Abstract
Type II CRISPR systems defend against invasive DNA by using Cas9 as an RNA-guided nuclease that creates double-stranded DNA breaks. Dual RNAs [CRISPR RNA (crRNA) and tracrRNA] are required for Cas9’s targeting activities observed to date. Targeting requires a protospacer adjacent motif (PAM) and crRNA-DNA complementarity. Cas9 orthologues [including Neisseria meningitidis Cas9 (NmeCas9)] have also been adopted for genome engineering. Here we examine the DNA cleavage activities and substrate requirements of NmeCas9, including a set of unusually complex PAM recognition patterns. Unexpectedly, NmeCas9 cleaves single-stranded DNAs in a manner that is RNA-guided but PAM- and tracrRNA-independent. Beyond the need for guide-target pairing, this “DNase H” activity has no apparent sequence requirements, and the cleavage sites are measured from the 5′ end of the DNA substrate’s RNA-paired region. These results indicate that tracrRNA is not strictly required for NmeCas9 enzymatic activation, and expand the list of targeting activities of Cas9 endonucleases.
Keywords: Neisseria meningitidis, CRISPR, Cas9 endonucleases, NmeCas9, DNA targeting, crRNA, sgRNA, tracrRNA
Graphical Abstract
INTRODUCTION
CRISPR (clustered, regularly interspaced, short palindromic repeats) loci and CRISPR-associated (Cas) proteins provide an RNA-guided adaptive immune system for bacteria and archaea (Barrangou and Marraffini, 2014; van der Oost et al., 2014; Sontheimer and Barrangou, 2015). CRISPRs consist of ~24–48 base pair (bp) repeats separated by similarly sized, non-repetitive spacers, which often match sequences from phage genomes or plasmids (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). Genetic interference specified by CRISPR-Cas pathways can protect against phage infection (Barrangou et al., 2007), and can also limit horizontal gene transfer (Marraffini and Sontheimer, 2008; Bikard et al., 2012; Zhang et al., 2013). CRISPR RNAs (crRNAs) (Brouns et al., 2008; Hale et al., 2008) are usually processed from a longer crRNA precursor (pre-crRNA). Each crRNA associates with one or more Cas proteins to form an interference complex that locates complementary “protospacer” regions in the foreign nucleic acids, and makes sequence-specific cuts in the invasive genetic element, preventing its establishment or expression (Barrangou and Marraffini, 2014; van der Oost et al., 2014; Sontheimer and Barrangou, 2015).
CRISPR-Cas systems are classified into five major types (I–V), based primarily on the identities of the Cas proteins (Makarova et al., 2015). Type II systems are distinguished partially by the involvement of a single protein (Cas9), which includes RuvC and HNH nuclease domains, for interference (Sapranauskas et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012), along with a trans-activating CRISPR RNA (tracrRNA) that functions in pre-crRNA processing (Deltcheva et al., 2011), interference (Jinek et al., 2012; Zhang et al., 2013), and new spacer acquisition (Heler et al., 2015). The type II systems are further divided into subtypes II-A, -B, and -C, based in part on the presence or absence of additional spacer acquisition factors Csn2 and Cas4 (Makarova et al., 2015).
Interest in type II CRISPR-Cas systems has increased dramatically due to its adoption as an RNA-guided, locus-specific, genome editing and DNA binding platform (Doudna and Charpentier, 2014; Hsu et al., 2014). Cas9 functions as an RNA-programmable DNA endonuclease using the HNH and RuvC nuclease domains to cleave the crRNA-complementary and non-complementary strands, respectively (Gasiunas et al., 2012; Jinek et al., 2012). The crRNA and tracrRNA cofactors can be fused into a single-guide RNA (sgRNA) without loss of activity (Jinek et al., 2012). Target cleavage by Cas9 requires the presence of a short protospacer adjacent motif (PAM) that flanks the target region (Deveau et al., 2008; Garneau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012), and the PAM sequence varies among Cas9 orthologs. CrRNA/target complementarity must be nearly perfect in the 7–12 nt “seed” region proximal to the PAM (Sapranauskas et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012).
The application of CRISPR/Cas9 in genome editing has sparked interest in characterizing different type II systems to identify Cas9 orthologs with distinct and perhaps improved genome targeting capabilities. To date, the editing functions of Cas9s from Streptococcus pyogenes (SpyCas9, Type II-A) (Jinek et al., 2012; Cho et al., 2013; Cong et al., 2013; Hwang et al., 2013; Jiang et al., 2013; Jinek et al., 2013; Mali et al., 2013), Streptococcus thermophilus (Sth1Cas9 and Sth3Cas9, both Type II-A) (Gasiunas et al., 2012; Cong et al., 2013; Esvelt et al., 2013; Chen et al., 2014), and N. meningitidis (NmeCas9, Type II-C) (Esvelt et al., 2013; Hou et al., 2013) are best characterized. NmeCas9 (Zhang et al., 2013) is of interest because it is almost 300 amino acids smaller than the commonly used SpyCas9, and this reduced size may facilitate its delivery via virus- or mRNA-based vectors. More recently, a second Cas9 in this smaller size range from Staphylococcus aureus (SauCas9, Type II-A) has also been adopted for genome editing (Ran et al., 2015). Certain pairs of Cas9 proteins and their respective guide RNAs are orthogonal (i.e., a guide RNA loads into the intended cognate Cas9 but not into the orthologous Cas9) (Esvelt et al., 2013; Briner et al., 2014; Fonfara et al., 2014), facilitating multiplexed applications. The structures of apo SpyCas9 (Jinek et al., 2014), and its complexes with an sgRNA (Jiang et al., 2015) or an sgRNA and a single-stranded (ss) DNA target (Nishimasu et al., 2014), show that the protein consists of a recognition lobe and a nuclease lobe, and that the RNA-DNA heteroduplex is bound at the interface of the two lobes. The structure of SpyCas9 with an sgRNA and a partially duplexed target DNA revealed the molecular basis for SpyCas9’s recognition of its 5′-NGG-3′ PAM (Anders et al., 2014). The structure of a more compact type II-C Cas9 from Actinomyces naeslundii (AnaCas9) has been reported (Jinek et al., 2014), but relevant functional information for that protein (e.g. editing efficiency and PAM specificity) is limited. Accordingly, there is a significant need to understand better the properties of the more compact Cas9s such as NmeCas9 and others from Type II-C.
Here we have defined mechanistic requirements and features of NmeCas9 for target DNA cleavage and cellular interference. Our results show that NmeCas9 has many characteristics in common with SpyCas9, Sth1Cas9, Sth3Cas9, and SauCas9, including cleavage site specificity, mismatch sensitivity, and protospacer-PAM linker length dependence. In contrast, NmeCas9 differs from SpyCas9 in that mutation of either strand of the target PAM inhibits double-stranded (ds) DNA target cleavage. Most strikingly, even in the absence of its tracrRNA cofactor, NmeCas9 can efficiently cleave ssDNA in an RNA-guided fashion, and this “DNase H-like” activity depends upon an intact HNH domain. Thus, NmeCas9 has target recognition capabilities that have not been observed previously in other orthologs and that could be useful for engineering applications.
RESULTS
Double-stranded DNA cleavage activity of NmeCas9 in vitro
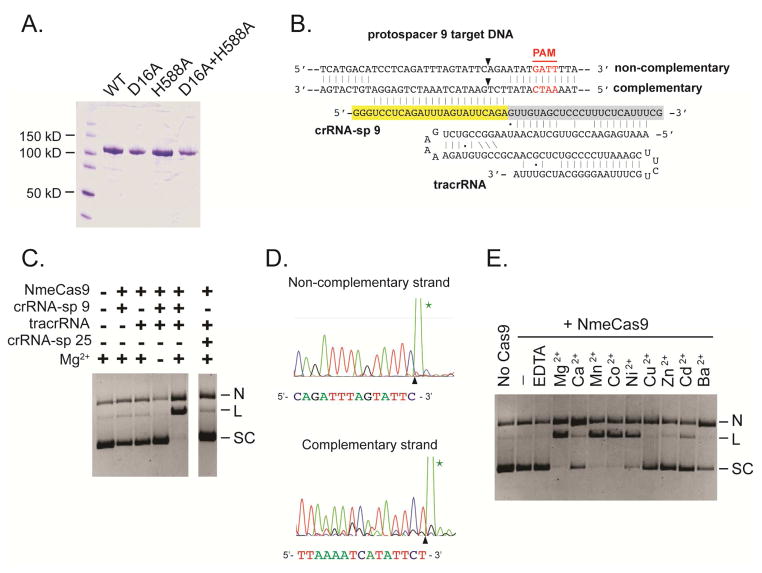
To define the biochemical properties of NmeCas9, we purified the protein from E. coli (Fig. 1A). We generated tracrRNA and a crRNA containing spacer 9 (sp 9) via in vitro transcription to complex with target DNA (see Fig. 1B). Plasmid cleavage assays showed that NmeCas9 cleaves dsDNA containing the matched protospacer and the reaction requires the cognate crRNA, tracrRNA, and magnesium (Fig. 1C). Cleavage occurred primarily between the 3rd and 4th nts of the protospacer (counting from the PAM-proximal end) (Fig. 1D). Plasmid cleavage is substantially complete after 5 min of incubation (Fig. S1A). NmeCas9 is most active when salt (KCl) concentration is lower than 300 nM (Fig. S1B). Divalent metals such as Mg2+, Mn2+, Co2+, and Ni2+ support cleavage of both strands, whereas Ca2+ and Ba2+ only support plasmid nicking, suggesting that they can function with only one of the endonuclease domains. Other metals such as Cu2+, Zn2+ and Cd2+ support little or no NmeCas9 cleavage (Fig. 1E).
Figure 1. NmeCas9 Functions as an RNA-guided DNA Endonuclease.
(A) A Coomassie-stained 10% denaturing PAGE of wildtype and mutant NmeCas9 proteins. WT: wildtype NmeCas9; D16A, H588A, and D16A+H558A: active site NmeCas9 mutants D16A, H588A, and D16A+H558A. The predicted molecular weight of NmeCas9 is ~110 kD. Weights of molecular markers are indicated.
(B) A schematic showing the complex of the sp 9 crRNA, the tracrRNA, and the target dsDNA. Yellow, crRNA spacer; grey, crRNA repeat; red, PAM; black arrows, predicted NmeCas9 cleavage sites.
(C) Plasmid cleavage by NmeCas9 requires tracrRNA, the cognate crRNA and Mg2+. N, nicked; L, linearized; SC: supercoiled.
(D) Sequencing of the NmeCas9 linearized pGCC2-ps 9 plasmid. Black arrows indicate predicted cleavage sites and the green stars indicate the A overhangs added by the sequencing reactions.
(E) Divalent metal ion specificity of NmeCas9. Plasmid cleavage assay was performed as in (C), except that dual RNAs were used for all lanes.
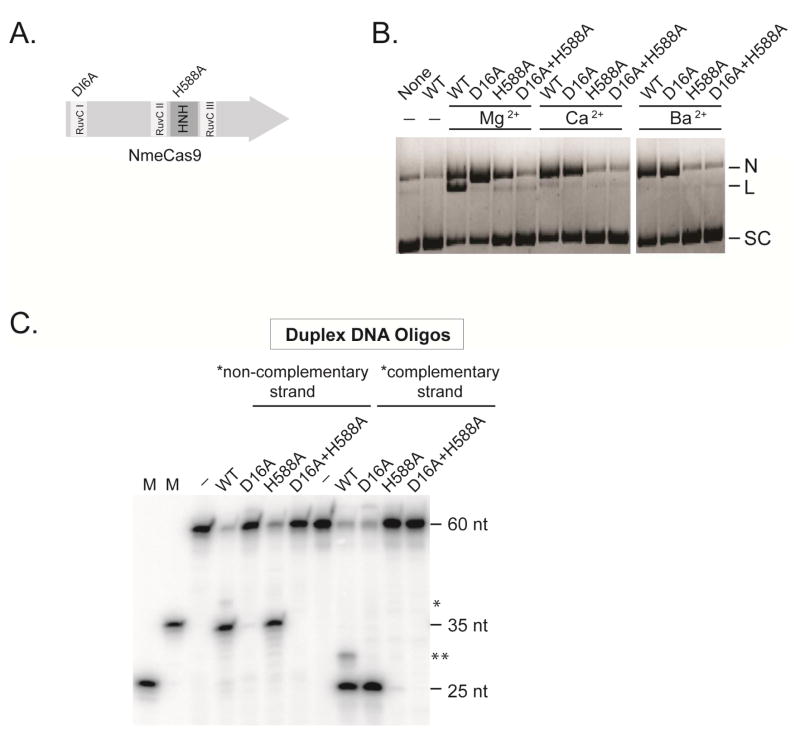
Residues D16 (in the RuvC-I domain) and H588 (in the HNH domain) of NmeCas9 (Fig. 2A) are required for cellular DNA targeting (Esvelt et al., 2013; Zhang et al., 2013). To test the involvement of these residues in NmeCas9 catalytic activity in vitro, mutant proteins D16A, H588A, and the double mutant (D16A, H588A) were expressed and purified (Fig. 1A). Plasmid cleavage assays showed that D16A and H588A mutants could each nick the plasmid, whereas the double mutant was inactive (Fig. 2B). Mg2+ supported the activity of both RuvC and HNH domains, but divalent metals such as Ca2+ and Ba2+ only supported the catalytic activity of the HNH domain (Fig. 2B). To determine the strand specificities of the catalytic domains, we radioactively labeled either the crRNA-complementary or non-complementary strand of another target duplex (sp 25). The D16A mutant only cleaved the complementary strand, whereas the H588A mutant only cleaved the non-complementary strand (Fig. 2C).
Figure 2. Strand Specificity of NmeCas9’s Two Nuclease Domains.
(A) Schematic representation of the location of the RuvC and HNH domains in the primary sequence of NmeCas9.
(B) Plasmid cleavage assays were performed using wildtype NmeCas9, as well as active site mutants D16A, H588A, and D16A+H588A. N, nicked; L, linearized; SC: supercoiled.
(C) NmeCas9 uses the HNH and RuvC nuclease domains to cleave the crRNA-complementary and non-complementary strands, respectively. The oligonucleotide cleavage assay was performed using tracrRNA- and crRNA-programmed, wildtype and mutant NmeCas9 proteins at 37°C for 30 min. Duplex DNA substrates were 5′ 32P labeled on either strand. M, size markers. The sizes of the substrates, cleavage products and markers are indicated. The single and double asterisks denote non-complementary strand and complementary strand (respectively) minor cleavage products that are abolished when either active site is mutated. The significance (if any) of these minor cleavage products is not clear.
Collectively, these experiments confirm that NmeCas9, like all other Cas9 orthologs studied to date, is an RNA-guided DNA endonuclease that requires a tracrRNA, a cognate crRNA, and a divalent metal ion. The HNH and the RuvC domains each cleave the crRNA-complementary and non-complementary DNA strands, respectively, targeting 3–4 bp into the protospacer, and each domain has different metal cofactor requirements.
NmeCas9 cleaves ssDNA in an RNA-programmed but tracrRNA-independent fashion
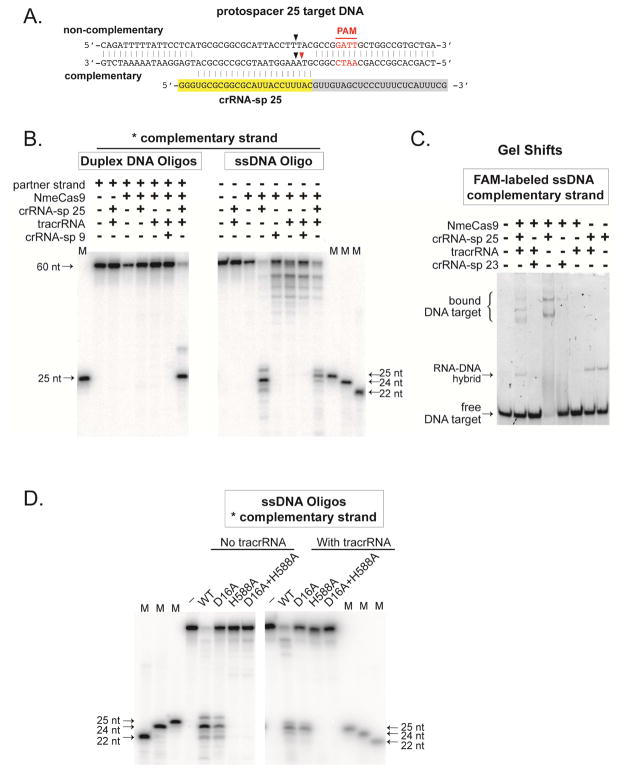
NmeCas9 requires both crRNA and tracrRNA to cleave either a plasmid DNA (Fig. 1C) or alternatively a dsDNA oligonucleotide substrate (Fig. 3A). No cleavage was observed on either strand when NmeCas9, tracrRNA, or cognate crRNA (sp 25) was omitted from the reaction (Fig. 3B and Fig. S2).
Figure 3. NmeCas9 Exhibits TracrRNA-Independent ssDNA Cleavage Activity.
(A) Schematic representation of ps 25-containing DNA oligonucleotides and the sp 25 crRNA used in (B) and (C). Red lettering, PAM; yellow, crRNA spacer; gray, crRNA repeat; black arrows, predominant cleavage sites for dual-RNA guided NmeCas9 cleavage; red arrow, predominant cleavage site for tracrRNA-independent NmeCas9 cleavage.
(B) NmeCas9 cleaves ssDNA efficiently in a tracrRNA-independent manner. NmeCas9 complexed with small RNAs was assayed for cleavage of double- (left) or single- (right) stranded DNA targets for sp 25. The complementary strand was 5′ 32P radiolabeled. M, size markers. The sizes of substrates, cleavage products and markers are indicated.
(C) NmeCas9 binds an ssDNA target in vitro in a tracrRNA-independent manner. Gel shifts were performed using a 5′ FAM-labeled ssDNA substrate (100nM), NmeCas9 (500nM), and various small RNAs (500nM) as indicated. Binding was performed in cleavage reaction buffer (but with Mg2+ omitted) at room temperature for 10 min, and then resolved by 5% native PAGE.
(D) An intact HNH domain is required for tracrRNA-independent cleavage of complementary strand ssDNA. Wildtype and active-site-mutant NmeCas9s were assayed for cleavage of ssDNA, in the absence (left) or presence (right) of tracrRNA. Reactions were performed as in Fig. 2C. M, size marker. The sizes of substrates, cleavage products and markers are indicated.
When using the complementary strand ssDNA substrate, NmeCas9 also catalysed cleavage (primarily between the third and fourth nts of the protospacer) when programmed with both cognate crRNA (sp 25) and tracrRNA (Fig. 3B). Surprisingly, the complementary strand was also robustly cleaved when NmeCas9 was programmed with the cognate crRNA alone (Fig. 3B). This tracrRNA-independent mode of ssDNA cleavage depended on the complementary crRNA spacer, since cleavage was not observed when a non-cognate crRNA (sp 9) or no crRNA was present (Fig. 3B). Interestingly, the cleavage pattern of the “no tracrRNA” reaction differed from the pattern observed when the tracrRNA was included in the reaction (Fig. 3B). In the former case, we observed several 22–26 nt products, with the 24 nt form dominating, whereas in the latter case we observe 24–26 nt products with the 25 nt form dominating. This cleavage pattern reflects a 1-nt shift of the primary cut site (Fig. 3A–B) in the PAM-proximal direction.
We used gel shift assays to investigate whether both RNAs were required for stable ssDNA binding by NmeCas9. All divalent metal ions were omitted to render NmeCas9 catalytically inactive. As expected, a 50-nt, fluorescently labelled, ssDNA (containing ps 25 and the PAM) was bound by NmeCas9 when a cognate crRNA (sp 25) and the tracrRNA were both present, but not when a non-cognate crRNA (sp 23) was used instead (Fig. 3C). Importantly, robust ssDNA target binding occurred when NmeCas9 was programmed with a cognate crRNA alone, but not with the tracrRNA alone, or with a non-cognate crRNA alone (Fig. 3C). These results indicate that sequence-specific binding of an ssDNA target by NmeCas9 can occur in a tracrRNA-independent manner. RNA binding studies demonstrated NmeCas9 binding to a variety of RNA substrates (e.g, crRNA, tracrRNA and even CRISPR-unrelated RNAs), showing that NmeCas9 has non-specific RNA-binding activity (Fig. S3). However, the tracrRNA-independent, crRNA-guided ssDNA cleavage (Fig. 3B) shows that crRNA is sufficient to engage NmeCas9 in an enzymatically productive fashion.
To investigate which of the two nuclease motifs is responsible for cleaving ssDNA in a tracrRNA-independent manner, we tested the D16A and H588A “nickases” in the ssDNA cleavage assay. Both modes of ssDNA cleavage, regardless of tracrRNA presence, were retained in the D16A nickase mutant but absent in the H588A nickase mutant and the D16A+H588A double mutant (Fig. 3D). These results indicate that NmeCas9 uses the HNH domain to cut the ssDNA targets.
Cleavage site selection and RNA guide requirements for ssDNA cleavage
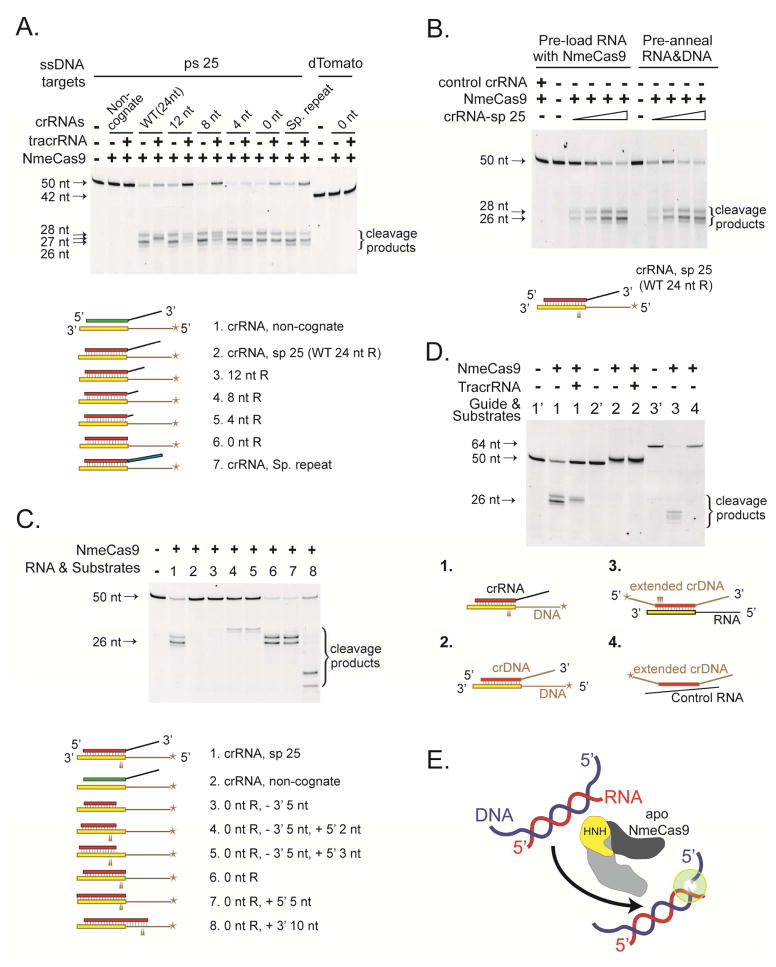
The mature crRNAs for NmeCas9 contain a 24 nt sequence derived from CRISPR spacers and a 24 nt sequence derived from the CRISPR repeats (Zhang et al., 2013) (Fig. 3A). To define the minimal region of CRISPR repeats required for tracrRNA-independent, ssDNA cleavage, we created serial 3′ deletions of the crRNA repeat and tested the abilities of these guides to cleave ssDNA, with or without tracrRNA (Fig. 4A). Surprisingly, in the absence of tracrRNA, the cleavage pattern of the ssDNA target was not affected by the repeat truncations, nor by replacement of the N. meningitidis repeat with a 20 nt S. pyogenes repeat (Fig. 4A). These results show that the CRISPR repeat is dispensable for the tracrRNA-independent ssDNA cleavage by NmeCas9. The sp 25-containing target was not cleaved by NmeCas9 with a non-cognate crRNA, and a truncated sp 25 guide failed to direct NmeCas9 cleavage of a non-cognate ssDNA target (Fig. 4A). These observations indicate that a base-paired RNA-DNA hybrid is necessary for the tracrRNA-independent activity of NmeCas9. This activity is reminiscent of the RNase H activity that cleaves the RNA strand of an RNA-DNA hybrid duplex (with little or no sequence preference) (Tadokoro and Kanaya, 2009), except that NmeCas9’s activity has the opposite nucleic acid specificity (i.e. it cleaves the DNA strand), and it cleaves at specific locations. Accordingly, we refer to this tracrRNA-independent, RNA-guided ssDNA cleavage activity as DNase H-like.
Figure 4. Guide Sequence Independence and Cleavage Site Selection for NmeCas9’s DNase H Activity.
(A) NmeCas9-catalyzed, crRNA-guided, tracrRNA-independent ssDNA cleavage does not require specific CRISPR repeats. Sp 25-specific crRNAs with repeat variants were assayed for NmeCas9-directed cleavage of an ssDNA target. Here and in the rest of this figure, cleavage reactions were performed as in Fig. 2C, using a 5′ FAM-labeled ssDNA bearing a sp 25 target. The sizes of substrates and cleavage products are indicated. The panel of RNAs and substrates examined are depicted below the gel image and are colored as follows: DNA, brown lines and boxes with brown borders; RNA, black lines and boxes with black borders; ps 25, yellow; sp 25 sequences, red; sp 23 sequences, green; S. pyogenes (Sp) repeat, blue; 5′ labels, stars. The non-cognate target is a 42-nt 5′ FAM-labeled ssDNA from the dTomato gene.
(B) ssDNA target cleavage by NmeCas9 is as efficient when crRNA is pre-annealed to DNA substrates (right), as compared to crRNA that is allowed to pre-load into NmeCas9 (left). Sp 25 crRNA (25, 50, 250, and 500 nM) is pre-annealed with ssDNA substrates (50 nM) in cleavage buffer at 37°C for 10 min. Pre-loading of crRNA (25, 50, 250, and 500 nM) with NmeCas9 (500 nM) is done in cleavage buffer at 37°C for 10 min.
(C) NmeCas9 determines the DNase H cleavage sites by a ruler mechanism that measures from the 5′ end of the DNA in the RNA-DNA hybrid, and requires a minimum 17–18 base-paired region. The guide-substrate pairs are depicted and named at the bottom of the panel. In the guides for samples 3–5, 7, and 8, additional nts (complementary to the ssDNA target) were added to (or removed from) either the 5′ or 3′ end of the guide, with the specific end and the number of added or subtracted nts indicated. All RNA guides have a 5′-terminal GGG trinucleotide to facilitate in vitro transcription, so the lengths of the DNA-RNA hybrid duplexes are 3 nt shorter than the RNA guides.
(D) Inverting the backbone composition of the guide-substrate duplex also inverts the strand asymmetry of the cleavage activity. 1′, 2′ and 3′ denote the control reactions including only the labeled DNA component for substrate pairs 1, 2, and 3, respectively. The basis for the slightly retarded mobility of the labeled target upon incubation with crDNA (substrate pair 2) is not known.
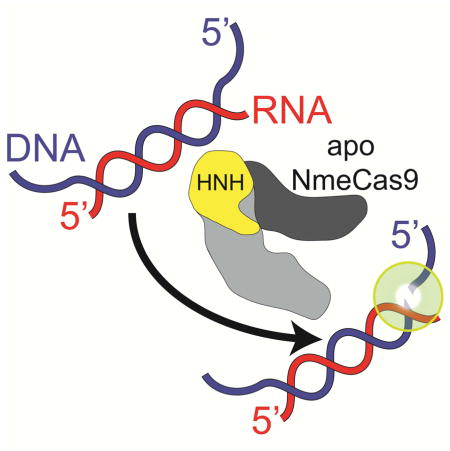
(E) DNase H activity of apo NmeCas9. The depiction of NmeCas9 is based on the structure of a different Type II-C Cas9 (AnaCas9; Jinek et al., 2014). Recognition lobe, dark grey; nuclease lobe, light grey; HNH domain, yellow.
In contrast to the tracrRNA-independent activity, progressive truncations of the CRISPR repeat in the presence of the tracrRNA lead to marked changes in the cleavage pattern. 3′ deletion of 12–16 nts of the repeat caused a shift from a pattern with one dominating product (indicating cleavage between the 3rd and 4th nts of the protospacer) to a pattern with three to four species (25–28 nts) (Fig. 4A). Further shortening of the repeat, or replacement by the S. pyogenes repeat, resulted in a pattern identical to that of the tracrRNA-independent cleavage (Fig. 4A). These data suggest that the repeat/anti-repeat pairing within a crRNA/tracrRNA duplex is important for tracrRNA influence on ssDNA cleavage site selection by NmeCas9.
To determine whether NmeCas9’s DNase H activity requires a pre-loaded guide RNA, we tested whether it can cleave the ssDNA in an RNA-DNA hybrid that was pre-formed in solution, in the absence of the enzyme. We assayed tracrRNA-independent ssDNA cleavage using guide RNAs that were either pre-annealed with the ssDNA target before the addition of NmeCas9 (Fig. 4B, right), or pre-incubated with NmeCas9 before the addition of ssDNA target (Fig. 4B, left). The cleavage patterns were similar under these two conditions (Fig. 4B), reinforcing the idea that NmeCas9 has a DNase H activity that cleaves the DNA strand of a RNA-DNA hybrid.
We next investigated the rules governing DNase H cleavage site selection. We assayed a panel of RNA guides with extensions or truncations compared to the sp 25 guide that lacks all repeat residues (0 nt R) (Fig. 4C). We found that the cleavage sites move in concert with the 5′ end of the ssDNA’s RNA-paired region (Fig. 4C, compare substrates 4 and 5 vs. 6 and 7, and vs. 8), but not with the 3′ end of the RNA-paired region (Fig. 4C, compare substrates 4 vs. 5, and 6 vs. 7). This indicates that the cleavage sites of NmeCas9’s DNase H activity are set by a ruler mechanism that measures from the 5′ end of the ssDNA’s RNA-paired region. This activity requires a minimum of 17–18 bps of RNA-DNA hybrid duplex, since the activity was lost when a 16 nt guide RNA was used (Fig. 4C, compare substrates 3, 4, and 5). A DNA guide that contains sequences identical to the sp 25 crRNA did not support cleavage, regardless of the tracrRNA’s presence (Fig. 4D, substrate 2). Finally, when the nucleic acid identities of the guide and the target were reversed (i.e. with a crDNA “guide” and a ps 25-containing ssRNA “target”), it was the crDNA that was cleaved, at positions close to the 5′ end of the RNA-paired region (Fig. 4D, substrate 3). Our results reveal an RNA-directed ssDNA cleavage activity of NmeCas9 that does not depend upon tracrRNA, or on any guide sequences that are specific to the N. meningitidis crRNAs (Fig. 4E).
Meningococcal cells are constitutively competent for natural transformation, which requires the degradation of one of the two DNA strands during uptake into the cytoplasm (Rotman and Seifert, 2014). The internalized ssDNA is then thought to associate with RecA protein, single-strand binding protein (SSB), or both (Johnston et al., 2014). We demonstrated that meningococcal transformation is subject to CRISPR interference (Zhang et al., 2013), though our tests have not addressed the question of whether DNA targeting occurred during uptake, during or after recombination into the chromosome, or some combination of these. To determine whether ssDNA bound by SSB or RecA can serve as a substrate for DNase H activity, we added increasing amounts of SSB (Fig. S4A) or RecA (Fig. S4B) to the reactions. We observed no inhibition of cleavage by the RecA protein at any concentration (Fig. S4B), and only modest inhibition of cleavage at the highest concentration of SSB (Fig. S4A). Gel shifts confirmed that the ssDNA was bound by both SSB and RecA proteins under these conditions (Fig. S4C). These results suggest that tracrRNA-independent, crRNA-guided cleavage can proceed with substrates that more closely approximate the protein-bound ssDNAs thought to exist within cells.
NmeCas9 PAM Specificity
Previously, we used bioinformatics to define the PAM for NmeCas9 as 5′-NNNNGATT-3′ (e.g. in Fig. 1B) and showed that a double mutation (GATT to GTAT) caused a partial loss of interference during transformation (Zhang et al., 2013). We also observed that a single A-to-C mutation (GATT to GCTT) is well tolerated for NmeCas9-mediated genome editing in human ES cells (Hou et al., 2013). Furthermore, a library depletion experiment in E. coli indicated that the NmeCas9 PAM has a very strong preference for guanine at the first position (Esvelt et al., 2013).
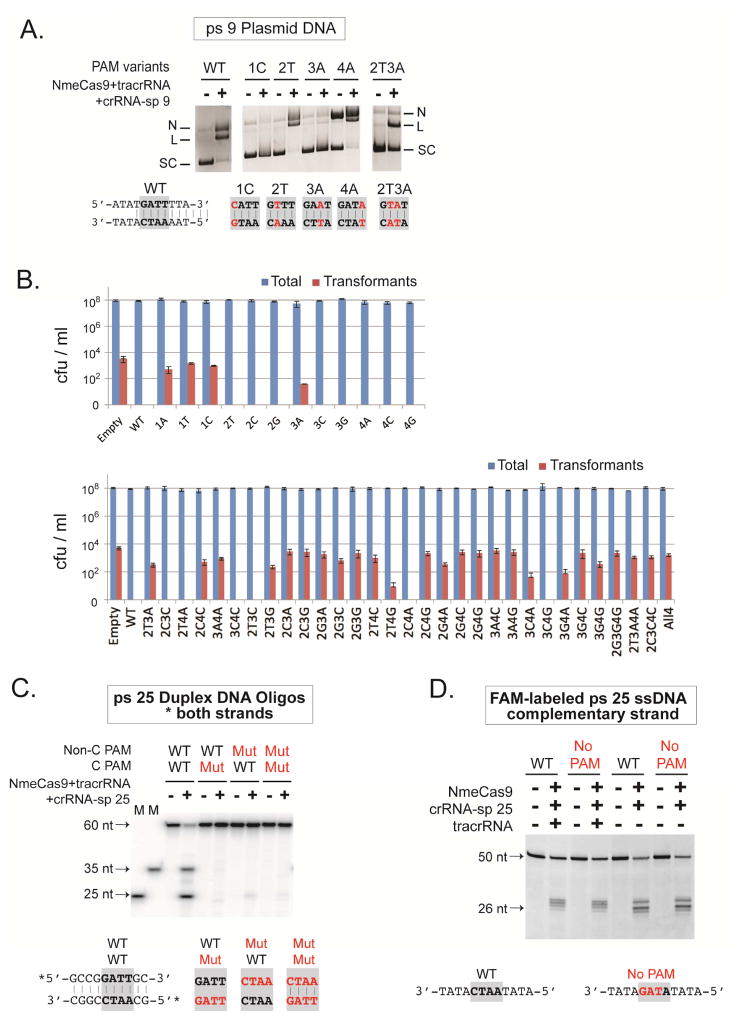
To understand better the rules governing NmeCas9 PAM specificity, we engineered and tested variant PAMs using both in vitro and cellular approaches. First, we mutated each single nt of the GATT PAM to its Watson-Crick complement in the context of a ps 9 plasmid (Fig. 5A), and assayed cleavage in vitro. The 2A to 2T and 4T to 4A mutations were readily cleaved, while the 1G to 1C and 3T to 3A mutations abolished cleavage (Fig. 5A). The 2A3T to 2T3A partial cleavage defect is in agreement with the partial loss of cellular interference observed previously (Zhang et al., 2013) (and confirmed below), indicating that our in vitro and cellular results are consistent. Next, we turned to cellular transformation interference to study all possible 1 nt PAM variants in the context of ps 25. The ps 25 plasmid with the wild-type PAM elicited interference, as reflected by the complete loss of transformants compared to the empty plasmid (Fig. 5B, upper panel). All three variants at the first guanine of the PAM led to major interference defects. In contrast, eight of the nine single-nt mutations in the other PAM nts were permissive for interference. Only the T-to-A mutation at position 3 exhibited a modest defect (Fig. 5B, upper panel). These results are largely consistent with our in vitro cleavage assay in Fig. 5A and confirm that NmeCas9 has a stringent preference for the guanine within the GATT PAM, and a high degree of tolerance for most single-nt variants at the 2nd, 3rd and 4th PAM positions. The same series of PAM variants in the context of a different protospacer (ps 24) yielded similar results (Fig. S5).
Figure 5. NmeCas9 Functions with a Range of PAM Variants.
(A) NmeCas9 cleavage of plasmids containing ps 9 and wildtype or mutant PAMs. NmeCas9 was programmed with tracrRNA and the sp 9 crRNA. Reactions were performed as in Fig. 1C. Mutations in the PAM are indicated in red, and PAMs are in bold.
(B) NmeCas9 targets a range of PAM variants in a cellular interference assay. Plasmids containing ps 25 and various PAM mutants were tested by natural transformation assays using wildtype MC8013 cells as recipients. The bar graphs are log-scale plots of colony-forming units (CFU)/ml (mean ± s.e.m.) for total cells (blue bars) and erythromycin-resistant (ErmR) transformants (red bars) from three independent experiments. The positions and nt identities of the mutations are indicated for each PAM variant.
(C) PAM residues are required on both strands to license NmeCas9 cleavage on a duplex DNA substrate. NmeCas9 programmed with tracrRNA and sp 25 crRNA were tested for oligonucleotide cleavage as in Fig. 2C. Non-C, non-complementary strand; C, complementary strand. Duplex DNA oligos were 5′ 32P-labeled on both strands. Mutant PAM sequences are shown (below the gel) in red, with the PAM region in bold.
(D) NmeCas9 cleavage of ssDNA targets does not require a PAM. The oligonucleotide cleavage assay was performed as in Fig. 2C, except that 5′ FAM-labeled complementary ssDNA oligos (100 nM) were used. NmeCas9 was programmed either with sp 25 crRNA alone or together with tracrRNA. PAM mutations are indicated (below the gel) in red, and PAMs are in bold. The “No PAM” oligonucleotide carries a triple mutation in the PAM.
Although the guanine in the GATT PAM is clearly the most important nt, additional tests revealed some dependence on the other positions. We constructed all 27 possible 2-nt variants in the other three PAM nts, and assayed their abilities to license CRISPR interference of transformation. Interestingly, two-thirds (18/27) of these 2-nt variants exhibited intermediate to severe interference defects (Fig. 5B, lower panel). In contrast, 6/27 (GCCT, GTTA, GACC, GTCT, GCTA and GACG) double mutants are fully functional, and three more (GTTG, GACA, GAGA) exhibited a partial decrease in transformation efficiency (Fig. 5B, lower panel). All three representative triple mutants that we generated (GGGG, GTAA, and GCCC) displayed severe defects (Fig. 5B, lower panel). All ps 25 plasmids tested efficiently transformed an isogenic strain carrying a disruption of cas9 (Fig. S6) (Zhang et al., 2013), indicating that the PAM variations affect interference, not transformation itself. Previously, the functional PAM specificities derived from a library depletion approach proved to be complex (GANN, GTTN, GNNT > GTNN, GNTN) (Esvelt et al., 2013). Our individual tests help to reinforce most of these previously defined PAM specificities and also reveal further complexities, as some PAM variants that match one of those motifs (e.g. GAAG, GAAC, and GAGC) are completely defective.
Extensive biochemical and structural studies of SpyCas9 revealed that its PAM is required only on the crRNA-noncomplementary strand of dsDNA targets (Jinek et al., 2012; Anders et al., 2014; Sternberg et al., 2014). To address the strand specificity of PAM recognition by NmeCas9, we tested three DNA duplexes where all 4 nts of the PAM were mutated in the complementary strand, the non-complementary strand, or both (Fig. 5C). Cleavage of either strand was lost or greatly diminished when the PAM was disrupted on either target strand, or on both (Fig. 5C). This observation demonstrated that, unlike SpyCas9, NmeCas9 requires the PAM element on both strands of the duplex DNA, or that recognition elements on one strand or the other can only be engaged in the context of a base-paired configuration.
To address the PAM requirement for ssDNA target recognition, we compared ssDNA cleavage using two substrates, one with a wildtype PAM and the other with no PAM (Fig. 5D). The “No PAM” substrate was cleaved as efficiently by NmeCas9 as the wildtype counterpart, regardless of whether the tracrRNA is present (Fig. 5D). This observation suggests that the PAM is not necessary for NmeCas9-mediated ssDNA cleavage, in line with previous reports for SpyCas9 and SthCas9 (Gasiunas et al., 2012; Jinek et al., 2012; Nishimasu et al., 2014).
Limited tolerance for protospacer/PAM linker length variation for NmeCas9
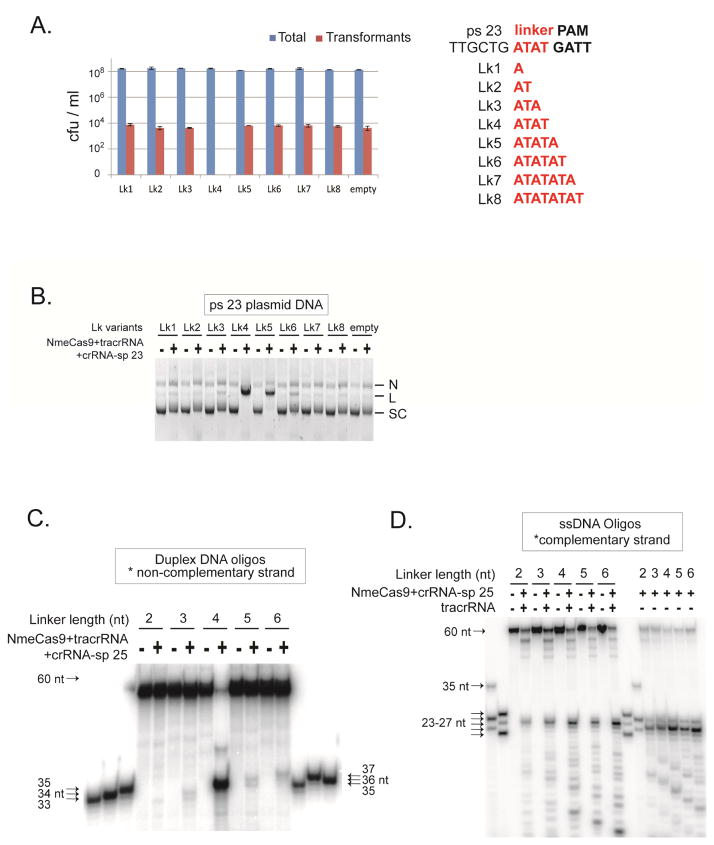
A short (1–4 nts), non-conserved linker usually separates the PAM and the adjacent protospacer (Deveau et al., 2008; Gasiunas et al., 2012; Jinek et al., 2012; Esvelt et al., 2013; Zhang et al., 2013; Chen et al., 2014; Fonfara et al., 2014; Ran et al., 2015). The length of this linker varies among individual Cas9 orthologs. So far there have been only two examples of flexibility of linker length in vivo – Sth1Cas9 of strain DGCC7710 (Briner et al., 2014; Ran et al., 2015) and SthCas9 of strain LMG18311 (Chen et al., 2014) tolerate extension from 2 nt to 3 nt. We tested whether NmeCas9 tolerates linker length variations during interference in N. meningitidis. First, a target for sp 23 of MC8013 was constructed in which the PAM and ps 23 were separated by a 4 nt “ATAT” linker (Fig. 6A, right panel). Then we varied the alternating A/T linker to separate the PAM from the protospacer by 1–8 nts (Fig. 6A, right panel). Because the first nt of the GATT PAM is especially critical, the lack of guanine in the linker ensures that the linker variants are all deprived of any potential functional PAM variants. All of the mutant plasmids transformed the cells as efficiently as the empty plasmid, indicating that they were unable to elicit NmeCas9-mediated interference (Fig. 6A, left panel). We conclude that cellular interference by NmeCas9 stringently requires a 4 nt linker.
Figure 6. NmeCas9 has Minimal Tolerance for Protospacer-PAM Linker Length Variation.
(A) NmeCas9 only functions with a 4 bp linker in bacterial cells. Left, cellular interference assay. Plasmids containing ps 23 and its linker length mutant derivatives were tested in natural transformation assays as in Fig. 5B. Sequences of the linkers between ps 23 and its PAM in the mutants used in (A) are shown on the right. The linkers are in red.
(B) The same plasmids tested in (A) were assayed for cleavage in vitro by NmeCas9 programmed with sp 23 crRNA and tracrRNA. Reactions were performed as in Fig. 1C. N, nicked; L, linearized; SC: supercoiled.
(C) NmeCas9 cleavage of the non-complementary strand in DNA duplexes is much less efficient when the linker length varies, and the cut site moves in concert with the PAM. The duplex DNAs were 5′ 32P labeled on the non-complementary strand only, and contain ps 25, a flanking GATT PAM and a 2–6 nt linker in between. NmeCas9 was programmed with tracrRNA and sp 25 crRNA. Reactions were performed as in Fig. 2C. M, size markers. The sizes of substrates, cleavage products and markers are indicated.
(D) NmeCas9 cleavage of the complementary strand ssDNA is as efficient when linker length varies, and the cut sites are at a fixed position. The ssDNA targets were 5′ 32P labeled, and contain ps 25, a flanking GATT PAM, and a 2–6 nt linker in between. NmeCas9 was programmed with sp 25 crRNA and with or without tracrRNA, as indicated. Reactions were performed as in Fig. 2C. M, size markers. The sizes of substrates, cleavage products and markers are indicated.
To determine whether linker length variations affect in vitro plasmid cleavage by NmeCas9, we analysed the same set of linker mutant plasmids. The 4 nt linker allowed efficient cleavage, as did the 5 nt linker (Fig. 6B). The remaining linker mutants permitted only weak (3, 6 nt) or background (1, 2, 7, 8 nt) cleavage (Fig. 6B). This observation suggests that the stringent 4 nt linker length requirement observed during cellular interference is not an intrinsic feature of NmeCas9’s target recognition activity in vitro.
The HNH and RuvC nuclease domains from the Type II-A SthCas9 (from strain LMG18311) use different mechanisms to determine their cleavage sites on the two strands: the RuvC cut is measured from the PAM, whereas the HNH cut is at a fixed position within the protospacer, without reference to the PAM (Chen et al., 2014). To determine whether the Type II-C NmeCas9 employs a similar mechanism, we mapped the cleavage sites with strand-specific end-labelled oligonucleotide substrates bearing ps 25 and its GATT PAM, separated by different linker lengths (2–6 bp). When the label was on the non-complementary strand, the RuvC domain efficiently cleaved the 4 bp linker substrate (Fig. 6C). Although cleavage efficiency was much lower with the other variants, the RuvC cleavage site moved in concert (Fig. 6C). These results revealed that the RuvC domain of NmeCas9 determines the cleavage site on the non-complementary strand by a “ruler” mechanism, cutting primarily between the 7th and 8th nts 5′ of the GATT PAM.
To study the HNH domain, we used radiolabeled, ssDNA oligonucleotides corresponding to the complementary strand of sp 25 (with 2–6 nt linkers) as substrates. For the dual RNA-mediated reactions, the most prominent products are consistently 25 nt (Fig. 6D), suggesting that neither extension nor shortening of the 4-nt linker altered the HNH domain cleavage site. The same holds true for the tracrRNA-independent mode of ssDNA cleavage, since the dominant products were consistently 24 nt for all substrates (Fig. 6D). We also observed several smaller products, potentially reflecting minor cleavage events 5′ of the predominant sites. Intriguingly, as the length of the linker increases, the sizes of these minor products decrease accordingly (Fig. 6D).
Targeting by NmeCas9 has relaxed stringency for seed complementarity in vitro and in bacteria
SpyCas9 requires near-perfect complementarity in the seed region for CRISPR interference and efficient genome editing (Jinek et al., 2012; Cho et al., 2013; Cong et al., 2013; Hwang et al., 2013; Jiang et al., 2013; Jinek et al., 2013; Mali et al., 2013). Our previous work showed that a 2-nt mismatch at the cleavage site of a ps abrogated interference, while a 2-nt mismatch further from the PAM (outside the seed) did not (Zhang et al., 2013). We also observed a strong requirement for perfect seed complementarity during NmeCas9-catalyzed genome editing in human ES cells (Hou et al., 2013). To understand better NmeCas9 targeting requirements in the seed region, we created serial, single-nt mutations in the 12 nt seed in the context of ps 9 (Fig. S6A). Plasmids containing these mutant protospacers were transformed into N. meningitidis and assayed for CRISPR interference. Unexpectedly, the majority (11/12) of these seed mutations had minimal impact on interference, while only one seed mutation (in the fourth bp, counting from the PAM-proximal end) exhibited an intermediate interference defect (Fig. S6B, upper panel). This same set of mutant plasmids was also subjected to in vitro NmeCas9 cleavage. Corroborating the in vivo results, only the seed mutation at the fourth bp of the protospacer led to a significant cleavage defect (Fig. S6C, “27”). To further investigate seed requirements, eleven double seed mutants (Fig. S6A) were tested by both the natural transformation assay and in vitro plasmid cleavage. The majority (8/11) of them resulted in loss of interference (Fig. S6B, lower panel) and defective plasmid cleavage (Fig. S6C). Analogous experiments in the context of a different ps (ps 25) yielded similar results (Fig. S7). Taken together, the above results suggest that NmeCas9 seed requirements are somewhat relaxed in N. meningitidis cells and in vitro compared to those observed during genome editing in mammalian cells (Hou et al., 2013).
Functional anatomy of NmeCas9’s RNA guides
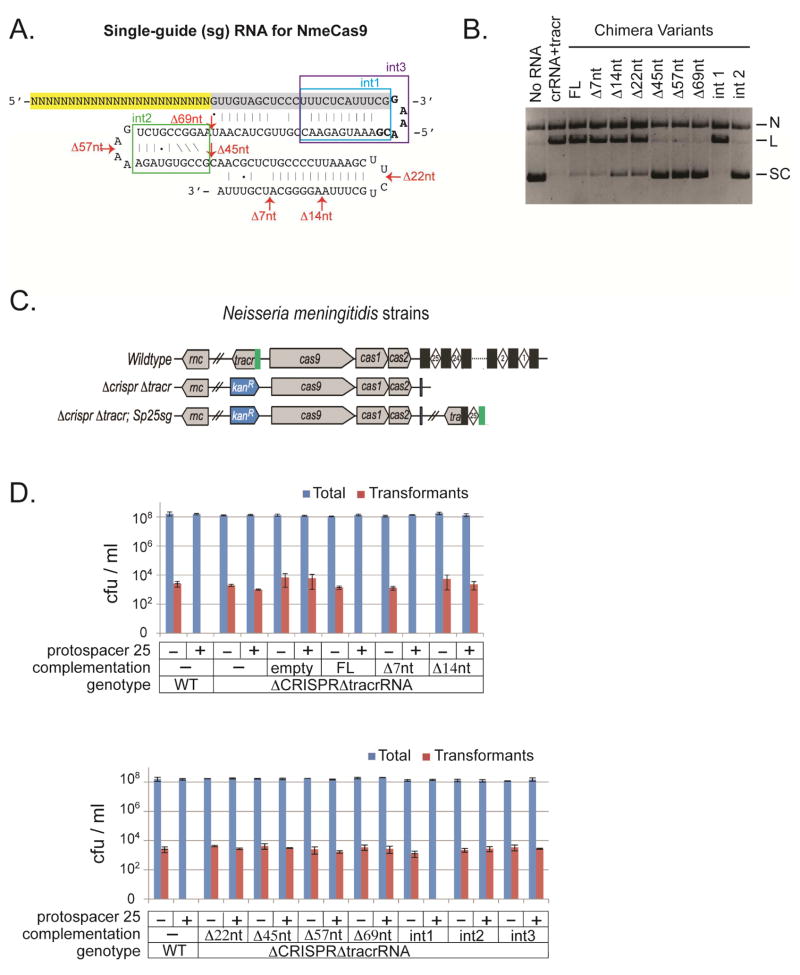
Our previous work in human stem cells showed that an sgRNA works as efficiently as dual crRNA and tracrRNA in NmeCas9-directed genome editing (Hou et al., 2013). To further dissect sgRNA structural and sequence requirements for NmeCas9, we established sgRNA function in vitro and then analysed a series of 3′-terminal and internal deletions (Fig. 7A). A full-length 145-nt sgRNA (corresponding to sp 9) directed plasmid cleavage, with efficiency comparable to a parallel reaction with separate crRNA and tracrRNA (Fig. 7B, 3 left-most lanes).
Figure 7. NmeCas9 sgRNA Requirements in vitro and in Bacteria.
(A) A schematic of the NmeCas9 sgRNA, a chimeric fusion in which the 3′ end of the mature crRNA and the 5′ end of the mature tracrRNA are connected by a GAAA tetraloop. Highlighted in yellow, crRNA spacer; highlighted in gray, crRNA repeat. Six 3′ terminal truncations are indicated by red arrows, and three internal deletions are indicated by colored boxes.
(B) Full-length and variant sgRNAs were assayed for NmeCas9 cleavage of ps 9-containing plasmid in vitro. NmeCas9 was programmed with tracrRNA and sp 9 crRNA. Reactions were performed as in Fig. 1C. The full-length tracrRNA used in in vitro assays had 6 additional 3′-terminal nts [compared to the sequence shown in (A)], whereas all the other RNAs are as shown. N, nicked; L, linearized; SC: supercoiled.
(C) Schematics of representative MC8013 isogenic strains used in (D). Relevant genotypes are given on the left. Grey boxes, genes; blue boxes, kanamycin-resistance marker; black rectangles, CRISPR repeats; white diamonds, CRISPR spacers; green boxes, tracrRNA promoter.
(D) Definition of sgRNA regions that are required for transformation interference. pYZEJS040 (−) and its ps 25-containing derivative (+) were introduced into MC8013-derived strains by natural transformation. Relevant genotypes and sgRNA complementation variants are indicated at the bottom. Data are presented as in Fig. 5B.
A recent report established six structural modules – spacer, lower stem, bulge, upper stem, nexus, and hairpins - within the sgRNAs for SpyCas9 and other type II-A systems (Briner et al., 2014). The sgRNA for NmeCas9 contains equivalents of these features. To begin defining the importance of these features in type II-C, we created serial sgRNA deletions with 7, 14, 22, 45, 57, and 69 nts removed from the 3′ end (Fig. 7A), and tested plasmid cleavage in vitro. The Δ7nt substrate showed no defects and the Δ14nt and Δ22nt RNAs exhibited modest defects, whereas the Δ45nt, Δ57nt and Δ69nt sgRNAs were completely defective (Fig. 7B). These results suggest that the 3′ terminal 7 nts are dispensable for cleavage, partial or total disruption (Δ14nt, Δ22nt) of the terminal hairpin modestly impedes function, and complete removal (Δ45nt, Δ57nt and Δ69nt) of the hairpin, or nexus plus the hairpin, abolishes cleavage. To further define sub-regions of the sgRNA that are required for targeting, we generated variants with most of the nexus (int2) or upper stem (int1) deleted (Fig. 7A). Severe cleavage defects were observed for the nexus deletion (int2), whereas no defects were detected for the upper stem deletion (int1) (Fig. 7B). This result shows that the nexus of the NmeCas9 sgRNA is necessary for NmeCas9 function, while the upper stem is dispensable, similar to the type II-A systems (Briner et al., 2014).
To determine whether the same modular requirements apply during cellular interference, we created a “clean” strain by deleting the crispr and tracrRNA loci (Fig. 7C), both of which are essential for interference [(Zhang et al., 2013) and Fig. 7D]. An sp 25-specific full-length sgRNA under the control of the tracrRNA promoter was then integrated into the chromosomal nics locus in the ΔcrisprΔtracr strain background (Fig. 7C). This sgRNA complementation restored interference against a ps 25-containing plasmid, whereas control complementation using the empty vector did not (Fig. 7D), indicating that the full-length sgRNA compensates for the loss of both cognate crRNA and tracrRNA. 3′ truncations and internal deletions parallel to those analysed in vitro (Fig. 7B) were created in this cellular context, and their abilities to restore interference were assayed. Among all the variants tested, only two (Δ7nt and int1) rescued interference (Fig. 7D), suggesting that in vivo, NmeCas9 is only tolerant of small 3′ terminal deletions or the removal of the upper stem. The GAAA tetraloop inserted to connect the crRNA and tracrRNA was retained in the int1 variant, and further deletion of this tetraloop from int1 (to generate int3, Fig. 7A) abolished sgRNA function (Fig. 7D). These results suggest that the structure of the bulge region is important for function, since forcing it to comprise a terminal loop results in a non-functional sgRNA.
DISCUSSION
NmeCas9 PAM and seed specificity
Successful dsDNA targeting by type II CRISPR-Cas systems requires not only crRNA/ps complementarity, but also the presence of a PAM. SpyCas9 requires a NGG PAM in both bacterial and eukaryotic cells, although the less optimal NAG variant could also function (Hsu et al., 2013; Jiang et al., 2013; Zhang et al., 2014). Here we confirm that NmeCas9 has a strong preference for G at the first position of the GATT PAM, both in its native cellular environment and in vitro, corroborating findings from an earlier study in E. coli (Esvelt et al., 2013). For the non-guanosine PAM residues, we carried out comprehensive mutagenesis in Neisseria cells and identified all functional PAM variants that carry 1- or 2-nt deviations from the consensus (Fig. 5). Some variants fit one of the minimal consensus PAMs defined previously (Esvelt et al., 2013) yet were non-functional, indicating that PAM recognition rules may be even more complex than previously thought, and illustrating the value of deconvoluting the specificities defined by library depletion. A previous study of NmeCas9-mediated genome editing in human ES and iPS cells identified one functional PAM variant (GCTT) (Hou et al., 2013); it remains to be reported how many additional NmeCas9 variants are tolerated during genome editing in mammalian cells.
A subset of Cas9 orthologs, including examples from S. pyogenes, S. mutans, S. thermophilus and N. meningitidis, use PAMs that begin with one or two guanosines (Deveau et al., 2008; Jinek et al., 2012; Zhang et al., 2013; Chen et al., 2014; Fonfara et al., 2014). SthCas9 from LMG18311 prefers a GYAAA PAM and, like NmeCas9, tolerates single-nt mutations in all positions except the first G (Chen et al., 2014). However, the effects of 2-nt (or more) variants from this PAM consensus were not defined. Structural analyses showed that SpyCas9 uses two conserved arginine residues to recognize the two guanosines in the GG PAM (Anders et al., 2014). These two arginines are not well conserved in Cas9s from many type II-C and some type II-A systems. Similar mechanistic analyses of other Cas9 orthologues will be required to elucidate general rules governing PAM recognition and PAM degeneracy.
We also observe a surprising insensitivity to seed mismatches during transformation (Figs. S7B and S8B), in stark contrast to the stringent requirement for seed complementarity during NmeCas9-mediated mammalian genome editing (Hou et al., 2013). With SpyCas9, single seed mismatches disrupt transformation interference by the native CRISPR-Cas system (Jinek et al., 2012), though much greater seed mismatch tolerance was observed with SpyCas9 in vitro, especially at elevated enzyme concentrations (Pattanayak et al., 2013). This latter observation, along with the stringent requirement for seed pairing in human cells (Hou et al., 2013), suggests that the mismatch tolerance we observe in bacteria could reflect cellular NmeCas9 expression levels rather than a greater intrinsic promiscuity of the enzyme.
TracrRNA-independent DNase H activity of NmeCas9
To our knowledge, in all Cas9 studies and applications reported to date, the tracrRNA (or the tracrRNA-derived portion of an sgRNA) is an essential component for activity (Bernick et al., 2012; Barrangou and Marraffini, 2014; Doudna and Charpentier, 2014; Hsu et al., 2014; van der Oost et al., 2014; Sontheimer and Barrangou, 2015). The tracrRNA-equivalent part of an sgRNA pairs with the crRNA repeat, forms multiple 3′ hairpins, and is contacted extensively by SpyCas9 (Anders et al., 2014; Nishimasu et al., 2014; Jiang et al., 2015). The tracrRNA also contains the “nexus” stem-loop that imparts specificity and enforces orthogonality among Cas9/tracrRNA pairs (Briner et al., 2014). Strikingly, our biochemical characterization of NmeCas9 revealed its ability to cleave efficiently ssDNA molecules even in the absence of tracrRNA (Figs. 3 and 4). Overall, NmeCas9-directed ssDNA cleavage appears to be as efficient as cleavage of dsDNAs (Figs. 1, 3 and 4). These observations are in stark contrast to other Cas9 orthologs biochemically characterized to date. For the well characterized SpyCas9 and Sth3Cas9, ssDNA cleavage (in the presence of tracrRNA) is much less efficient than dsDNA cleavage (Gasiunas et al., 2012; Jinek et al., 2012; Sternberg et al., 2014).
Structural analyses provide hints of the possible enzymatic basis for the tracrRNA-independent DNase H activity that we observe for NmeCas9. The structure of NmeCas9 is not known, and the only Type II-C Cas9 to be structurally characterized is AnaCas9 (in the apo form only, i.e. without bound guides or targets) (Jinek et al., 2014). In contrast, crystal structures of SpyCas9 have been solved in the apo state (Jinek et al., 2014), with bound sgRNA (Jiang et al., 2015), and with sgRNA recognizing a fully ssDNA (Nishimasu et al., 2014) or partially dsDNA (Anders et al., 2014) target. The structures reveal that the apo enzyme is found in an unproductive conformation and that sgRNA binding to SpyCas9 induces extensive domain movements (including the HNH domain) that lead to catalytic activation. The HNH domain of apo SpyCas9 faces outwards away from the body of the nuclease lobe, and it appears to be autoinhibited (in addition to being poorly ordered in the crystal) (Jinek et al., 2014). Although the HNH domain of apo AnaCas9 is located in an equivalent position to that of apo SpyCas9, it has fewer contacts with the C-terminal domain and it is more ordered. In SpyCas9 the guide/target heteroduplex is trapped in a tunnel formed by the RuvC, HNH and REC domains, at the interface between the two lobes of the protein (Anders et al., 2014; Nishimasu et al., 2014). Furthermore, residues in the tunnel do not interact with any of the tracrRNA regions in the sgRNA/SpyCas9 structures, suggesting that an RNA/DNA hybrid stem could easily be recognized and accommodated by NmeCas9 even in the absence of tracrRNA and positioned in a manner such that the HNH domain can engage the DNA strand (Anders et al., 2014; Nishimasu et al., 2014; Jiang et al., 2015). The known flexibility of the HNH domains, together with the largely modular nature of the interactions with crRNA and tracrRNA, could explain the DNase H activity that we observe.
These biochemical features of NmeCas9 are intriguing in light of known modes of genetic exchange in meningococci and many other bacteria. Natural transformation proceeds through the internalization of exogenous DNAs in ssDNA form without strand preference, followed by integration into host chromosomes by homologous recombination (Johnston et al., 2014). Internalized ssDNAs are coated with RecA and ssDNA-binding proteins that facilitate homology search, and we show that these proteins do not inhibit NmeCas9 activity in vitro. It is not known which of these stages of transformation are subject to CRISPR interference, though genomic dsDNA is clearly susceptible (Bikard et al., 2012; Jiang et al., 2013; Vercoe et al., 2013). In addition, either DNA strand can be randomly internalized during transformation, yet a crRNA that is complementary to only one strand completely blocks transformation (Bikard et al., 2012; Zhang et al., 2013). This result indicates that targeting during the pre-recombination ssDNA phases, if it occurs, is not obligatory. Nonetheless, our finding that NmeCas9 can potentially target ssDNA regardless of tracrRNA availability could provide a route toward sequence-specific transformation interference in a manner that does not require a lethal chromosome breakage event. We previously found that a Δtracr strain exhibited a complete rather than partial loss of transformation interference (Zhang et al., 2013), indicating that conditions under which tracrRNA-independent ssDNA interference could occur, if any, have yet to be identified. Another potential natural ssDNA target could be the genomic ssDNA of filamentous phages, such as those reported previously in meningococci (Kawai et al., 2005).
The tracrRNA-independent nature of the ssDNA cleaving activity is also intriguing, though little is known about the regulation of tracrRNA expression. To our knowledge, Type II CRISPR-cas loci generally have only a single tracrRNA transcription unit, whereas there are much larger numbers of CRISPR repeats (up to 105 in a strain of Mycoplasma gallisepticum). Accordingly, complete use of a CRISPR locus’ transcriptional output would require a substantial molar excess of tracrRNA, which could be challenging to generate, especially for large CRISPR loci. This problem could be exacerbated even further in the case of N. meningitidis (and at least some other Type II-C systems), because pre-crRNAs are generated from numerous promoters (one for each CRISPR repeat) (Zhang et al., 2013), potentially increasing the stoichiometric imbalance between the tracrRNA and the crRNA repeats. Thus, it is straightforward to envision potential benefits of tracrRNA-independent functions, such as the RNA-guided ssDNA cleaving activity that we report here, during interference with natural ssDNAs. However, given the lack of evidence thus far for RNA guide sequence specificity during tracrRNA-independent ssDNA cleavage, it is not clear how this activity could specifically employ pre-crRNAs, as might be expected.
The DNase H activity of NmeCas9 could serve as a programmable, RNA-guided “restriction enzyme” that cleaves ssDNA. To our knowledge, site-specific and ssDNA-specific DNases are uncommon. This activity could also have utility within eukaryotic cells, for instance in destroying ssDNA regions of the genomes of certain DNA viruses such as Hepatitis B virus (Dienstag, 2008). In the absence of the tracrRNA, NmeCas9 could be used for such a purpose with little or no risk of collateral damage to the host cell’s dsDNA genome.
EXPERIMENTAL PROCEDURES
Detailed experimental and analytical procedures are found in the supplemental information.
Bacterial Strains, Plasmids and Oligonucleotides
N. meningitidis 8013 (MC8013) and mutant derivatives thereof that were used in this study are listed in supplementary information, as are lists of all plasmids and oligonucleotides.
Mutant Strain Construction
All mutant strains were generated by plasmid transformation and confirmed by PCR and sequencing. The unmarked Δcrispr strain was created by two-step transformation as previously described for creating the unmarked Δcas9 strain (Zhang et al., 2013). For generation of the ΔcrisprΔtracr strain, we transformed the Δcrispr strain with genomic DNA (gDNA) isolated from the Δtracr strain (Zhang et al., 2013), followed by KanR selection. For complementation of sgRNA variants, we cloned wt and variant copies of the sgRNA into pGCC2 and transformed the resulting plasmids into the ΔcrisprΔtracr strain.
Natural Transformation
Natural transformation assays were performed as previously described (Zhang et al., 2013). Antibiotic-resistant cfu/ml and total cfu/ml were reported from three independent experiments (mean +/− s.e.m.), except where noted.
In Vitro Transcription
RNAs were generated by in vitro transcription using a MEGAscript T7 kit (Ambion) and gel purified. Transcription templates were gel-purified PCR products, linearized plasmids, or annealed DNA oligonucleotides carrying the T7 promoter sequence.
Recombinant NmeCas9 Expression and Purification
NmeCas9 was cloned with an N-terminal His6-tag, overexpressed in E. coli, and purified as described in the supplementary methods.
Plasmid DNA Cleavage
Plasmid DNA (300 ng, ~9 nM) was incubated with NmeCas9 protein (50–500 nM) and pre-annealed crRNA:tracrRNA duplex (1:1, 50–500 nM) in cleavage buffer [20 mM HEPES (pH 7.5), 150 mM KCl, 10% glycerol, 1 mM DTT, and 10 mM MgCl2] at 37°C for 5–30 min. Other divalent metals were also tested at 10 mM.
Oligonucleotide Cleavage Assay
10 pmol of gel-purified DNA oligonucleotides (IDT) were 32P-labeled with T4 PNK (NEB) and [γ-32P]-ATP (PerkinElmer), and cleaned up by MicroBio Spin 6 columns (BioRad). Duplex DNA (100 nM) substrates were generated by annealing of equimolar amounts of two oligos. DNA oligonucleotides (IDT) [5–10 nM for 32P-labeled, or 25–50 nM for 6-carboxyfluorescein (FAM)-labeled oligos] were incubated with NmeCas9 proteins (500 nM) and pre-annealed crRNA:tracrRNA duplex (1:1, 500 nM) in cleavage buffer at 37°C for 30 min. Reactions were resolved on 15% denaturing PAGE and visualized with a Phosphorimager or ImageQuant LAS 4000 imager. Reactions with 32P-labeled oligos were purified by phenol/chloroform extraction and ethanol precipitation before electrophoresis.
Gel Shift Assay
FAM-labeled ssDNA oligonucleotides (IDT, 25–50 nM) were incubated with NmeCas9 (500 nM) and various small RNAs (500 nM) in cleavage buffer (without Mg2+) at room temperature for 8–10 min. The reactions were resolved by 6% native PAGE at 4°C and visualized by an ImageQuant LAS 4000 imager.
Supplementary Material
Acknowledgments
We thank Carl Gunderson for help and guidance with mutant strain construction and bacterial transformation, Amy Osterman for technical assistance, and Jennifer Doudna for communicating unpublished results. We also thank members of the Sontheimer laboratory, as well as Craig Mello and members of his laboratory, for helpful discussions. This work was supported by American Heart Association postdoctoral fellowships to Y.Z. and R.R., and by NIH grants R01 GM051350 to A.M., R37 AI033493 and R01 AI044239 to H.S.S., and R01 GM093769 to E.J.S.
Footnotes
Author contributions: Y.Z., R.R., H.S.S., A.M. and E.J.S. designed experiments; Y.Z. constructed strains; R.R. expressed and purified NmeCas9; Y.Z. and R.R. performed experiments; Y.Z., R.R., H.S.S., A.M. and E.J.S. analysed data; Y.Z., R.R., and E.J.S. wrote the manuscript; and all authors edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernick DL, Cox CL, Dennis PP, Lowe TM. Comparative genomic and transcriptional analyses of CRISPR systems across the genus Pyrobaculum. Front Microbiol. 2012;3:251. doi: 10.3389/fmicb.2012.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host & Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Choi J, Bailey S. Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J Biol Chem. 2014;289:13284–13294. doi: 10.1074/jbc.M113.539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486–1500. doi: 10.1056/NEJMra0801644. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Martin B, Fichant G, Polard P, Claverys JP. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 2014;12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- Kawai M, Uchiyama I, Kobayashi I. Genome comparison in silico in Neisseria suggests integration of filamentous bacteriophages by their own transposase. DNA Res. 2005;12:389–401. doi: 10.1093/dnares/dsi021. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, et al. An updated evolutionary classification scheme for CRISPR-Cas systems. Nat Rev Microbiol. 2015 doi: 10.1038/nrmicro3569. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman E, Seifert HS. The genetics of Neisseria species. Annu Rev Genet. 2014;48:405–431. doi: 10.1146/annurev-genet-120213-092007. [DOI] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Barrangou R. The bacterial origins of the CRISPR genome-editing revolution. Hum Gene Ther. 2015;26:413–424. doi: 10.1089/hum.2015.091. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ge X, Yang F, Zhang L, Zheng J, Tan X, Jin ZB, Qu J, Gu F. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.