Abstract
Cupriavidus sp. strain AMP6 is an aerobic, motile, Gram-negative, non-spore-forming rod that was isolated from a root nodule of Mimosa asperata collected in Santa Ana National Wildlife Refuge, Texas, in 2005. Mimosa asperata is the only legume described so far to exclusively associates with Cupriavidus symbionts. Moreover, strain AMP6 represents an early-diverging lineage within the symbiotic Cupriavidus group and has the capacity to develop an effective nitrogen-fixing symbiosis with three other species of Mimosa. Therefore, the genome of Cupriavidus sp. strain AMP6 enables comparative analyses of symbiotic trait evolution in this genus and here we describe the general features, together with sequence and annotation. The 7,579,563 bp high-quality permanent draft genome is arranged in 260 scaffolds of 262 contigs, contains 7,033 protein-coding genes and 97 RNA-only encoding genes, and is part of the GEBA-RNB project proposal.
Keywords: Root-nodule bacteria, Nitrogen fixation, Betaproteobacteria, Texas, Mimosa asperata, GEBA-RNB
Introduction
Cupriavidus is one of two known genera of Betaproteobacteria that include legume root-nodule symbionts [1]. The other genus, Burkholderia, has multiple species associated with diverse legume host plants indigenous to North and South America, South Africa and Australia [2–8]. Cupriavidus, by contrast, has only been isolated from four species in two legume genera in the tribe Mimoseae (Mimosa, Parapiptadenia), at a few locations in the native geographic ranges of their host plants (south Texas, the Caribbean, central America, French Guiana, and Uruguay; [2, 9–12]). However, both Cupriavidus and Burkholderia have now spread to many new regions along with species of Mimosa that are invasive weeds [10, 13–17]. In South America, Cupriavidus was uncommon in French Guiana and Uruguay (3-10 % of nodule isolates; [9, 11]), and was not detected at all in extensive surveys of Mimosa in central Brazil [5, 6]. However, it has been isolated from two cultivated legumes in Minas Gerais, Brazil [18]. This suggests that further surveys in South America may discover additional wild legume hosts that utilize Cupriavidus symbionts.
The only legume studied to date that is exclusively associated with Cupriavidus nodule symbionts is Mimosa asperata, from which Cupriavidus strain AMP6 was isolated in 2005 [12]. The range of M. asperata is centered in Mexico and extends slightly into south Texas, Cuba, and northern Central America [19]. Based on both housekeeping loci and symbiotic loci, strain AMP6 represents an early-diverging lineage of nodule-symbiotic Cupriavidus [10, 12], whose genome may provide insights about how legume nodule symbiosis became established in this group.
Strain AMP6 was collected at the Santa Ana National Wildlife Refuge in Hidalgo County, Texas. Cupriavidus nodule bacteria resembling strain AMP6 are currently known only from M. asperata populations in the lower Rio Grande valley of Texas, and have not been detected in surveys of Mimosa species in other geographic locations [2, 9–11]. Nevertheless, inoculation tests have indicated that Cupriavidus strain AMP6 has the capacity to develop an effective nitrogen-fixing symbiosis with three other species of Mimosa [12]. M. asperata occurs mainly along the margins of seasonally flooded wetlands [20], a habitat characterized by heavy silt/clay soils with neutral to moderately alkaline pH (pH 7.0 - 8.4; [21]).
The first completed genome for a betaproteobacterial legume symbiont was that of Cupriavidus taiwanensisLMG 19424T [22]. Here we provide an analysis of the high-quality permanent draft genome sequence of Cupriavidus strain AMP6, enabling comparative analyses of symbiotic trait evolution in this genus.
Organism information
Classification and features
Cupriavidus sp. strain AMP6 is a motile, Gram-negative, non-spore-forming rod (Fig. 1 Left, Center) in the order Burkholderiales of the class Betaproteobacteria. The rod-shaped form varies in size with dimensions of 0.4-0.6 μm in width and 1.2-1.7 μm in length (Fig. 1 Left). It is fast growing, forming 1.2-1.6 mm diameter colonies after 24 h when grown on YMA [23] at 28 °C. Colonies on YMA are white-opaque, slightly domed, moderately mucoid with smooth margins (Fig. 1 Right).
Fig. 1.
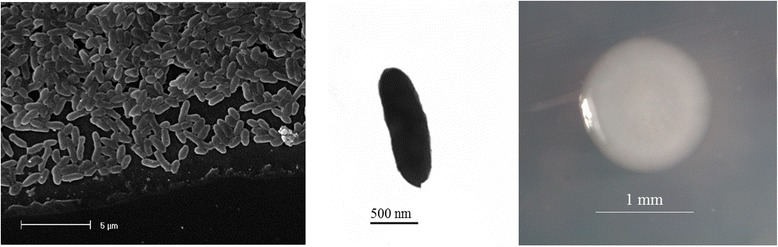
Images of Cupriavidus sp. strain AMP6 using scanning (Left) and transmission (Center) electron microscopy and the appearance of colony morphology on solid media (Right)
Figure 2 shows the phylogenetic relationship of Cupriavidus sp. strain AMP6 in a 16S rRNA gene sequence based tree. This strain is phylogenetically most related to Cupriavidus taiwanensisLMG 19424T, Cupriavidus alkaliphilusASC-732T and Cupriavidus necator N-1T (deposited as ATCC43291T) with sequence identities to the AMP6 16S rRNA gene sequence of 99.11 %, 99.04 % and 98.69 %, respectively, as determined using the EzTaxon-e server [24]. Cupriavidus taiwanensisLMG 19424T is a plant symbiont and was isolated from root nodules of Mimosa pudica collected from three fields at Ping-Tung Country in the southern part of Taiwan [25]. Both ASC-732T and N-1T are soil bacteria that are not able to nodulate or fix nitrogen with legumes [26, 27]. Minimum Information about the Genome Sequence (MIGS) [28] of AMP6 is provided in Table 1.
Fig. 2.
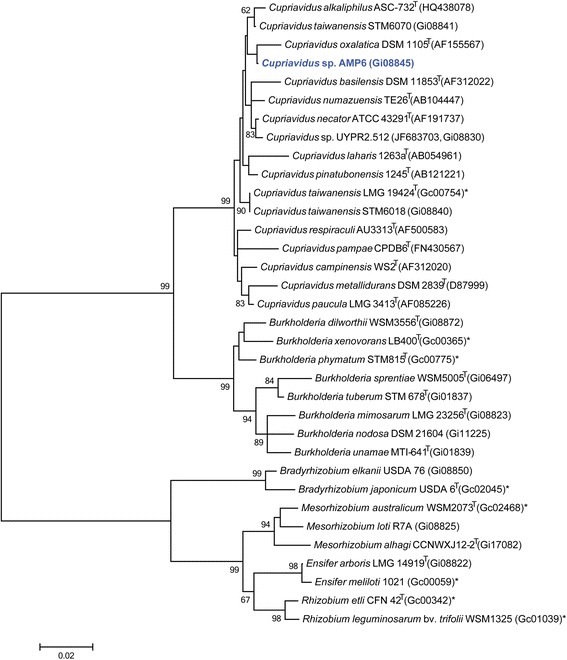
Phylogenetic tree highlighting the position of Cupriavidus sp. strain AMP6 (shown in blue print) relative to other type and non-type strains in the Cupriavidus genus using a 1,024 bp internal region of the 16S rRNA gene. Several Alpha-rhizobia sequences were used as an outgroup. All sites were informative and there were no gap-containing sites. Phylogenetic analyses were performed using MEGA, version 5.05 [46]. The tree was build using the maximum likelihood method with the General Time Reversible model. Bootstrap analysis with 500 replicates was performed to assess the support of the clusters. Type strains are indicated with a superscript T. Strains with a genome sequencing project registered in GOLD [30] have the GOLD ID mentioned after the strain number, otherwise the NCBI accession number is provided. Finished genomes are designated with an asterisk
Table 1.
Classification and general features of Cupriavidus sp. strain AMP6 in accordance with the MIGS recommendations [28] published by the Genome Standards Consortium [47]
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [48] | |
| Phylum Proteobacteria | TAS [49, 50] | ||
| Class Betaproteobacteria | TAS [51] | ||
| Order Burkholderiales | TAS [52] | ||
| Family Burkholderiaceae | TAS [53] | ||
| Genus Cupriavidus | TAS [54] | ||
| Species Cupriavidus sp. | TAS [12] | ||
| (Type) strain AMP6 | TAS [12] | ||
| Gram stain | Negative | TAS [54] | |
| Cell shape | Rod | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Non-sporulating | TAS [54] | |
| Temperature range | Mesophile | TAS [54] | |
| Optimum temperature | 28 °C | IDA | |
| pH range; Optimum | Not reported | ||
| Carbon source | Not reported | ||
| MIGS-6 | Habitat | Soil, root nodule on host | IDA |
| MIGS-6.3 | Salinity | Not reported | |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Symbiotic | IDA |
| MIGS-14 | Pathogenicity | Non-pathogenic | NAS |
| MIGS-4 | Geographic location | Texas, USA | TAS [12] |
| MIGS-5 | Nodule collection date | 2005 | TAS [12] |
| MIGS-4.1 | Longitude | −98.138 | TAS [12] |
| MIGS-4.2 | Latitude | 26.0794 | TAS [12] |
| MIGS-4.4 | Altitude | 30 m | IDA |
Evidence codes – IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [55]
Symbiotaxonomy
Cupriavidus sp. strain AMP6 was isolated from Mimosa asperata nodules collected at the Santa Ana National Wildlife Refuge in Hidalgo County, Texas [12]. Cupriavidus sp. strain AMP6 was assessed for nodulation and nitrogen fixation on five mimosa species, including M. pigra, M. pudica, M. invisia, M. strigillosa and M. quadrivalvis [12]. Strain AMP6 could nodulate all hosts apart from M. quadrivalvis [12]. Additional acetylene reduction assays provided information on the nitrogenase activity of strain AMP6 on those hosts. These test showed substantial nitrogenase activity with M. pudica and M. invisia but only a small amount with M. pigra [12]. The absence of nodule nitrogenase activity was also observed for M. strigillosa and M. quadrivalvis [12].
Genome sequencing information
Genome project history
This organism was selected for sequencing on the basis of its environmental and agricultural relevance to issues in global carbon cycling, alternative energy production, and biogeochemical importance, and is part of the Genomic Encyclopedia of Bacteria and Archaea, The Root Nodulating Bacteria chapter project at the U.S. Department of Energy, Joint Genome Institute [29]. The genome project is deposited in the Genomes OnLine Database [30] and the high-quality permanent draft genome sequence in IMG [31]. Sequencing, finishing and annotation were performed by the JGI using state of the art sequencing technology [32]. A summary of the project information is shown in Table 2.
Table 2.
Genome sequencing project information for Cupriavidus sp. strain AMP6
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality permanent draft |
| MIGS-28 | Libraries used | Illumina Std PE |
| MIGS-29 | Sequencing platforms | Illumina HiSeq 2000 |
| MIGS-31.2 | Fold coverage | 117.0x Illumina |
| MIGS-30 | Assemblers | Velvet 1.1.04, ALLPATHS V.r42328 |
| MIGS-32 | Gene calling methods | Prodigal 1.4 |
| Locus Tag | K309 | |
| Genbank ID | AUFE00000000 | |
| Genbank Date of Release | December 12, 2013 | |
| GOLD ID | Gp0009812 | |
| BIOPROJECT | PRJNA195776 | |
| MIGS-13 | Source Material Identifier | AMP6 |
| Project relevance | Symbiotic N2fixation, agriculture |
Growth conditions and genomic DNA preparation
Cupriavidus sp. strain AMP6 was grown on YMA solid medium [23] for 3 days, a single colony was selected and used to inoculate 5 ml TY broth medium. The culture was grown for 48 h on a gyratory shaker (200 rpm) at 28 °C. Subsequently 1 ml was used to inoculate 60 ml TY broth medium and grown on a gyratory shaker (200 rpm) at 28 °C until OD 0.6 was reached. DNA was isolated from 60 mL of cells using a CTAB bacterial genomic DNA isolation method [33]. Final concentration of the DNA was 0.6 mg/ml.
Genome sequencing and assembly
The genome of Cupriavidus sp. AMP6 was generated at the DOE Joint genome Institute [32]. An Illumina Std shotgun library was constructed and sequenced using the Illumina HiSeq 2000 platform which generated 15,823,344 reads totaling 2,373.5 Mbp. All general aspects of library construction and sequencing performed at the JGI can be found at the JGI web site [34]. All raw Illumina sequence data was passed through DUK, a filtering program developed at JGI, which removes known Illumina sequencing and library preparation artifacts (Mingkun L, Copeland A, Han J. unpublished). Following steps were then performed for assembly: (1) filtered Illumina reads were assembled using Velvet (version 1.1.04) [35] (2) 1–3 Kbp simulated paired end reads were created from Velvet contigs using wgsim [36] (3) Illumina reads were assembled with simulated read pairs using Allpaths–LG (version r42328) [37]. Parameters for assembly steps were: 1) Velvet (velveth: 63 –shortPaired and velvetg: −very clean yes –exportFiltered yes –min contig lgth 500 –scaffolding no–cov cutoff 10) 2) wgsim (−e 0 –1 100 –2 100 –r 0 –R 0 –X 0) 3) Allpaths–LG (PrepareAllpathsInputs: PHRED 64 = 1 PLOIDY = 1 FRAG COVERAGE = 125 JUMP COVERAGE = 25 LONG JUMP COV = 50, RunAllpathsLG: THREADS = 8 RUN = std shredpairs TARGETS = standard VAPI WARN ONLY = True OVERWRITE = True). The final draft assembly contained 262 contigs in 260 scaffolds. The total size of the genome is 7.6 Mbp and the final assembly is based on 886.3 Mbp of Illumina data, which provides an average of 117.0× coverage of the genome.
Genome annotation
Genes were identified using Prodigal [38], as part of the DOE-JGI genome annotation pipeline [39, 40] followed by a round of manual curation using GenePRIMP [41] for finished genomes and Draft genomes in fewer than 10 scaffolds. The predicted CDSs were translated and used to search the NCBI non-redundant database, UniProt, TIGRFam, Pfam, KEGG, COG, and InterPro databases. The tRNAScanSE tool [42] was used to find tRNA genes, whereas ribosomal RNA genes were found by searches against models of the ribosomal RNA genes built from SILVA [43]. Other non–coding RNAs such as the RNA components of the protein secretion complex and the RNase P were identified by searching the genome for the corresponding Rfam profiles using INFERNAL [44]. Additional gene prediction analysis and manual functional annotation was performed within the Integrated Microbial Genomes-Expert Review (IMG-ER) system [45] developed by the Joint Genome Institute, Walnut Creek, CA, USA.
Genome properties
The genome is 7,579,563 nucleotides with 65.46 % GC content (Table 3) and comprised of 260 scaffolds and 262 contigs. From a total of 7,130 genes, 7,033 were protein encoding and 97 RNA only encoding genes. The majority of genes (80.24 %) were assigned a putative function whilst the remaining genes were annotated as hypothetical. The distribution of genes into COG functional categories is presented in Table 4.
Table 3.
Genome statistics for Cupriavidus sp. AMP6
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 7,579,563 | 100.00 |
| DNA coding (bp) | 6,545,489 | 86.36 |
| DNA G + C (bp) | 4,961,426 | 65.46 |
| DNA scaffolds | 260 | 100.00 |
| Total genes | 7,130 | 100.00 |
| Protein-coding genes | 7,033 | 98.64 |
| RNA genes | 97 | 1.36 |
| Pseudo genes | 0 | 0.00 |
| Genes in internal clusters | 538 | 7.55 |
| Genes with function prediction | 5,721 | 80.24 |
| Genes assigned to COGs | 4,791 | 67.19 |
| Genes with Pfam domains | 5,837 | 81.87 |
| Genes with signal peptides | 681 | 9.55 |
| Genes with transmembrane helices | 1,477 | 20.72 |
| CRISPR repeats | 1 |
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | % age | COG Category |
|---|---|---|---|
| J | 182 | 3.37 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.02 | RNA processing and modification |
| K | 527 | 9.76 | Transcription |
| L | 188 | 3.48 | Replication, recombination and repair |
| B | 4 | 0.07 | Chromatin structure and dynamics |
| D | 32 | 0.59 | Cell cycle control, Cell division, chromosome partitioning |
| V | 59 | 1.09 | Defense mechanisms |
| T | 210 | 3.89 | Signal transduction mechanisms |
| M | 275 | 5.09 | Cell wall/membrane/envelope biogenesis |
| N | 96 | 1.78 | Cell motility |
| U | 119 | 2.20 | Intracellular trafficking, secretion, and vesicular transport |
| O | 164 | 3.04 | Posttranslational modification, protein turnover, chaperones |
| C | 447 | 8.28 | Energy production and conversion |
| G | 256 | 4.74 | Carbohydrate transport and metabolism |
| E | 501 | 9.28 | Amino acid transport and metabolism |
| F | 90 | 1.67 | Nucleotide transport and metabolism |
| H | 185 | 3.43 | Coenzyme transport and metabolism |
| I | 344 | 6.37 | Lipid transport and metabolism |
| P | 272 | 5.04 | Inorganic ion transport and metabolism |
| Q | 235 | 4.35 | Secondary metabolite biosynthesis, transport and catabolism |
| R | 659 | 12.21 | General function prediction only |
| S | 552 | 10.23 | Function unknown |
| - | 2339 | 32.81 | Not in COGS |
The total is based on the total number of protein coding genes in the genome
Conclusion
Cupriavidus sp. AMP6 belongs to a group of Beta-rhizobia isolated from Mimosa asperata. Phylogenetic analysis revealed that AMP6 is most closely related to Cupriavidus taiwanensisLMG 19424T, which was isolated from Mimosa pudica, and is able to nodulate and fix nitrogen in association with several Mimosa species [13]. In total five Cupriavidus strains (AMP6, LMG 19424T, STM6018, STM6070 and UYPR2.512), which can form a symbiotic association have now been sequenced. A comparison of these strains reveals that AMP6 has the second largest genome (7.6 Mbp), with the highest KOG count (1398) and the second lowest GC (65.46 %) and signal peptide (9.55 %) percentages in this group. All of these genomes share the nitrogenase-RXN MetaCyc pathway characterized by the multiprotein nitrogenase complex. Out of five Cupriavidus strains (AMP6, LMG 19424T, STM6018, STM6070 and UYPR2.512), which contain the N-fixation pathway, only Cupriavidus sp. AMP6 has been shown to fix effectively with Mimosa asperata. The genome attributes of Cupriavidus sp. AMP6, in conjunction with other Cupriavidus genomes, will be important for ongoing molecular analysis of the plant microbe interactions required for the establishment of Mimosa symbioses.
Acknowledgements
This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396.
Abbreviations
- GEBA-RNB
Genomic Encyclopedia of Bacteria and Archaea – Root Nodule Bacteria
- JGI
Joint Genome Institute
- TY
Trypton Yeast
- CTAB
Cetyl trimethyl ammonium bromide
- WSM
Western Australian Soil Microbiology
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
MP supplied the strain and background information for this project, PVB supplied DNA to JGI, TR performed all imaging, SDM and WR drafted the paper, JH provided financial support and all other authors were involved in sequencing the genome and editing the final manuscript. All authors read and approved the final manuscript.
References
- 1.Gyaneshwar P, Hirsch AM, Moulin L, Chen WM, Elliott GN, Bontemps C, et al. Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol Plant Microbe Interact. 2011;24:1276–88. [DOI] [PubMed]
- 2.Barrett CF, Parker MA. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp nodule bacteria on two Mimosa spp. in Costa Rica. Appl Environ Microbiol. 2006;72:1198–1206. doi: 10.1128/AEM.72.2.1198-1206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garau G, Yates RJ, Deiana P, Howieson JG. Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol Biochem. 2009;41:125–134. doi: 10.1016/j.soilbio.2008.10.011. [DOI] [Google Scholar]
- 4.Howieson JG, De Meyer SE, Vivas-Marfisi A, Ratnayake S, Ardley JK, Yates RJ. Novel Burkholderia bacteria isolated from Lebeckia ambigua - A perennial suffrutescent legume of the fynbos. Soil Biol Biochem. 2013;60:55–64. doi: 10.1016/j.soilbio.2013.01.009. [DOI] [Google Scholar]
- 5.Bontemps C, Elliott GN, Simon MF, Dos Reis FBD, Gross E, Lawton RC, et al. Burkholderia species are ancient symbionts of legumes. Mol Ecol. 2010;19:44–52. [DOI] [PubMed]
- 6.dos Reis Jr FB, Simon MF, Gross E, Boddey RM, Elliott GN, Neto NE, et al. Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol. 2010;186:934–46. [DOI] [PubMed]
- 7.Beukes CW, Venter SN, Law IJ, Phalane FL, Steenkamp ET. South african papilionoid legumes are nodulated by diverse Burkholderia with unique nodulation and nitrogen-fixation Loci. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0068406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker R, Watkin E, Tian R, Brau L, O'Hara G, Goodwin L, et al. Genome sequence of the acid-tolerant Burkholderia sp. strain WSM2230 from Karijini National Park, Australia. Stand Genomic Sci. 2014;9:551–61. [DOI] [PMC free article] [PubMed]
- 9.Mishra RP, Tisseyre P, Melkonian R, Chaintreuil C, Miche L, Klonowska A, et al. Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol Ecol. 2012;79:487–503. [DOI] [PubMed]
- 10.Andrus AD, Andam C, Parker MA. American origin of Cupriavidus bacteria associated with invasive Mimosa legumes in the Philippines. FEMS Microbiol Ecol. 2012;80:747–750. doi: 10.1111/j.1574-6941.2012.01342.x. [DOI] [PubMed] [Google Scholar]
- 11.Taule C, Zabaleta M, Mareque C, Platero R, Sanjurjo L, Sicardi M, et al. New betaproteobacterial rhizobium strains able to efficiently nodulate Parapiptadenia rigida (Benth.) Brenan. Appl Environ Microbiol. 2012;78:1692–700. [DOI] [PMC free article] [PubMed]
- 12.Andam CP, Mondo SJ, Parker MA. Monophyly of nodA and nifH genes across Texan and Costa Rican populations of Cupriavidus nodule symbionts nodule symbionts. Appl Environ Microbiol. 2007;73:4686–4690. doi: 10.1128/AEM.00160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WM, Moulin L, Bontemps C, Vandamme P, Bena G, Boivin-Masson C. Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J Bacteriol. 2003;185:7266–7272. doi: 10.1128/JB.185.24.7266-7272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verma SC, Chowdhury SP, Tripathi AK. Phylogeny based on 16S rDNA and nifH sequences of Ralstonia taiwanensis strains isolated from nitrogen-fixing nodules of Mimosa pudica, in India. Can J Microbiol. 2004;50:313–322. doi: 10.1139/w04-020. [DOI] [PubMed] [Google Scholar]
- 15.Parker MA, Wurtz AK, Paynter Q. Nodule symbiosis of invasive Mimosa pigra in Australia and in ancestral habitats: A comparative analysis. Biol Invasions. 2007;9:127–138. doi: 10.1007/s10530-006-0009-2. [DOI] [Google Scholar]
- 16.Elliott GN, Chou J-H, Chen W-M, Bloemberg GV, Bontemps C, Martínez-Romero E, et al. Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ Microbiol. 2009;11:762–78. [DOI] [PubMed]
- 17.Liu XY, Wei S, Wang F, James EK, Guo XY, Zagar C, et al. Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol Ecol. 2012;80:417–26. [DOI] [PubMed]
- 18.Florentino LA, Duque Jaramillo PM, Silva KB, da Silva JS, de Oliveira SM, de Souza Moreira FM. Physiological and symbiotic diversity of Cupriavidus necator strains isolated from nodules of Leguminosae species. Scientia Agricola. 2012;69:247–258. doi: 10.1590/S0103-90162012000400003. [DOI] [Google Scholar]
- 19.Barneby RC. Sensitive Censitae: a description of the genus Mimosa Linnaeus (Mimosaceae) in the New World. New York: The New York Botanical Garden; 1991. [Google Scholar]
- 20.Vora RS. Plant communities of the Santa Ana National Wildlife Refuge, Texas. Tex J Sci. 1990;42:115–128. [Google Scholar]
- 21.Jacobs JL. Soil Survey of Hidalgo County, Texas. Washington DC: United States Soil Conservation Service; 1981. [Google Scholar]
- 22.Amadou C, Pascal G, Mangenot S, Glew M, Bontemps C, Capela D, et al. Genome sequence of the beta-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res. 2008;18:1472–1483. doi: 10.1101/gr.076448.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincent JM. International Biological Programme Handbook. 15. Oxford, UK: Blackwell Scientific Pubilications; 1970. A manual for the practical study of the root-nodule bacteria. [Google Scholar]
- 24.Kim O-S, Cho Y-J, Lee K, Yoon S-H, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–21. [DOI] [PubMed]
- 25.Chen WM, Laevens S, Lee TM, Coenye T, De Vos P, Mergeay M, et al. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol. 2001;51:1729–35. [DOI] [PubMed]
- 26.Makkar NS, Casida LE. Cupriavidus necator gen. nov., sp. nov. - a nonobligate bacterial predator of bacteria in soil. Int J Syst Evol Microbiol. 1987;37:323–326. [Google Scholar]
- 27.Estrada-de los Santos P, Martínez-Aguilar L, López-Lara IM, Caballero-Mellado J. Cupriavidus alkaliphilus sp. nov., a new species associated with agricultural plants that grow in alkaline soils. Syst Appl Microbiol. 2012;35:310–314. doi: 10.1016/j.syapm.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. Towards a richer description of our complete collection of genomes and metagenomes "Minimum Information about a Genome Sequence " (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeve W, Ardley J, Tian R, Eshragi L, Yoon J, Ngamwisetkun P, et al. A genomic encyclopedia of the root nodule bacteria: Assessing genetic diversity through a systematic biogeographic survey. Stand Genomic Sci. 2015;10:14. [DOI] [PMC free article] [PubMed]
- 30.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, et al. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–579. [DOI] [PMC free article] [PubMed]
- 31.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–7. [DOI] [PMC free article] [PubMed]
- 32.Mavromatis K, Land ML, Brettin TS, Quest DJ, Copeland A, Clum A, et al. The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS ONE. 2012;7: e48837. [DOI] [PMC free article] [PubMed]
- 33.CTAB DNA extraction protocol [http://jgi.doe.gov/collaborate-with-jgi/pmo-overview/protocols-sample-preparation-information/]
- 34.JGI Website [http://www.jgi.doe.gov]
- 35.Zerbino D, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.wgsim [https://github.com/lh3/wgsim]
- 37.Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 2011;108:1513–8. [DOI] [PMC free article] [PubMed]
- 38.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mavromatis K, Ivanova NN, Chen IM, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard Operating Procedure for the annotations of microbial genomes. Stand Genomic Sci. 2009;1:63–67. doi: 10.4056/sigs.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen IM, Markowitz VM, Chu K, Anderson I, Mavromatis K, Kyrpides NC, Ivanova NN. Improving microbialgenome annotations in an integrated database context. PLoS ONE. 2013;8:e54859 [DOI] [PMC free article] [PubMed]
- 41.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, et al. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods. 2010;7:455–7. [DOI] [PubMed]
- 42.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. [DOI] [PMC free article] [PubMed]
- 44.INFERNAL. Inference of RNA alignments [http://infernal.janelia.org]
- 45.Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Field D, Amaral-Zettler L, Cochrane G, Cole JR, Dawyndt P, Garrity GM, et al. The Genomic Standards Consortium. PLoS Biol. 2011;9, e1001088. [DOI] [PMC free article] [PubMed]
- 48.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proceedings of the National Academy of Sciences USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen WX, Wang ET, Kuykendall LD. The Proteobacteria. New York: Springer - Verlag; 2005. [Google Scholar]
- 50.Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int J Syst Evol Microbiol. 2005; 55:2235–38. [DOI] [PubMed]
- 51.Garrity GM, Bell JA, Lilburn TE. Class II. Betaproteobacteria. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. 2. New York: Springer - Verlag; 2005. [Google Scholar]
- 52.Garrity GM, Bell JA, Lilburn TE. Order 1. Burkholderiales. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology, vol. 2. 2nd ed. New York: Springer - Verlag; 2005.
- 53.Garrity GM, Bell JA, Lilburn TE. Family I. Burkholderiaceae. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. 2. New York: Springer - Verlag; 2005. [Google Scholar]
- 54.Balkwill DL. Genus I. Cupriavidus. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. 2. New York: Springer - Verlag; 2005. [Google Scholar]
- 55.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. [DOI] [PMC free article] [PubMed]


