Summary
Various cell death mechanisms are integral to host defense in both plants and mammals. Plant defense against biotrophic pathogens is associated with programmed cell death (PCD) of the infected cell. This effector-triggered PCD is partly analogous to pyroptosis, an inflammatory host cell death process that plays a crucial role in defense against microbial infections in mammals. Plant effector-triggered PCD also shares with mammalian apoptosis the involvement of cell cycle regulators as signaling components. Here we explore the similarities between these different cell death programs as they relate to host defense and their relationship to the cell-cycle.
Introduction
Plant defense against biotrophic pathogens is strongly associated with programmed cell death (PCD) of infected cells, which occurs as a result of host recognition of one or more pathogen effectors (Jones and Dangl, 2006). This effector-triggered PCD is an active process that is thought to limit pathogen proliferation by eliminating its growth niche (Jones and Dangl, 2006). In mammals, when specialized innate immune cells are compromised by intracellular pathogens, they are eliminated by a process of cell death termed pyroptosis, which in some aspects is analogous to the effector-triggered PCD in plants. Significant advances have been made in the field of pyroptosis research due to detailed studies in many pathogen systems (Jorgensen and Miao, 2015). However, the signaling events leading to pyroptotic caspase activation are less clear compared to those of other mammalian death processes, such as apoptosis.
Effector-triggered PCD in plants is regulated by plant homologs of the canonical cell cycle regulators Retinoblastoma (Rb) and E2F (Wang et al., 2014). Apoptosis in animals is also reported to be regulated by homologous cell cycle regulators (Polager and Ginsberg, 2009). Extending this comparison, here we review the involvement of cell cycle regulators in these different modes of cell death and discuss their relevance to immunity in plants and animals. For a systematic comparison of plant and animal cell death mechanisms, the reader is directed to the excellent review by Coll et al. (2011).
Comparing Effector-Triggered PCD in Plants and Pyroptosis in Mammals: Immune Receptors and Consequence
The ability to distinguish self from non-self is fundamental to recognition of and defense against pathogens. Most plant pathogens proliferate in the intercellular space called the apoplast, perhaps due to the presence of plant cell walls and the lack of phagocytosis, which make cell invasion difficult. Detection of extracellular pathogens in plants is through cell surface pattern recognition receptors (PRRs) that recognize conserved microbe-associated molecular patterns (MAMPs). Successful pathogens, however, can often bypass MAMP-triggered surface defense by directly delivering a collection of effectors that perturb host cell metabolic activities and promote infection (Jones and Dangl, 2006). The host has evolved to use some of these effectors as signals for intracellular nucleotide-binding leucine-rich repeat proteins (NB-LRRs), which serve as receptors for effector-triggered immunity (ETI). Each plant genome encodes hundreds of NB-LRRs that are responsible for activating ETI (Hofberger et al., 2014). Regardless of pathogen type (viral, bacterial, fungal or even small animals such as nematodes), ETI is commonly accompanied by PCD at the infection site, which is often visible to the naked eye. Even though its contribution to immunity at the molecular level is still up for debate, the ubiquitous presence of PCD during ETI suggests the importance of this defense strategy. In contrast to plants, where ETI is cell autonomous, mammals recognize non-self-signals by specialized immune cells such as dendritic cells and macrophages. Conserved MAMP signals from extracellular and intracellular pathogens are first detected by these immune cells through their membrane-bound Toll-like receptors (TLRs) or the intracellular NOD-like receptors (NLRs) before pathogen-specific antigens are presented to the adaptive immune system (Jorgensen and Miao, 2015). However, when pathogens manage to hijack the function of these antigen-presenting immune cells to allow for intracellular infection, these cells signal to induce a unique form of PCD, pyroptosis, which leads to the release of damage-associated molecular patterns (DAMPs) to stimulate inflammation (Jorgensen and Miao, 2015).
One remarkable similarity between effector-triggered PCD in plants and MAMP-triggered pyroptosis in mammals are the immune receptors involved (Table 1). NB-LRRs and NLRs are both members of the STAND (Signal Transduction ATPases with Numerous Domains) family of proteins with conserved protein architecture and a similar ability to trigger PCD, even though they arose from distinct evolutionary origins (Ausubel, 2005). Both immune receptors recognize intracellular pathogen signals or activities, but the signals recognized by plant NB-LRRs are pathogen-specific effectors whereas the signals for mammalian NLRs are conserved MAMPs (Maekawa et al., 2011). Mammals have evolved adaptive immune mechanisms to recognize pathogen-specific antigens. Compared to the astronomical numbers of immunoglobulin specificities present in the mammalian system (>1011 in humans), which are created through somatic DNA rearrangement, plants inherit only hundreds of NB-LRRs to deal with the wide range of pathogens (Flajnik and Kasahara, 2010; Hofberger et al., 2014). According to the “Guard Hypothesis”, this is accomplished by NB-LRRs guarding a limited number of important host proteins (“hubs”) which are common targets of pathogen effectors, instead of binding to these effectors directly (Dangl and Jones, 2001; Wessling et al., 2014). For instance, Arabidopsis has two NB-LRRs binding to the RIN4 protein, which is known to be targeted by at least two bacterial effectors that modify RIN4 by proteolytic cleavage and phosphorylation, respectively (Axtell and Staskawicz, 2003; Mackey et al., 2002).
Table 1.
Summary of comparable factors involved in regulating cell death as discussed in this review
| Effector-triggered PCD | Pyroptosis | Apoptosis | |
|---|---|---|---|
| Signal | Pathogen Effectors | MAMPs, DAMPs | |
| DNA Damage, External Signals | |||
| Signal Transduction | Nucleotide-Binding Leucine-rich Repeat proteins (NB-LRRs) | Nod-like Receptors (NLRs) | |
| Absent in Melanoma 2 (AIM2) | |||
| Retinoblastoma-related 1 (RBR1) | Retinoblastoma (Rb) | ||
| E2FA, E2FB, E2FC | E2F1 | ||
| Execution | Metacaspase MC1 | Caspases 1, 11 | Caspases 3, 8, 9 |
| Inhibition | Metacaspase MC2 | ||
| B-cell lymphoma 2 (BCL2)-associated athanogene domain-containing (BAG6) | B-cell lymphoma 2 (BCL2) | ||
| BAX-inhibitor 1 (BI1) | BAX-inhibitor 1 (BI1) | ||
| Inhibitor of Apoptosis-like protein (IAP-l) | Inhibitor of Apoptosis (IAP) family |
The rows represent the different factors and their roles, and the columns represent the different modes of cell death. Spaces left blank indicate gaps in knowledge or lack of conserved factors (between plants and animals). For relevant references and details, see main text.
Activation of NB-LRRs in plants and NLRs in mammals leads to PCD with parallel immune outcomes (Figure 1). A hallmark of the effector-triggered PCD mediated by plant NB-LRRs is the leakage of ions due to cytoplasmic shrinkage, which can be measured using a conductivity meter, and production of the immune signal, salicylic acid, for systemic acquired resistance (Jones and Dangl, 2006; Mackey et al., 2002). Activation of NLRs in mammals leads to assembly of the inflammasome and execution of pyroptosis, which involves membrane rupture and leakage of cytosolic contents, resulting in a full inflammatory response (Martin et al., 2012). Pyroptosis is distinct from apoptosis. During apoptosis, the cellular contents remain sealed in an apoptotic body, which is then cleared away by phagocytosis. It is believed that death by apoptosis has little to no immunological consequence (Jorgensen and Miao, 2015). In contrast to mammals, plant cells are fixed in space with no mechanism for the clearance of apoptotic bodies as there is in animals. The currently described effector-triggered PCD in plants has an outcome similar to pyroptosis, in which cellular contents are released into the surrounding tissue, possibly leading to the release of DAMPs and immune signals to trigger systemic defense responses (Boller and Felix, 2009).
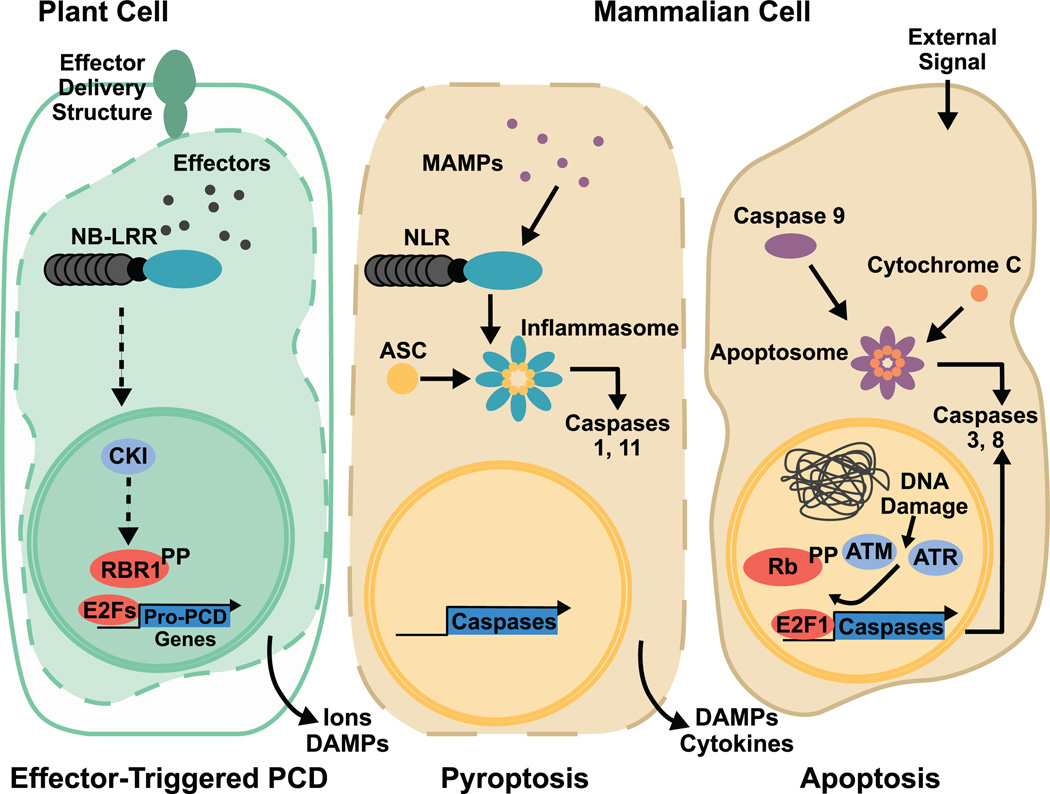
Figure 1. Diagram of relevant similarities and differences between plant effector-triggered PCD (left) mammalian pyroptosis (center) and mammalian apoptosis (right).
In plants, effector-triggered PCD is initiated by NB-LRR-mediated perception of the presence of specific intracellular effectors secreted by pathogens. Effector recognition signals CKI release, which promotes RBR1 hyperphosphorylation, release of E2Fs, and transcription of pro-PCD genes. PCD results in cytoplasmic shrinkage and leakage of ions and DAMPs, and production of the immune signal salicylic acid to stimulate the immune response. In mammals, during pyroptosis, NLRs recognize intracellular pathogens through conserved MAMPs, triggering assembly of the inflammasome and activation of pyroptotic caspases. Pyroptotic death results in cell lysis and the release of DAMPs and inflammatory cytokines, leading to a full inflammatory response. DNA damage checkpoint-induced apoptosis includes an accumulation of hyperphosphorylated Rb as well as phosphorylation and stabilization of E2F1 by ATM and ATR, deregulating the Rb/E2F cell cycle pathway. The apoptosome is formed by interaction of procaspase 9 and cytochrome c, leading to activation of apoptotic caspases and a PCD phenotype that includes the formation of an apoptotic body.
Comparing Effector-Triggered PCD in Plants and Pyroptosis in Mammals: Downstream Events
A defining factor for a programmed, rather than passive necrotic, cell death in mammals is the activation of caspases, which execute PCD through proteolytic cleavage of cellular substrates. In pyroptosis, the inflammasome forms a platform for activation of a caspase, canonically caspase-1, which serves a dual role in processing pro-inflammatory cytokines interleukin-1b (IL-1b) and IL-18 and in executing PCD (Schroder and Tschopp, 2010). Analogous to the apoptosome, which is comprised of initiator procaspase 9 and cytochrome c and activates apoptotic caspases, an inflammasome either contains a caspase activation and recruitment domain (CARD) or a pyrin domain, for example NLRC4 and NLRP3, respectively (Schroder and Tschopp, 2010). In addition to NLRs, an inflammasome can also be formed by absent in melanoma 2 (AIM2), which recruits caspase-1 similarly to NLRP3 (Schroder and Tschopp, 2010). In plants, no close homologs of caspases have been found, suggesting that effector-triggered PCD in plants is executed through a distinct mechanism. The involvement of the distantly related metacaspases in regulating PCD is not clear. Two Arabidopsis metacaspases have been shown to function antagonistically. Knocking out AtMC1 (METACASPASE 1) or mutating its caspase-like catalytic residues moderately reduced effector-triggered PCD, whereas an activity inhibitory of PCD and independent of any known caspase-like domains was detected for AtMC2 (METACASPASE 2) (Coll et al., 2010). In some aspects, effector-triggered PCD in plants is similar to mammalian necroptosis, yet another form of PCD that occurs in the absence of caspase activity but is regulated by the death domain kinase RIPK1 (Silke et al., 2015). In addition to the immune inducing properties of both cell death programs, plant effector-triggered PCD is also similar to necroptosis with regard to mitochondrial swelling and significant ROS production (Coll et al., 2011).
Despite the limited involvement of caspases in plant effector-triggered PCD, certain inhibitors of mammalian apoptosis are conserved in plants. For instance, anti-apoptotic Bax-inhibitor 1 attenuates effector-triggered PCD caused by recognition of fungal and bacterial pathogens in a number of plants (Kawai-Yamada et al., 2009). Overexpression of the Arabidopsis B-cell lymphoma 2 (BCL2)-associated athanogene domain-containing BAG6 causes PCD-like phenotypes in plants, demonstrating a role in PCD regulation similar to pro-apoptotic BCL2-domain containing BH3 proteins (Kang et al., 2006). Finally, the mammalian Inhibitor of Apoptosis (IAP) family, which inhibit caspase-dependent PCD signaling, has at least one Arabidopsis homolog, IAP-like protein, which inhibits bacterial effector-triggered PCD when overexpressed (Kim et al., 2011). The conservation of these inhibitors suggests the existence of an apoptosis-like signaling cascade in plants, although the precise function of these regulators during effector-triggered PCD requires further investigation.
Effector-Triggered PCD in Plants and Apoptosis in Mammals: Control by Cell Cycle Regulators
Recently it was shown that although effector-triggered PCD in plants is similar in activation and phenotype to pyroptosis and necroptosis in mammals, the signaling for PCD induction is in fact partially analogous to that of apoptosis (Figure 1) (Wang et al., 2014). Plant homologs of E2Fs, a family of mammalian cell cycle checkpoint transcription factors, were found to be essential for full development of PCD upon pathogen challenge. Furthermore, RETINOBLASTOMA-RELATED 1 (RBR1), the plant homolog of the E2F regulator Rb, was hyperphosphorylated during ETI. Increased hyperphosphorylation of RBR1 leads over-activation of E2F, resulting in PCD instead of cell cycle progression (Wang et al., 2014). There is ample evidence that mammalian Rb, first identified as a tumor suppressor, and E2Fs, especially E2F1, are involved in the induction of apoptosis caused by a variety of stimuli (Polager and Ginsberg, 2009). E2F1 targets regulators of apoptosis at both the transcriptional and post-translational levels, including activation of pro-apoptotic caspase genes and BH3 genes and repression of anti-apoptotic BCl2 (Polager and Ginsberg, 2009). As some of the apoptosis-related targets of E2F1 in mammals are conserved in plants, such as the BCL2-family genes mentioned previously (Kang et al., 2006), the role of E2F1 as a central regulator of PCD may be directly comparable between apoptosis and effector-triggered PCD.
Despite the usual association of inflammasome activation with pyroptosis, there is evidence indicating that the inflammasome can activate not only pyroptotic but also apoptotic caspases (caspases 1, 11 and caspases 3, 8, 9, respectively) under different conditions. In caspase-1 knockout cells, apoptosis occurs in the presence of pyroptotic stimuli, suggesting that apoptosis may be employed as a back-up to the more immunologically useful pyroptotic death (Pierini et al., 2012). Alternatively, recent work on DNA-dependent inflammasome activation suggests that the induction of apoptosis over pyroptosis is biased by dose, with lower doses of the stimulus leading to the immunologically subtle apoptotic response (Sagulenko et al., 2013). It is currently unknown whether Rb and E2F play any regulatory role in this inflammasome-mediated apoptosis. However, there is recent evidence for Rb and E2F playing a role in innate immunity in mammals in a different context. Loss of Rb in epithelial and liver cells has been found to significantly reduce responsiveness to a number of TLR ligands. This effect probably results from removal of Rb inhibition on E2F1, which is a repressor of TLR expression in these cells (Taura et al., 2012).
Control of Rb and E2F during Cell Death
Rb and E2F are core cell cycle components and are activated during the cell cycle by upstream cyclin/cyclin-dependent kinase (CDK) complexes. Their mechanism of activation during PCD is less clear. In the processes of apoptosis during mammalian brain development and apoptosis induced by mitochondrial dysfunction, Rb is hyperphosphorylated by the metabolic and oxidative stress sensor AMP-activated protein kinase, indicating that noncanonical Rb-inactivating kinases can play a role in the induction of death in certain contexts (Dasgupta et al., 2012; Raimundo et al., 2012). However, in the case of antigen-induced death of T-cells, which occurs due to the absence of inflammatory cytokines, it has been shown that cells need to enter into the cell cycle up to the G1-S transition before they can undergo apoptosis (Lissy et al., 1998). Additionally, during antigen-stimulated T-cell proliferation, NF-κB biases E2F1 regulation toward proliferation and away from apoptosis in a CDK-dependent manner (Wan and DeGregori, 2003). Together these results suggest that in T cells, Rb is regulated to induce apoptosis in a cell cycle-dependent manner.
Upstream cell cycle regulators are also involved in effector-triggered PCD and immunity in plants (Bao et al., 2013; Chandran et al., 2014; Wang et al., 2014). Bao et al. reported that repressors of anaphase promoting complex/cyclosome (APC/C), a ubiquitin E3 ligase complex that regulates cell-cycle progression, facilitate PCD through upregulation of NB-LRR gene expression (Bao et al., 2013). The involvement of APC/C is also supported by Chandran et al., who found that the Arabidopsis atypical DP-E2F-like 1 (DEL1) is a repressor of immunity (Chandran et al., 2014). These authors attribute the DEL1 immune effect to a reduction in basal expression of Enhanced Disease Susceptibility 5, a transporter of the plant immune hormone salicylic acid. However, since DEL1 is known to block endoreduplication and promote cell proliferation through repression of APC/C activator CCS52A2, a homolog of mammalian CDH1 (Lammens et al., 2008), DEL1 may also influence plant immunity through the APC/C. In the Wang et al. study, mutants of CDK inhibitors (CKIs), sim (siamese) and smr1 (siamese-related 1), which were originally found to affect development of trichomes (Walker et al., 2000), were identified as genetic suppressors of effector-triggered PCD. The loss-of-function sim smr1 double mutant blocks, rather than stimulates, RBR1 hyperphosphorylation during PCD, a result opposite of that which would be expected according to the canonical cell cycle pathway. How do the same CKIs block RBR1 phosphorylation during cell cycle but promote RBR1 phosphorylation during effector-triggered PCD? During PCD, SIM and SMR1 stabilize the CDKA1 protein, a homolog of mammalian CDK2, which is known to regulate Rb (van den Heuvel and Dyson, 2008), suggesting that these CKIs inhibit CDKA1 kinase activity during the cell cycle but stabilize the CDKA1 protein during PCD. Further investigation is required to elucidate the underlying molecular mechanism. However, it is possible that a novel kinase is activated by SIM and SMR1, either directly or indirectly, during PCD to hyperphosphorylate RBR1 independent of the activity of CDKA1 in cell cycle control.
Why Engage Cell Cycle Regulators in Immune-Related Cell Death?
The biological significance of engaging cell cycle regulators Rb and E2F in plant effector-triggered PCD has yet to be fully elucidated. During cell cycle in animals and plants, Rb and E2Fs function at the G1-S phase checkpoint, where they exert critical control over a cell’s commitment to replicate DNA (van den Heuvel and Dyson, 2008). If too much DNA damage has accumulated during the last round of the cell cycle, Rb and E2F1 are able to form both activator and repressor complexes that work to repress the progression of the cell cycle and activate pro-apoptotic signaling at a transcriptional level (Biswas and Johnson, 2012). Additionally, E2F1 is phosphorylated by Ataxia Telangiectasia-Mutated (ATM) and ATM- and Rad3-related (ATR) kinases and targeted to the sites of double-stranded breaks to assist the recruitment of DNA repair enzymes (Biswas and Johnson, 2012). The role of E2Fs in DNA damage repair is particularly interesting for researchers studying innate immunity, as DNA damage can serve as a signal for innate immune receptor activation and it has been demonstrated that the DNA damage repair machinery is utilized during immune responses in both plants and mammals (Fontes et al., 2015; Yan et al., 2013).
Due to the importance of DNA damage responses at the G1-S transition, the status of a cell, whether it is actively dividing, in a Gap phase, or endocycling, is integrally related to its commitment to DNA repair. Vegetative growth in plants occurs predominantly in endoreduplicating cells. These cells have made the switch from normal cell division to DNA replication without mitosis, resulting in increased ploidy (John and Qi, 2008). Endoreduplication seems to be employed in Arabidopsis to avoid apoptosis during normal cell cycles in response to DNA damage, and in Drosophila endocycles also repress apoptosis due to DNA damage (Adachi et al., 2011; Mehrotra et al., 2008). Therefore, the ability to endocycle may lessen the need for an Rb/E2F-regulated cell-cycle checkpoint in plants, which mainly contain cells of high ploidy. In animal immune cells, active cell cycles may prohibit Rb and E2F from taking on a role as positive regulators of pyroptosis. We hypothesize that in addition to DNA damage, other signals are required to activate RBR1 and E2F to trigger PCD in plants.
Intriguingly, ploidy has been shown to further increase in Arabidopsis leaf mesophyll cells interacting with the feeding structure of the powdery mildew fungal pathogen Golovinomyces orontii, and many mutants resistant to this pathogen have reduced basal or pathogen-induced cell ploidy (Chandran et al., 2013). This pathogen-induced increase in ploidy has been suggested to be necessary to support the high metabolic activity required in cells being utilized by a biotrophic pathogen, and ploidy has also been found to be a determinant for susceptibility to many other biotrophic pathogens and symbionts that utilize a sustained site of nutrient acquisition in plants (Wildermuth, 2010). It is reasonable to speculate that the biological significance of using cell cycle regulators that sit at the crux of the decision to divide or to endoreduplicate as regulators of effector-triggered PCD is to turn cells from endoreduplication, which favors pathogen infection, to cell death, a part of this major defense mechanism in plants.
Conclusion
Although effector-triggered PCD in plants is analogous to inflammasome-mediated pyroptosis in mammals with regard to the immune receptors involved and immune outcomes, plant PCD is controlled by the same cell cycle regulators RBR1 and E2F as those controlling apoptosis in animals. These cell cycle components are worth investigating in relation to other immune cell death programs, including mammalian pyroptosis and necroptosis. Careful consideration of the age, growth stage, and perhaps even cell-cycle synchrony of cells used in immune research may lead to new discoveries in this arena. The identification of E2F as a required transcription factor for effector-triggered PCD in plants may serve as a starting point to identify the genes that execute PCD in plants.
Acknowledgments
We thank Dr. Qijing Li and Dr. Bernard Mathey-Prevot for their helpful discussions, and Dr. Paul Zwack for critical reading of the manuscript. X.D. is supported by a grant from NIH (2R01-GM069594-09) and by the Howard Hughes Medical Institute-Gordon and Betty Moore Foundation (through grant GBMF3032).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:10004–10009. doi: 10.1073/pnas.1103584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Bao Z, Yang H, Hua J. Perturbation of cell cycle regulation triggers plant immune response via activation of disease resistance genes. Proc Natl Acad Sci U S A. 2013;110:2407–2412. doi: 10.1073/pnas.1217024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–17. doi: 10.1158/0008-5472.CAN-11-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Chandran D, Rickert J, Cherk C, Dotson BR, Wildermuth MC. Host cell ploidy underlying the fungal feeding site is a determinant of powdery mildew growth and reproduction. Mol Plant Microbe Interact. 2013;26:537–545. doi: 10.1094/MPMI-10-12-0254-R. [DOI] [PubMed] [Google Scholar]
- Chandran D, Rickert J, Huang Y, Steinwand MA, Marr SK, Wildermuth MC. Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe. 2014;15:506–513. doi: 10.1016/j.chom.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P. Arabidopsis type I metacaspases control cell death. Science. 2010;330:1393–1397. doi: 10.1126/science.1194980. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Dasgupta B, Ju JS, Sasaki Y, Liu X, Jung SR, Higashida K, Lindquist D, Milbrandt J. The AMPK beta2 subunit is required for energy homeostasis during metabolic stress. Mol Cell Biol. 2012;32:2837–2848. doi: 10.1128/MCB.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes FL, Pinheiro DM, Oliveira AH, Oliveira RK, Lajus TB, Agnez-Lima LF. Role of DNA repair in host immune response and inflammation. Mutat Res Rev Mutat Res. 2015;763:246–257. doi: 10.1016/j.mrrev.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Hofberger JA, Zhou B, Tang H, Jones JD, Schranz ME. A novel approach for multi-domain and multi-gene family identification provides insights into evolutionary dynamics of disease resistance genes in core eudicot plants. BMC Genomics. 2014;15:966. doi: 10.1186/1471-2164-15-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John PC, Qi R. Cell division and endoreduplication: doubtful engines of vegetative growth. Trends Plant Sci. 2008;13:121–127. doi: 10.1016/j.tplants.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev. 2015;265:130–142. doi: 10.1111/imr.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CH, Jung WY, Kang YH, Kim JY, Kim DG, Jeong JC, Baek DW, Jin JB, Lee JY, Kim MO, et al. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 2006;13:84–95. doi: 10.1038/sj.cdd.4401712. [DOI] [PubMed] [Google Scholar]
- Kawai-Yamada M, Hori Z, Ogawa T, Ihara-Ohori Y, Tamura K, Nagano M, Ishikawa T, Uchimiya H. Loss of calmodulin binding to Bax inhibitor-1 affects Pseudomonas-mediated hypersensitive response-associated cell death in Arabidopsis thaliana. J Biol Chem. 2009;284:27998–28003. doi: 10.1074/jbc.M109.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Lee SY, Jung YJ, Chae HB, Nawkar GM, Shin MR, Kim SY, Park JH, Kang CH, Chi YH, et al. Inhibitor of apoptosis (IAP)-like protein lacks a baculovirus IAP repeat (BIR) domain and attenuates cell death in plant and animal systems. J Biol Chem. 2011;286:42670–42678. doi: 10.1074/jbc.M111.262204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci U S A. 2008;105:14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissy NA, Van Dyk LF, Becker-Hapak M, Vocero-Akbani A, Mendler JH, Dowdy SF. TCR antigen-induced cell death occurs from a late G1 phase cell cycle check point. Immunity. 1998;8:57–65. doi: 10.1016/s1074-7613(00)80458-6. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 2011;12:817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Henry CM, Cullen SP. A perspective on mammalian caspases as positive and negative regulators of inflammation. Mol Cell. 2012;46:387–397. doi: 10.1016/j.molcel.2012.04.026. [DOI] [PubMed] [Google Scholar]
- Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008;22:3158–3171. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierini R, Juruj C, Perret M, Jones CL, Mangeot P, Weiss DS, Henry T. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 2012;19:1709–1721. doi: 10.1038/cdd.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Raimundo N, Song L, Shutt TE, McKay SE, Cotney J, Guan MX, Gilliland TC, Hohuan D, Santos-Sacchi J, Shadel GS. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–726. doi: 10.1016/j.cell.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko V, Thygesen SJ, Sester DP, Idris A, Cridland JA, Vajjhala PR, Roberts TL, Schroder K, Vince JE, Hill JM, et al. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 2013;20:1149–1160. doi: 10.1038/cdd.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Silke J, Rickard JA, Gerlic M. The diverse role of RIP kinases in necroptosis and inflammation. Nat Immunol. 2015;16:689–697. doi: 10.1038/ni.3206. [DOI] [PubMed] [Google Scholar]
- Taura M, Suico MA, Koyama K, Komatsu K, Miyakita R, Matsumoto C, Kudo E, Kariya R, Goto H, Kitajima S, et al. Rb/E2F1 regulates the innate immune receptor Toll-like receptor 3 in epithelial cells. Mol Cell Biol. 2012;32:1581–1590. doi: 10.1128/MCB.06454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127:3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- Wan YY, DeGregori J. The survival of antigen-stimulated T cells requires NFkappaB-mediated inhibition of p73 expression. Immunity. 2003;18:331–342. doi: 10.1016/s1074-7613(03)00053-0. [DOI] [PubMed] [Google Scholar]
- Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X. A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe. 2014;16:787–794. doi: 10.1016/j.chom.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessling R, Epple P, Altmann S, He Y, Yang L, Henz SR, McDonald N, Wiley K, Bader KC, Glasser C, et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe. 2014;16:364–375. doi: 10.1016/j.chom.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC. Modulation of host nuclear ploidy: a common plant biotroph mechanism. Curr Opin Plant Biol. 2010;13:449–458. doi: 10.1016/j.pbi.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Yan S, Wang W, Marques J, Mohan R, Saleh A, Durrant WE, Song J, Dong X. Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol Cell. 2013;52:602–610. doi: 10.1016/j.molcel.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]