis over time, the main axis in Vcl-KO MEFs frequently changed direction. Consistent with this finding, Vcl-KO MEFs showed a significantly wider cumulative probability distribution of the main axis reorientation angle than did control MEFs (Fig. 1H). Together, these results show that vinculin promotes the elongation and stable orientation of primary MEFs migrating in 3D collagen.
Vinculin is dispensable for lamellipodium formation in 3D collagen ECM
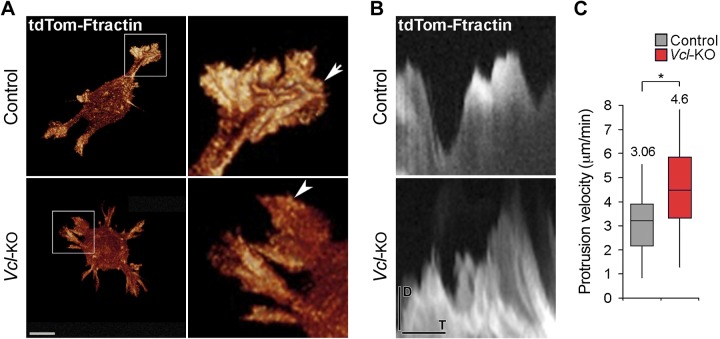
We next tested whether the impaired elongation of Vcl-KO MEFs in 3D collagen is caused by a defect in actin-based cell protrusion. We imaged lamellipodia of live control and Vcl-KO MEFs expressing the F-actin marker tdTomato–F-tractin in 3D collagen at 30 s time intervals by 2-photon Bessel beam plane illumination microscopy (24). Both control and Vcl-KO MEFs displayed multiple sheet-like lamellipodial protrusions that were comparable in size and morphology (Fig. 2A, arrow, arrowhead; Supplemental Movie 2). We then monitored the protrusion and retraction dynamics of lamellipodia in live control and Vcl-KO MEFs expressing tdTomato–F-tractin at 5 s time intervals using SDC microscopy. Kymograph analyses (Fig. 2B) revealed a slight increase in lamellipodium protrusion velocity in Vcl-KO vs. control MEFs (Fig. 2C). These data show that the formation of cell protrusions was not obviously affected by the loss of vinculin, suggesting that impaired cell elongation, orientation, and migration of Vcl-KO MEFs in 3D collagen are not caused by a cell protrusion formation defect.
Figure 2.
Vinculin is dispensable for lamellipodia formation and leading-edge protrusion of primary MEFs in 3D collagen. A) Projections of Bessel beam image stacks of live control and Vcl-KO MEFs expressing tdTomato–F-tractin, to label actin-based cell protrusions (insets). Note the presence of sheetlike, lamellipodial protrusions of similar size and morphology in control (arrow) and Vcl-KO MEFs (arrowhead). Scale bar, 10 μm. B) Kymographs placed along the direction of protrusion through lamellipodia of live control and Vcl-KO MEFs in 3D collagen I ECM, showing lamellipodium dynamics. Cell exterior: top, cell interior: bottom. SDC confocal time lapses, 5 s frame rate. D, distance; T, time. C) Kymograph-based quantification of lamellipodium protrusion velocity of control and Vcl-KO MEFs in 3D collagen I. n = 35 (control) and 26 (Vcl-KO) kymographs from 12 (control) and 9 (Vcl-KO) MEFs. Numbers indicate means; *P < 0.005, Student’s t test.
Vinculin is essential for persistent cell protrusion and efficient traction force generation of primary MEFs in 3D collagen ECM
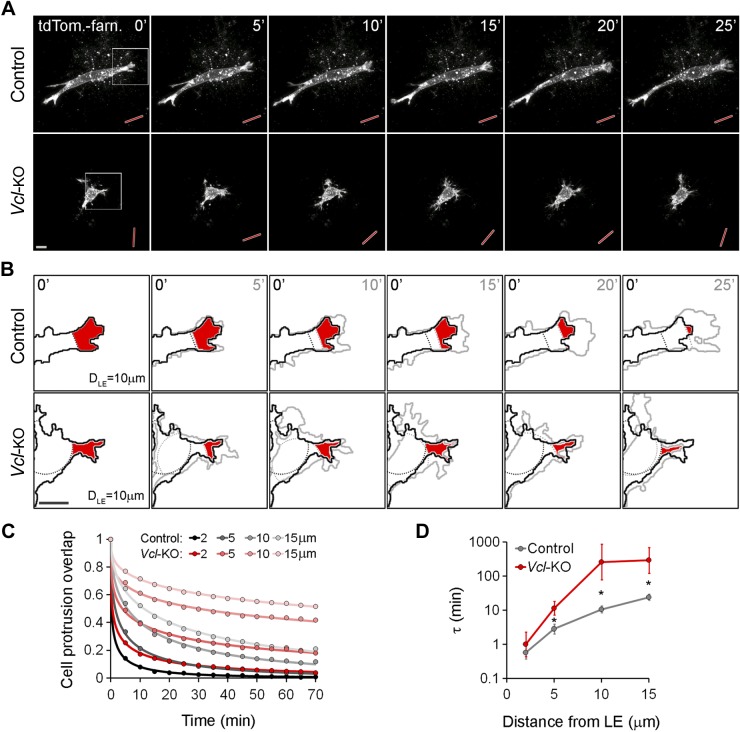
Vinculin has been shown to strengthen cell adhesion on planar ECM substrates (17, 26), and we hypothesized that vinculin promotes cell elongation, stable orientation, and directionally persistent migration in 3D collagen by strengthening cell adhesion to the collagen fiber network. To test this possibility, we first examined the stability of cell protrusions. On our confocal time-lapse image series of control and Vcl-KO MEFs expressing tdTomato-farnesyl (Supplemental Movie 3), we quantified the decay of spatial overlap between defined peripheral cell regions (cell area in the first 2, 5, 10, and 15 μm from the cell poles; Supplemental Fig. S1) at increasing time intervals, as a measure of the persistence of cell protrusions (Fig. 3B). Control MEFs typically showed a persistent advance of leading-edge protrusion over time (Fig. 3A), resulting in a rapid decay of the protrusion overlap for different distances from the leading edge (Fig. 3B, red areas, C). In contrast, Vcl-KO MEFs showed repeated protrusion and retraction, with less net advance of the cell edge (Fig. 3A), resulting in a significantly slower decay of the protrusion overlap (Fig. 3B, red areas, C). Stretched exponential fitting of the protrusion overlap decay (Fig. 3C) revealed an increasing time constant with increasing distance from the leading edge for both control and Vcl-KO MEFs (Fig. 3D), demonstrating that the turnover time of protrusive cell regions increases with distance from the leading edge. Vcl-KO MEFs showed significantly slower protrusion turnover times than control MEFs at leading-edge distances of more than 2 μm (Fig. 3D). These data demonstrate that vinculin, although it is dispensable for protrusion formation, promotes persistent cell edge protrusion in 3D collagen ECM, suggesting that vinculin-mediated cell adhesion to the ECM stabilizes protrusions against retraction in 3D collagen, thus promoting a net advance of the leading edge.
Figure 3.
Vinculin promotes persistent protrusion and stable orientation of primary MEFs migrating in 3D collagen. A) Projections of confocal image stacks of live tdTomato-farnesyl–expressing control and Vcl-KO MEFs in 3D collagen; 5 min frame rate. Persistent protrusion and stable main axis orientation (red rods) were observed in control MEFs, but repeated protrusion–retraction and frequent change in main axis orientation in Vcl-KO MEFs. Scale bar, 10 μm. B) Magnification of insets from (A) showing overlay of cell segments at t = 0 min (black outline) and at the indicated time points (gray outlines). Dashed lines: 10 μm distance from the leading edge (DLE; minimum bounding circle at DLE = 0 μm is not shown) for both time points in each panel (black: t = 0 min; gray: t = 5, 10, 15, 20, and 25 min). Red areas: overlap of peripheral cell area at DLE = 10 μm limits between t = 0 min and the respective time points. There was a persistent decrease in overlap in control MEFs, but a repeated decrease–increase of overlap in Vcl-KO MEFs. C) Decay of peripheral cell area (protrusion) overlap at increasing time intervals for DLE = 2, 5, 10, and 15 μm in control and Vcl-KO MEFs. Dots: original data from 12 control and 10 Vcl-KO MEFs with multiple overlap measurements; lines: stretched exponential fits. Decay of protrusion overlap was slower in Vcl-KO vs. control MEFs for all DLE. D) Quantification of time constant (τ), representing protrusion turnover, for DLE = 2, 5, 10, and 15 μm, derived from stretched exponential fits. n = 12 control and 10 Vcl-KO MEFs; error bars, sem; *P < 0.05, Student’s t test.
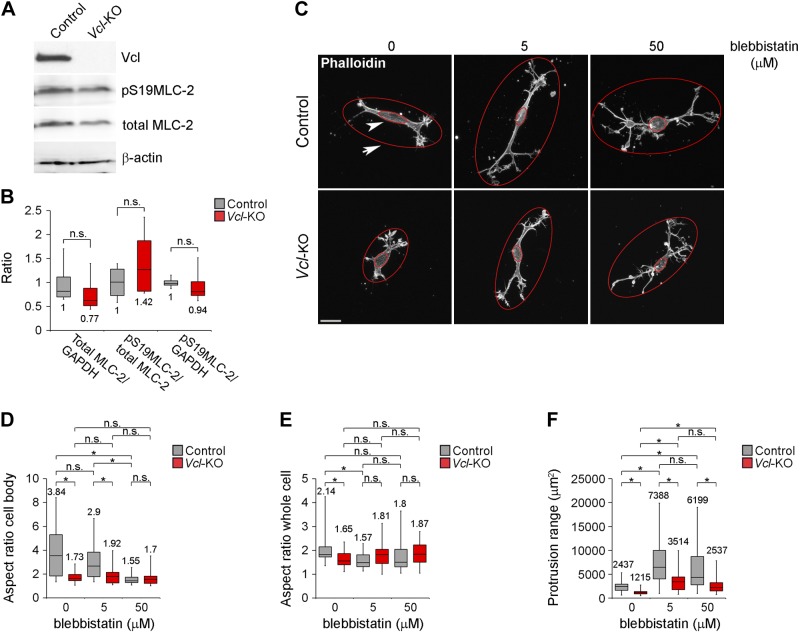
We have reported that vinculin-deficient clonal MEFs have a softer, more fluid cytoskeleton than do wild-type clonal MEFs (16). In addition, myosin II inhibition perturbs cell polarization and directional migration in a manner similar to vinculin depletion (27). Hence, we sought to test whether the protrusion and elongation defects of Vcl-KO MEFs were caused by reduced cellular contractility, rather than reduced cell adhesion to the ECM. To this end, we first assessed the contractile status of primary MEFs plated on collagen I ECM by Western blot analysis of MLC-2 expression and Ser19 phosphorylation, as an indicator of the activation of myosin II (Fig. 4A, B). Quantitative analysis showed that loss of vinculin had no effect on the level of either MLC-2 expression or phosphorylated MLC-2, relative to total MLC-2, (Fig. 4B), indicating that global myosin II contractility was not significantly affected by the loss of vinculin. Next, we tested whether inhibition of cellular contractility in control MEFs phenocopied the protrusion and elongation defects induced by vinculin deficiency and whether inhibition of contractility affected the protrusion and elongation defects of Vcl-KO MEFs. We treated control and Vcl-KO MEFs embedded in 3D collagen for 1 h with low (5 μM) or high (50 μM) concentrations of blebbistatin, to inhibit myosin II contractility, and imaged the cells after fixation and phalloidin staining by laser scanning confocal microscopy (Fig. 4C). We fitted minimum outer bounding ellipses around the whole cell (Fig. 4C, arrow), to quantify the protrusive range and elongation of the cell, and maximum inner bounding ellipses around the cell body (Fig. 4C, arrowhead) to quantify its elongation. Whereas untreated control MEFs showed a significantly higher cell body aspect ratio than untreated Vcl-KO MEFs, increasing concentrations of blebbistatin successively reduced the cell body aspect ratio of control MEFs (Fig. 4D), such that the aspect ratio of control cells treated with 50 μM blebbistatin was no different than that of untreated Vcl-KO MEFs, indicating that myosin II contributes to the elongation of the cell body. In contrast, whereas the whole-cell aspect ratio of control cells was significantly higher than that of Vcl-KO MEFs, blebbistatin treatment abrogated this difference, with the ratio in blebbistatin-treated control cells at either concentration not significantly different from that of untreated Vcl-KO MEFs (Fig. 4F). In contrast to control MEFs, blebbistatin treatment had no effect on the aspect ratio of either the cell body or the whole cell in Vcl-KO MEFs. The loss of the polarity and elongation induced by blebbistatin in control MEFs, however, was not the result of reduced cell protrusion, but was associated with an increased protrusion range in both control and Vcl-KO MEFs (Fig. 4C, F), suggesting that reducing cellular contractility in control MEFs did not induce a protrusion phenotype similar to loss of vinculin (Fig. 3). Vcl-KO MEFs showed a notable significantly reduced protrusion range compared with control MEFs, with values of ∼50% compared with control MEFs under all conditions: untreated and 5 and 50 μM blebbistatin treatment (Fig. 4F). Together, these data demonstrate that reduced cell contractility is not the primary cause of the protrusion and polarization defects in the absence of vinculin and, together with our previous results, suggest that vinculin could promote cell protrusion and elongation by strengthening cell–ECM adhesion.
Figure 4.
Vinculin promotes protrusion of primary MEFs in 3D collagen independent of myosin II contractility. A) Western blot analysis of vinculin, phosphor-Ser19 MLC-2, total MLC-2, and β-actin (loading control) in control and Vcl-KO MEFs. B) Quantification of Western blot analyses as shown in (A); *P > 0.05, Student’s t test, n = 4. C) Maximum projections of confocal image stacks from control and Vcl-KO MEFs treated with 5 or 50 μM blebbistatin, or with DMSO (0). Large ellipses outside of cells (arrow) denote smallest outer-bounding ellipses for quantification of whole-cell aspect ratio and protrusion range. Small ellipses inside the cell body (arrowhead) denote largest inner-bounding ellipses for quantification of cell body aspect ratio. With increasing blebbistatin concentrations, the aspect ratio of the inner ellipse area decreased in control MEFs and increased for the outer ellipse area in control and Vcl-KO MEFs. Scale bar, 20 μm. D) Quantification of the inner ellipse aspect ratio, representing the cell body aspect ratio. *P < 0.05, Student’s t test, n = 20 cells per condition. E) Quantification of outer ellipse aspect ratio, representing the whole-cell aspect ratio. *P < 0.05, Student’s t test; n = 20 cells per condition. F) Quantification of outer ellipse area, representing protrusion range. *P < 0.05, Student’s t test, n = 20 cells per condition.
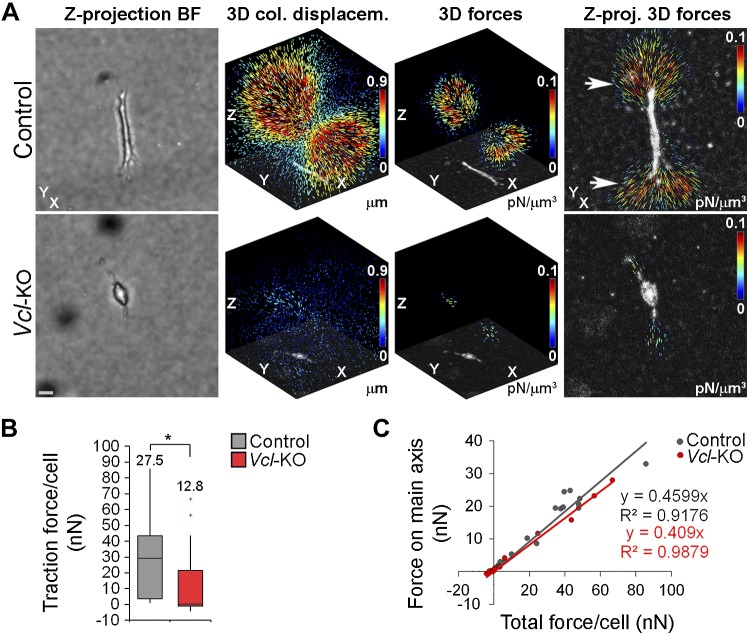
To begin to explore the role of vinculin in polarizing cells through its effects on adhesion, we tested whether vinculin is necessary for an efficient interaction of the cells with the 3D collagen network. We quantitatively assessed the effect of vinculin on the local deformation of collagen and performed 3D traction force microscopy of control and Vcl-KO MEFs embedded in collagen. We imaged the local collagen fiber network around the cells by confocal microscopy in the reflection mode and tracked the collagen fiber displacement after relaxation of cellular forces with cytochalasin D, to disassemble the actin cytoskeleton. Control cells showed 3D displacement fields that were elongated and extended over tens of micrometers (Fig. 5A). Consistent with this observation, reconstruction of 3D traction force fields revealed high traction exerted by control MEFs on the collagen ECM. The traction forces were highest at the cell poles (Fig. 5A, arrows), consistent with a role for vinculin in stabilizing cell protrusions in 3D collagen (Fig. 3D). In contrast, Vcl-KO MEFs showed much less overall collagen displacement and strongly reduced traction forces (Fig. 5B). Displacement and force fields of Vcl-KO MEFs appeared elongated, similar to those of control MEFs (Fig. 5A), despite their rounder cell shape (Fig. 1F, G). To characterize the geometry of the 3D traction force fields, we plotted the amount of traction force directed along the main force axis as a function of the total force for individual cells (Fig. 5C). This revealed that in both control and Vcl-KO MEFs, 40–45% of the total forces were directed along the main axis, indicating that the geometry of the force fields was not significantly altered in the absence of vinculin. Together, these data demonstrate that vinculin is required for the generation of high-traction forces in 3D collagen, suggesting that vinculin-mediated cytoskeleton–ECM force coupling is necessary to stabilize cell protrusions and hence to promote stable cell polarization and directionally persistent migration in 3D collagen.
Figure 5.
Vinculin promotes traction force generation of primary MEFs in 3D collagen. A) Traction force microscopy of control and Vcl-KO MEFs in 3D collagen, using confocal reflection (collagen) and bright-field (cell) imaging. The x/y/z axes are labeled for orientation. Left to right: bright-field projections of respective cells. Scale bar, 20 μm. 3D collagen displacement fields overlaid with negative minimum projections of the cells. 3D traction force reconstructions overlaid with negative minimum projections of the cells. Z-projections of reconstructed 3D forces, overlaid with negative minimum projections of the cells, showing localization of the highest forces at the cell poles (arrows). B) Box-and-whisker plot of total traction force per cell generated by control and Vcl-KO MEFs in 3D collagen. n = 18 (control) and 15 (Vcl-KO) MEFs; numbers indicate means; *P < 0.05, Mann-Whitney U test. C) Scatterplot of the fraction of cellular forces directed along a main force axis vs. total traction force for individual control and Vcl-KO MEFs. n = 18 (control) and 15 (Vcl-KO) MEFs.
Vinculin promotes FN remodeling and is critical for fiber alignment and contraction of 3D collagen
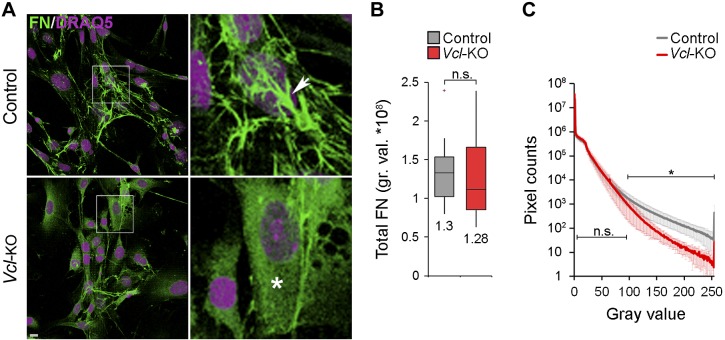
Cellular force generation can also affect the directionality of cell migration through tension-dependent remodeling and alignment of the ECM (7–10). We next investigated whether vinculin also affects ECM remodeling. FN is a critical ECM component that can affect the directionality of cell migration in vivo, and tensional forces promote its assembly into fibrils (28–30). To test the effect of vinculin on FN assembly in the absence of other ECM proteins, we applied a 2D FN assembly assay, where soluble FN was added to 2D cultures, bound by the cells, and assembled into an insoluble ECM, which we immunostained and quantified. Control and Vcl-KO MEFs assembled similar total amounts of FN, demonstrating that vinculin is not necessary for binding of soluble FN on the cell surface (Fig. 6A, B). However, whereas FN assembled by control MEFs appeared fibrous (Fig. 6A, arrow), FN assembled by Vcl-KO MEFs appeared smooth and evenly distributed across the cell (Fig. 6A, asterisk). Quantification of confocal image stack histograms revealed a progressive decrease in the number of pixels with high fluorescence intensities, representing bright, thick fibrils (Fig. 6A, arrow, C) that reached significance above 95 of 255 gray values (Fig. 6C). These data indicate that vinculin, although it is dispensable for the assembly of soluble FN by primary MEFs, is necessary for the subsequent formation of thick FN fibrils.
Figure 6.
Vinculin promotes assembly of soluble FN into fibers. A) Projections of confocal image stacks of control and Vcl-KO MEFs after FN immunofluorescence staining (green) and DRAQ5 labeling of nuclei (purple), showing FN assembly over 15 h, showing fibrous FN morphology in control MEFs (arrow) and homogenous FN morphology in Vcl-KO MEFs (arrowhead). Scale bar, 10 μm. B) Box-and-whisker plot of total assembled FN per cell for control and Vcl-KO MEFs (15 h). n = 15 (control) and 12 (Vcl-KO) MEFs; numbers indicate means; P = 0.93, Student’s t test. C) FN-immunofluorescence intensity histograms across confocal stacks of control and Vcl-KO MEFs. There was a progressive decrease in the number of high-intensity pixels in Vcl-KO vs. control MEFs. n = 15 (control) and 12 (Vcl-KO) MEFs; *P < 0.05 for gray values >95, Student’s t test.
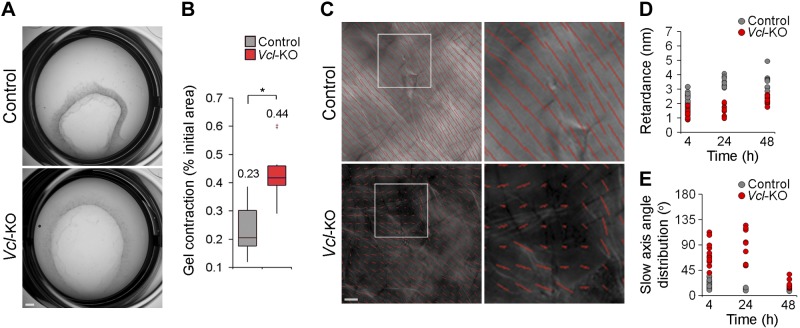
We next investigated whether vinculin affects the remodeling of 3D collagen ECM. To this end, we first tested the effect of vinculin on the macroscopic reorganization of collagen by measuring the contraction of collagen plugs containing control or Vcl-KO MEFs over 24 h. This test revealed that the Vcl-KO MEFs contracted the collagen matrix significantly less than did the control MEFs (Fig. 7A, B). This result was consistent with vinculin’s role in promoting 3D traction force generation, suggesting that vinculin-generated traction forces promote 3D collagen remodeling. To test this hypothesis, we analyzed whether vinculin also promotes the alignment of collagen fibers in 3D gels (12). We embedded control and Vcl-KO MEFs in collagen gels and imaged the gels 4, 24, and 48 h after gelation by polarized light-field microscopy (Fig. 7C), which enabled us to visualize the degree of fiber alignment (Fig. 7D, retardance) and the spatial distribution of the preferred fiber orientation (Fig. 7E, slow axis angle distribution) (25). Collagen gels containing control MEFs showed a significantly increased retardance and reduced slow axis angle distribution at 24 h after gelation (Fig. 7D, E). In contrast, gels containing Vcl-KO MEFs did not show a significant increase in retardance or decrease in slow axis angle distribution until 48 h after gelation. Together, these results demonstrate that vinculin promotes fibronectin fibrillization and the alignment and macroscopic contraction of 3D collagen ECM.
Figure 7.
Vinculin promotes macroscopic contraction and fiber alignment of 3D collagen. A) 3D collagen contraction by control and Vcl-KO MEFs after 24 h of culture. Collagen plugs containing Vcl-KO MEFs were increased compared with controls. Scale bar, 1 mm. B) Box-and-whisker plot of 3D collagen contraction by control and Vcl-KO MEFs after 24 h. n = 12 (control) and 12 (Vcl-KO) MEFs. Numbers indicate means; *P < 0.0001, Mann-Whitney U test. C) LCPolScope micrographs of collagen gels containing control or Vcl-KO MEFs, overlaid with rod maps showing spatial distribution of preferred collagen fiber orientation (rod orientation, insets) and degree of fiber alignment (rod length/image brightness, insets). Rod length was reduced and homogenous rod orientation lessened in gels containing Vcl-KO vs. control MEFs. Scale bar, 10 μm. D) Scatterplot of retardance (rod length/image brightness) of collagen containing control or Vcl-KO MEFs 4, 24, and 48 h after gelation. n = 22 (4 h), 16 (24 h), and 9 (48 h) control MEFs and 11 (4 h), 11 (24 h), and 13 (48 h) Vcl-KO MEFs. E) Quantification of slow axis angle distribution (rod orientation) of collagen containing control or Vcl-KO MEFs 4, 24, and 48 h after gelation. Number of cells at each time point is as in (D).
DISCUSSION
We report the effects of vinculin gene ablation on the morphodynamics and migratory behavior of primary MEFs in 3D collagen. Vinculin promoted traction force generation, persistent protrusion, stable polarization, and directionally persistent migration. In addition, vinculin promoted ECM fiber formation and alignment and the macroscopic contraction of 3D collagen. Our data suggest that vinculin-dependent traction forces promote and coordinate cell migration and ECM remodeling in complex 3D environments.
Our results demonstrate for the first time that vinculin is critical for cellular force generation in a 3D environment, consistent with its role in promoting force generation at FA on 2D substrates (13, 15, 16). However, whereas vinculin reduced cell migration velocity and directional persistence on 2D substrates (13, 16, 17), it promoted 3D cell migration by increasing the directional persistence, consistent with its role in promoting cell invasion into 3D collagen (16, 20). These data suggest that vinculin facilitates directionally persistent 3D cell motion by promoting high ECM traction force generation to overcome the steric hindrance of the complex collagen network (5). Consistent with this, perturbation of cellular force generation by myosin II inhibition or reducing α5β1 integrin expression similarly impaired the directional persistence of cell migration in 3D collagen (2, 27). In addition, vinculin was necessary for cell polarization in 3D collagen. Despite the rounded shape of the Vcl-KO MEFs, their traction force fields were normally polarized. Increased adhesion dynamics could explain this surprising finding. Faster adhesion growth and turnover rates, as described for vinculin-deficient cells on 2D substrates (13), could result in a polar distribution of adhesions in 3D collagen, despite a less elongated cell shape. Myosin II inhibition and low α5β1 integrin levels similarly impairs 3D cell polarization (2, 27). These data suggest that vinculin promotes cell polarization in 3D collagen by increasing traction force generation. Since vinculin depletion has been shown to soften the cytoskeleton of MEFs on 2D substrates (16), our data raise the question of whether vinculin promotes force generation and cell polarization in 3D collagen primarily by enabling cell adhesion or also by enhancing cell contraction. Our finding that vinculin was dispensable for the formation of cell protrusions, but necessary for their stabilization, suggests a key role for vinculin-mediated cell–ECM adhesion in 3D collagen, similar to its role in 2D cell adhesion (13–15, 26). In addition, we did not detect a substantial effect of vinculin depletion on the global Ser19 phosphorylation of MLC-2. Moreover, inhibition of myosin II contractility with low and high concentrations of blebbistatin did not affect the relative differences in cell protrusion between control and vinculin-deficient MEFs, suggesting that cell adhesion rather than contraction makes a critical contribution to vinculin-dependent protrusion stabilization. We propose that vinculin, by strengthening cell adhesion to the 3D collagen fiber network, stabilizes cell protrusions and promotes traction force generation, thus facilitating stable cell polarization and directionally persistent migration. Notably, 2D cell migration depends on an optimal balance of cell adhesion and contraction (31). Such a balance point may also determine 3D cell migration and could be shifted by loss of vinculin.
Loss of vinculin also increased lamellipodium protrusion velocity. Altered cell protrusion through effects on Rho-GTPases (27, 32, 33) or vinculin-binding actin regulators such as vasodilator-stimulated phosphoprotein (VASP) (34) or Arp2/3 (35) may also contribute to the polarization and persistence defects of Vcl-KO MEFs. However, vinculin depletion also increased lamellipodium protrusion velocity in 2D culture (data not shown), where loss of vinculin has been shown to increase directional persistence (16).
The reduced expression of vinculin in several human cancers (19) and the diminished capacity of cancer cells overexpressing vinculin to form metastases when injected into nude mice (18) suggest that vinculin inhibits cell migration during cancer metastasis in vivo, analogous to its role in regulating 2D cell migration. Our data are not consistent with such a model. However, migration and mechanical properties of mesenchymal fibroblasts differ from those of epithelial cancer cells. In addition, our study does not account for vinculin functions in cadherin-based cell–cell adhesion and regulation of proliferation and apoptosis, which are critical for cancer metastasis (36–39). It is noteworthy, though, that a recent study showed that vinculin also promotes the migration of premalignant mammary epithelial cells in 3D collagen (20).
Vinculin promoted not only cell polarization and directionally persistent migration in 3D collagen, but also homogenous fiber alignment and macroscopic contraction of 3D collagen. This observation is consistent with previous ones showing that directional cell migration is coupled to ECM alignment (7–10). Our data suggest a role for vinculin in coordinating the directionality of cell migration and ECM reorganization in complex 3D environments. Collagen fiber alignment and contraction are tension-dependent processes, indicating that vinculin may also promote collagen ECM remodeling through the generation of ECM traction forces. Control MEFs showed collagen displacement fields that often extended over several hundreds of micrometers and involved multiple individual collagen fibers. Tension-dependent collagen alignment through vinculin-dependent force generation in the cell vicinity could further support 3D cell polarization and migration by promoting collagen strain stiffening and hence force transmission through the otherwise compliant local collagen network (20, 40). Efficient force propagation through collagen may be important to overcome the steric hindrance of the 3D fiber network. Such a combinatorial effect of ECM compliance and dimensionality (41) could contribute to the opposing effects of vinculin on cell migration in compliant 3D collagen matrices and on stiff 2D substrates, where the major movement-resisting force that the migrating cells experience is ECM adhesion, per se. Quantifying the migration of vinculin-deficient cells under experimental conditions that allow for separate manipulation of collagen stiffness and pore size may help to further elucidate how vinculin couples 3D cell migration and collagen remodeling.
It is noteworthy that the role of vinculin in promoting ECM remodeling was not limited to collagen, but also applied to FN, a major ECM component in vivo, suggesting the possibility that vinculin also promotes remodeling of complex in vivo matrices consisting of multiple components. This interpretation is consistent with previous findings showing that the assembly of soluble FN into fibrils can be affected by tensional forces (29, 30). Together, our data assign a critical role to vinculin in regulating cell morphodynamics, migration, and ECM remodeling in 3D collagen ECM and emphasize the role of vinculin-dependent cellular force generation for integrating cell motility and ECM remodeling, a critical factor in many migratory processes during tissue morphogenesis and homeostasis.
Acknowledgments
The authors thank the U.S. National Institutes of Health (NIH), National Heart, Lung, and Blood Institute (NHLBI) Flow Cytometry Core Facility, for fluorescence-activated cell sorting, Silvio Gutkind for adenovirus, and Robert Fischer for helpful discussion. This work was supported by the NIH NHLBI Division of Intramural Research (C.M.W. and I.T.), Deutsche Forschungsgemeinschaft (B.F., I.T., and J.S.), the Human Frontier Science Program Fellowship (N.F.), and the U.S. National Science Foundation Graduate Research Fellowship Program (R.S.M.).
Glossary
- 2D
2-dimensional
- 3D
3-dimensional
- BSA
bovine serum albumin
- ECM
extracellular matrix
- EGFP
enhanced green fluorescent protein
- FA
focal adhesion
- F-actin
filamentous actin
- FN
fibronectin
- HRP
horseradish peroxidase
- LCPol
liquid crystal polarizing
- MLC
myosin light chain
- MEF
murine embryonic fibroblast
- SDC
spinning disk confocal
- tdTomato
tandem tomato
- Vcl-KO
vinculin knockout
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Parsons J. T., Horwitz A. R., Schwartz M. A. (2010) Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mierke C. T., Frey B., Fellner M., Herrmann M., Fabry B. (2011) Integrin α5β1 facilitates cancer cell invasion through enhanced contractile forces. J. Cell Sci. 124, 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradbury P., Fabry B., O’Neill G. M. (2012) Occupy tissue: the movement in cancer metastasis. Cell Adhes. Migr. 6, 424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lämmermann T., Bader B. L., Monkley S. J., Worbs T., Wedlich-Söldner R., Hirsch K., Keller M., Förster R., Critchley D. R., Fässler R., Sixt M. (2008) Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453, 51–55 [DOI] [PubMed] [Google Scholar]
- 5.Wolf K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A. L., Hoffman R. M., Figdor C. G., Weiss S. J., Friedl P. (2013) Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201, 1069–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedl P., Sahai E., Weiss S., Yamada K. M. (2012) New dimensions in cell migration. Nat. Rev. Mol. Cell Biol. 13, 743–747 [DOI] [PubMed] [Google Scholar]
- 7.Rozario T., DeSimone D. W. (2010) The extracellular matrix in development and morphogenesis: a dynamic view. Dev. Biol. 341, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson L. A., Dzamba B. D., Keller R., Desimone D. W. (2008) Live imaging of cell protrusive activity, and extracellular matrix assembly and remodeling during morphogenesis in the frog, Xenopus laevis. Dev. Dyn. 237, 2684–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzamba B. J., Jakab K. R., Marsden M., Schwartz M. A., DeSimone D. W. (2009) Cadherin adhesion, tissue tension, and noncanonical Wnt signaling regulate fibronectin matrix organization. Dev. Cell 16, 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigo S. L., Bilder D. (2011) Global tissue revolutions in a morphogenetic movement controlling elongation. Science 331, 1071–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conklin M. W., Eickhoff J. C., Riching K. M., Pehlke C. A., Eliceiri K. W., Provenzano P. P., Friedl A., Keely P. J. (2011) Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Provenzano P. P., Inman D. R., Eliceiri K. W., Trier S. M., Keely P. J. (2008) Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys. J. 95, 5374–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thievessen I., Thompson P. M., Berlemont S., Plevock K. M., Plotnikov S. V., Zemljic-Harpf A., Ross R. S., Davidson M. W., Danuser G., Campbell S. L., Waterman C. M. (2013) Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202, 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries J. D., Wang P., Streuli C., Geiger B., Humphries M. J., Ballestrem C. (2007) Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grashoff C., Hoffman B. D., Brenner M. D., Zhou R., Parsons M., Yang M. T., McLean M. A., Sligar S. G., Chen C. S., Ha T., Schwartz M. A. (2010) Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mierke C. T., Kollmannsberger P., Zitterbart D. P., Diez G., Koch T. M., Marg S., Ziegler W. H., Goldmann W. H., Fabry B. (2010) Vinculin facilitates cell invasion into three-dimensional collagen matrices. J. Biol. Chem. 285, 13121–13130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W., Baribault H., Adamson E. D. (1998) Vinculin knockout results in heart and brain defects during embryonic development. Development 125, 327–337 [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez Fernández J. L., Geiger B., Salomon D., Sabanay I., Zöller M., Ben-Ze’ev A. (1992) Suppression of tumorigenicity in transformed cells after transfection with vinculin cDNA. J. Cell Biol. 119, 427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng X., Nelson E. S., Maiers J. L., DeMali K. A. (2011) New insights into vinculin function and regulation. Int. Rev. Cell Mol. Biol. 287, 191–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubashkin M. G., Cassereau L., Bainer R., DuFort C. C., Yui Y., Ou G., Paszek M. J., Davidson M. W., Chen Y. Y., Weaver V. M. (2014) Force engages vinculin and promotes tumor progression by enhancing PI3K activation of phosphatidylinositol (3,4,5)-triphosphate. Cancer Res. 74, 4597–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch T. M., Münster S., Bonakdar N., Butler J. P., Fabry B. (2012) 3D Traction forces in cancer cell invasion. PLoS One 7, e33476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang N. R., Münster S., Metzner C., Krauss P., Schürmann S., Lange J., Aifantis K. E., Friedrich O., Fabry B. (2013) Estimating the 3D pore size distribution of biopolymer networks from directionally biased data. Biophys. J. 105, 1967–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin W., Fischer R. S., Kanchanawong P., Kim Y., Lim J., Myers K. A., Nishimura Y., Plotnikov S. V., Thievessen I., Yarar D., Sabass B., Waterman C. M. (2010) A Versatile, Multi-color Total Internal Reflection Fluorescence and Spinning Disk Confocal Microscope System for High-Resolution Live Cell Imaging, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA [Google Scholar]
- 24.Planchon T. A., Gao L., Milkie D. E., Davidson M. W., Galbraith J. A., Galbraith C. G., Betzig E. (2011) Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8, 417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oldenbourg R., Mei G. (1995) New polarized light microscope with precision universal compensator. J. Microsc. 180, 140–147 [DOI] [PubMed] [Google Scholar]
- 26.Carisey A., Tsang R., Greiner A. M., Nijenhuis N., Heath N., Nazgiewicz A., Kemkemer R., Derby B., Spatz J., Ballestrem C. (2013) Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 23, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer R. S., Gardel M., Ma X., Adelstein R. S., Waterman C. M. (2009) Local cortical tension by myosin II guides 3D endothelial cell branching. Curr. Biol. 19, 260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George E. L., Georges-Labouesse E. N., Patel-King R. S., Rayburn H., Hynes R. O. (1993) Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 119, 1079–1091 [DOI] [PubMed] [Google Scholar]
- 29.Legant W. R., Chen C. S., Vogel V. (2012) Force-induced fibronectin assembly and matrix remodeling in a 3D microtissue model of tissue morphogenesis. Integr. Biol. (Camb). 4, 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith M. L., Gourdon D., Little W. C., Kubow K. E., Eguiluz R. A., Luna-Morris S., Vogel V. (2007) Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 5, e268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupton S. L., Waterman-Storer C. M. (2006) Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 125, 1361–1374 [DOI] [PubMed] [Google Scholar]
- 32.Kutys M. L., Yamada K. M. (2014) An extracellular-matrix-specific GEF-GAP interaction regulates Rho GTPase crosstalk for 3D collagen migration. Nat. Cell Biol. 16, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrie R. J., Gavara N., Chadwick R. S., Yamada K. M. (2012) Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197, 439–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brindle N. P., Holt M. R., Davies J. E., Price C. J., Critchley D. R. (1996) The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 318, 753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeMali K. A., Barlow C. A., Burridge K. (2002) Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J. Cell Biol. 159, 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subauste M. C., Pertz O., Adamson E. D., Turner C. E., Junger S., Hahn K. M. (2004) Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. J. Cell Biol. 165, 371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subauste M. C., Nalbant P., Adamson E. D., Hahn K. M. (2004) Vinculin controls PTEN protein level by maintaining the interaction of the adherens junction protein b-catenin with the scaffolding protein MAGI-2. J. Biol. Chem. 280, 5676–5681 [DOI] [PubMed] [Google Scholar]
- 38.Huveneers S., Oldenburg J., Spanjaard E., van der Krogt G., Grigoriev I., Akhmanova A., Rehmann H., de Rooij J. (2012) Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J. Cell Biol. 196, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bays J. L., Peng X., Tolbert C. E., Guilluy C., Angell A. E., Pan Y., Superfine R., Burridge K., DeMali K. A. (2014) Vinculin phosphorylation differentially regulates mechanotransduction at cell-cell and cell-matrix adhesions. J. Cell Biol. 205, 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen Q., Janmey P. A. (2013) Effects of non-linearity on cell-ECM interactions. Exp. Cell Res. 319, 2481–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang N. R., Skodzek K., Hurst S., Mainka A., Steinwachs J., Schneider J., Aifantis K. E., Fabry B. (2015) Biphasic response of cell invasion to matrix stiffness in three-dimensional biopolymer networks. Acta Biomater. 13, 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]