Abstract
Background
Schizochytrium limacinum SR21 is a potential industrial strain for docosahexaenoic acid (DHA) production that contains more than 30–40 % DHA among its total fatty acids.
Methods
To resolve the DHA biosynthesis mechanism and improve DHA production at a systematic level, a genomescale metabolic model (GSMM), named iCY1170_DHA, which contains 1769 reactions, 1659 metabolites, and 1170 genes, was reconstructed.
Results
Based on genome annotation results and literature reports, a new DHA synthesis pathway based on a polyketide synthase (PKS) system was detected in S. limacinum. Similarly to conventional fatty acid synthesis, the biosynthesis of DHA via PKS requires abundant acetyl-CoA and NADPH. The in silico addition of malate and citrate led to increases of 24.5 % and 37.1 % in DHA production, respectively. Moreover, based on the results predicted by the model, six amino acids were shown to improve DHA production by experiment. Finally, 30 genes were identified as potential targets for DHA over-production using a Minimization of Metabolic Adjustment algorithm.
Conclusions
The reconstructed GSMM, iCY1170_DHA, could be used to elucidate the mechanism by which DHA is synthesized in S. limacinum and predict the requirements of abundant acetyl-CoA and NADPH for DHA production as well as the enhanced yields achieved via supplementation with six amino acids, malate, and citrate.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2042-y) contains supplementary material, which is available to authorized users.
Keywords: Schizochytrium limacinum SR21, Docosahexaenoic acid, Genome-scale metabolic model, Polyketide synthase system, Minimization of metabolic adjustment algorithm
Background
Docosahexaenoic acid (DHA), which is an n-3 polyunsaturated fatty acid (PUFA), has been shown to have a positive effect on diseases such as hypertension, arthritis, atherosclerosis, depression, adult-onset diabetes mellitus, myocardial infarction, thrombosis, and some cancers [1]. DHA is necessary for the development of the brain and retina of infants, and it is important in maintaining brain function in adults [2]. Oceanic fish and fish oil products are typical dietary sources of DHA [3]. However, because of emerging concerns over the sustainability of marine resources and the levels of environmental contaminants present in fish, major efforts have been made to identify or create alternative sources of DHA [4].
Schizochytrium limacinum SR21 (Aurantiochytrium limacinum ATCC MYA-1381) is a marine thraustochytrid that can synthesize lipids with a high content of DHA. In SR21, total fatty acids reportedly constitute more than 50 % of the dry cell weight (DCW) [5], and about 30–40 % of the fatty acids of this strain are DHA [6]. In addition, a number of studies have confirmed the safety of DHA-rich oil extracted from Schizochytrium sp. in recent years [7, 8]. For these reasons, DHA from microorganisms has been widely incorporated into infant formulas and health products for elderly persons [9]. In addition, industrial utilization of this organism as a commercial source of DHA is currently receiving much attention [10, 11].
The dry cell weight, lipid content, and DHA percentage of the total fatty acids are three important parameters for evaluating fermentations. Many efforts have been directed towards improving the DHA yield. (1) To optimize the culture medium, it was demonstrated that using glucose and glycerol as mixed carbon sources, the DHA productivity was 15.24 % higher than that obtained using glucose as single carbon source [12]. (2) To improve the fermentation process, a NH4-pH-auxostat system was developed that appears to be a promising technique for the first stage of production of Schizochytrium sp. biomass as a means of achieving the fastest possible growth rate [13]. A two-stage oxygen supply control strategy was applied to the DHA fermentation. With this protocol, the production of biomass and DHA improved to 37.9 g/L and 6.56 g/L, which increased 18.1 % and 9.88 %, respectively [10]. (3) To enhance metabolic regulation, a strategy was proposed that reinforces acetyl-CoA and NADPH supply. By adding 4 g/L malic acid, the DHA content among the total fatty acids increased from 35 % to 60 %. The total lipid content also showed an apparent increase of 35 % and reached 19 g/L when 40 mL ethanol/L were added [14]. Although many strategies have been applied to improve DHA production, some problems still exist during the fermentation process. For example, the growth of SR21 is unstable and cell viability is low. The mechanism of DHA synthesis is still unclear, and cell metabolism is difficult to regulate by genetic manipulation.
A genome-scale metabolic model (GSMM) represents the microbial metabolic genotype–phenotype relationships of an organism [15]. Such models have been widely used in many contexts, such as contextualizing high-throughput data, understanding complex biological phenomena, guiding metabolic engineering, directing hypothesis-driven discovery, interrogating multi-species relationships, and discovering network properties [16–19]. The release of the whole genome sequence of S. limacinum SR21 and corresponding literature reports have made the reconstruction of a GSMM possible.
In this study, a GSMM of S. limacinum SR21 was reconstructed. Using this model, we first made a comparison with two oleaginous fungi, Mortierella alpina and Yarrowia lipolytica, that are used for industrial production of arachidonic acid (ARA) and eicosapentaenoic acid (EPA), respectively. Then the pathway of DHA biosynthesis in SR21 was resolved based on genome annotation results and literature mining. Based on the GSMM, biochemical and genetic strategies were applied to improve DHA production.
Results and discussion
Genome-scale reconstruction and general features of model iCY1170_DHA
The reconstructed GSMM of Schizochytrium limacinum SR21, which contains 1769 reactions and 1659 metabolites, was named iCY1170_DHA (Additional file 1). Model iCY1170_DHA consists of 1386 intracellular metabolic reactions and 195 transport reactions. 7.9 % of the total open reading frames (ORFs), corresponding to 1170 genes of 14,859 ORFs, were incorporated into the model. In iCY1170_DHA, all 1769 reactions (including transport and exchange reactions) were classified into 10 different subsystems, according to the KEGG Pathway Database (http://www.genome.jp/kegg/pathway.html): carbohydrate metabolism, amino acid metabolism (20 common acid acids), other amino acids (D-type amino acids) metabolism, nucleotide metabolism, energy metabolism, lipid metabolism, metabolism of cofactors and vitamins, transport reactions, exchange reactions, and other metabolisms (Fig. 1a). Among them, lipid metabolism ranks as the largest subsystem in iCY1170_DHA (20.4 %), followed by amino acid metabolism (18.0 %), which agrees with model iCY1106 for Mortierella alpina (Fig. 1b). The sum of the three largest subsystems, lipid metabolism, amino acid metabolism, and carbohydrate metabolism, accounts for over half of the total number of reactions (51.4 %). The difference between the two models was that, in model iCY1170_DHA, the third largest subsystem is carbohydrate metabolism, while in model iCY1106, transport reactions rank third, which means that M. alpina has more transport mechanisms than S. limacinum.
Fig. 1.
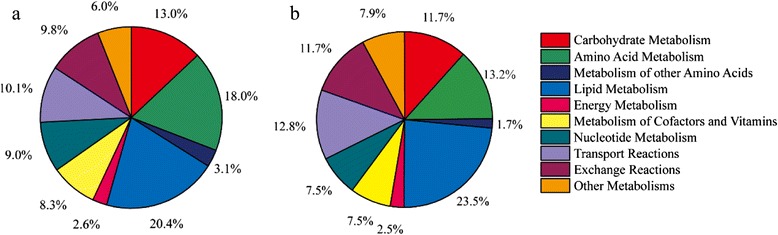
Metabolic subsystems distribution for in silico models. a, S limacinum model iCY1170_DHA, b, M. alpina model iCY1106. ‘Other metabolisms’ subsystem contains Glycan Biosynthesis and Metabolism, Metabolism of Terpenoids and Polyketides, Biosynthesis of other secondary metabolites, and Xenobiotics Biodegradation and Metabolism
The essentialities of individual genes of S. limacinum were analyzed under minimal glucose (MG) and yeast extract (YE) medium conditions using iCY1170_DHA by deleting each gene in turn. The genes were categorized into three classes: essential genes, partially essential genes and non-essential genes. The gene essentiality study revealed that a total of 56 genes were essential in both YE and MG medium. An additional 35 genes were essential only in MG medium (Additional file 2). When comparing the distribution of essential genes in different culture media, for amino acid metabolism most genes were only essential in MG medium (increased from 2.6 % to 15.5 %, Fig. 2a). Since YE medium is supplemented with all of the amino acids, some of the amino acids were directly consumed from the medium without utilizing their biosynthetic pathways (Fig. 2b). For example, Aurli1_48454 (argininosuccinate synthase, EC: 6.3.4.5) and Aurli1_69076 (argininosuccinate lyase, EC: 4.3.2.1), which convert aspartate into arginine, were essential only when grown in MG medium. Interestingly, the ‘other metabolism’ subsystem contained the same proportion of essential genes (11.9 % of total reactions in the other metabolism subsystem) in both media. That means that, in this subsystem, the pathway of squalene biosynthesis does not have many alternative routes and is quite rigid in S. limacinum.
Fig. 2.

The distribution of essential genes in different media. a the minimal glucose medium (MG); b the yeast extract medium (YE)
Similarly to model iCY1106, model iCY1170_DHA had more reactions, metabolites, and genes than model iYL619_PCP [20] for Yarrowia lipolytica (Table 1). However, the ORF coverage of iCY1170_DHA was a little lower than the other two models. When ignoring compartment information, not including transport and exchange reactions, models iCY1170_DHA, iCY1106, and iYL619_PCP contain 1195, 1124, and 748 reactions, respectively. And 457 reactions were present in all these three models (Additional file 3, Fig. 3). Models iCY1170_DHA and iCY1106 both shared 803 reactions, which account for 67.2 % and 71.4 % of total reactions, respectively. This means these two models are similar in biochemical reactions to some degree. However, 343 reactions (28.7 % of total reactions) were unique in model iCY1170_DHA, and 61.3 % of these unique reactions were distributed across lipid metabolism, amino acid metabolism, and carbohydrate metabolism. For example, it was reported that S. limacinum could grow with arabinose as the carbon source [21, 22], whereas Y. lipolytica and M. alpina could not use arabinose [20]. Reactions that can convert arabinose into glucose (involving 12 reactions) are thus unique in model iCY1107_DHA (Additional file 1). In addition, there were two pathways for synthesizing lysine, starting from aspartate or 2-oxoglutarate according to the genome annotation results, whereas in models iYL619_PCP and iCY1106 only the second pathway for lysine biosynthesis is present. Reactions involving the first pathway must also be unique in model iCY1170_DHA.
Table 1.
Features of the in silico genome-scale metabolic model of S. limacinum, M. alpina and Y. lipolytica
| Features | iCY1170_DHA | iCY1106 | iYL619_PCP |
|---|---|---|---|
| Genome feature | |||
| Genome size (Mb) | 60.93 | 38.38 | 20.50 |
| Open reading frames (ORFs) | 14,859 | 11,631 | 6453 |
| In silico metabolic model | |||
| Reactions included in the model | 1769 | 1854 | 1142 |
| Biochemical reactions | 1386 | 1391 | 781 |
| Transport reactions | 195 | 247 | 236 |
| Exchange reactions | 188 | 216 | 125 |
| Metabolites | 1659 | 1732 | 843 |
| ORFs assigned in metabolic model | 1170 | 1106 | 619 |
| ORF coveragea (%) | 7.9 | 9.5 | 9.6 |
aThe number of ORFs in the iCY1170_DHA model divided by the total number of ORFs in the genome
Fig. 3.
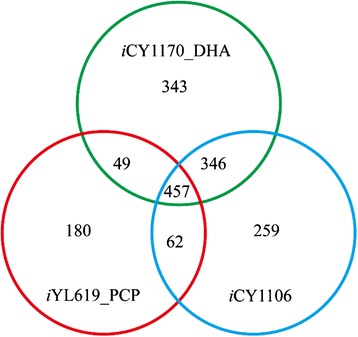
Comparison among three existing models of different oleaginous fungi, S. limacinum, M. alpina, and Y. lipolytica
Verification and simulation of model iCY1170_DHA
Data from batch cultures of S. limacinum grown in glucose and glycerol media were used to validate iCY1170_DHA. Both of the in silico media contain the basic elements, such as C, N, H, O, P, S (Table 2). When using glucose or glycerol as carbon source, their maximum uptake rates were 1.4 mmol/gDW/h and 1.6 mmol/gDW/h, respectively (Additional file 1) [23]. To simulate cellular growth in the different media, the biomass equation was maximized in the flux analysis simulations. Notably, simulation results were consistent with observed growth rates (Table 3). Without the constraints of production, for the glucose medium, the in silico simulation predicted cell growth of 0.0812/h, which was very close to the experimentally observed specific growth rate of 0.0887/h (8.5 %) [24]. And for the glycerol medium, the simulated result was only 1.1 % lower than the experimental result [25]. However, when the DHA synthesis rate was constrained at 0.03 mmol/gDW/h [24], the in silico result was 16.42 % lower than experimental result [24]. This means the synthesis of DHA may require other nutrients, apart from amino acids. And the replacement of yeast extract by inorganic nitrogen could lead to these nutritional deficiencies.
Table 2.
The in silico glucose minimal media composition of the model iCY1170_DHA
| Reaction description | Equation | LBa1 | UBb2 |
|---|---|---|---|
| Exchange of D-glucose | D-glucose[e] < => | −1.4 | 1000 |
| Exchange of Water | H2O[e] < => | −1000 | 1000 |
| Exchange of Oxygen | O2[e] < => | −1000 | 1000 |
| Exchange of Ammonia | NH3[e] < => | −10 | 1000 |
| Exchange of Orthophosphate | Pi[e] < => | −1000 | 1000 |
| Exchange of Sulfate | SO4[e] < => | −1000 | 1000 |
| Exchange of H+ | H[e] < => | −1000 | 1000 |
a1, LB, lower bound, whose unit is mmol/gDW/h. b2, UB, upper bound, whose unit is also mmol/gDW/h
Table 3.
Comparison of in silico and in vivo growth rates of S. limacinum
| Media condition | Growth rate (/h) | |
|---|---|---|
| (mmol/gDW/h) | In vivo | In silico |
| glucose (v = 1.4) | 0.0887 | 0.0812 |
| glucose (v = 1.4, DHA production = 0.03) | 0.0883 | 0.0738 |
| glycerol (v = 1.6) | 0.0620 | 0.0613 |
In vivo: experimental results, In silico: simulation results. In vivo growth rate was calculated from the growth curve of S. limacinum. In silico growth rate was the solution of S (m × n) matrix, when the biomass function was used as objective function
Apart from the growth rate simulation, the ability of iCY1170_DHA to utilize different carbon and nitrogen sources has been verified. According to literature reports, S. limacinum can grow using 15 kinds of carbon sources and 3 kinds of inorganic nitrogen sources, such as NH4+, NO3−, and urea. However, the simulation results show that the model could not grow using 5 carbon sources, including xylose, arabinose, lactose, starch, and trehalose (Table 4). It also could not grow on a medium using NO3− as the nitrogen source. After a series of gap filling and model debugging steps, ten reactions were added to the model. For example, lactose galactohydrolase (EC: 3.2.1.23), which can convert lactose into glucose, was not found during genome annotation. After filling this gap, the model could grow with lactose as the carbon source. All of these simulation results, including the growth rate simulations and usage of different carbon and nitrogen sources, indicated that the model iCY1170_DHA is reliable and can be used for further prediction and analysis.
Table 4.
Comparison between experimental results and simulated results about the usage of different carbon and nitrogen sources
| Carbon source | In vivo | In silico a | In silico b | Reference |
|---|---|---|---|---|
| Glucose | + | + | + | [6, 21, 22] |
| Fructose | + | + | + | [6, 21, 22] |
| Mannose | + | + | + | [21, 22] |
| Galactose | + | + | + | [21, 22] |
| Xylose | + | - | + | [21, 22] |
| Arabinose | + | - | + | [21, 22] |
| Ribose | + | + | + | [21, 22] |
| Lactose | + | - | + | [6, 21, 22] |
| Sucrose | + | + | + | [21, 22] |
| Maltose | + | + | + | [6, 21, 22] |
| Starch | + | - | + | [6, 21, 22] |
| Glycerol | + | + | + | [6, 21, 22] |
| Melibiose | + | + | + | [21] |
| Raffinose | + | + | + | [21] |
| Trehalose | + | - | + | [21] |
| Nitrogen Source | ||||
| NH4 + | + | + | + | [6] |
| NO3 − | + | - | + | [6] |
| Urea | + | + | + | [6, 21] |
aBefore filling gaps; bAfter filling gaps. To make the GSMM consistent with experimental data, some missing reactions were added to model
Resolving the DHA synthesis pathway based on model iCY1170_DHA
There are two pathways that can synthesize DHA, the conventional fatty acid synthesis (FAS) route and the polyketide synthase (PKS) system [26]. In the FAS route, fatty acids are biosynthesized in the form of either C16:0 or C18:0 saturated fatty acids. These fatty acids are then modified through a sequence of desaturations and elongations so that extended ranges of unsaturated fatty acids and PUFAs are produced [27]. In this route, DHA is synthesized by delta-4 desaturase, which can transform C22:5 to C22:6 (DHA). In the PKS pathway, acyl carrier protein (ACP), generated by CoA, is used as a covalent attachment point for chain synthesis, which proceeds with reiterative cycles. During the full fatty acid synthesis process, a series of enzymes including 3-ketoacyl synthase (KS), 3-ketoacyl-ACP reductase (KR), enoyl reducatase (ER), and dehydrase/isomerase (DH) are necessary (Fig. 4). However, the whole genome annotation results showed that S. limacinum does not contain delta-4 desaturase, in agreement with literature reports. On the other hand, some ORFs in S. limacinum were predicted to be potential PKS proteins (Additional file 4). This means S. limacinum could not synthesize DHA by the FAS route, but rather employs the PKS system for PUFA biosynthesis [26, 28, 29].
Fig. 4.
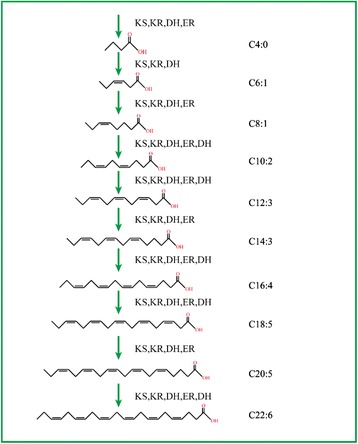
A scheme to account for the formation of DHA by the PKS route of synthesis in S. limacinum. KS, 3-ketoacyl synthase; KR, 3-ketoacyl-ACP reductase; ER, enoyl reducatase; DH, dehydrase/isomerase
When compared with M. alpina, which synthesizes arachidonic acid via the FAS route, the DHA synthesis in S. limacinum does not need much more oxygen in the PKS route. DoubleRobustnessAnalysis results showed that, when the growth rate was fixed, a low oxygen uptake rate (below 5 mmol/gDW/h) was beneficial for DHA accumulation (Fig. 5) while, at the cell-number-increasing stage, to acquire large quantities of cells for lipid accumulation, an abundant oxygen supply was necessary. This means that, during the DHA fermentation process, a two-stage oxygen supply strategy could improve DHA production efficiency [10, 30, 31].
Fig. 5.
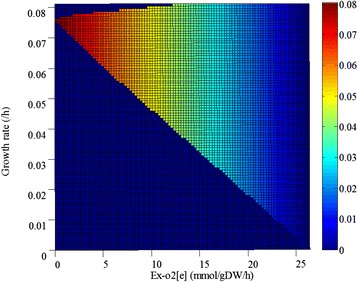
DoubleRobustnessAnalysis of the relationship among oxygen uptake rate, growth rate and DHA production
Similarly to the FAS pathway, two steps in the PKS cycle, catalyzed by KR and ER, also require NADPH [29]. Flux balance analysis shows that, in YE medium, a total of 57 reactions involving NADPH have fluxes (Additional file 5). And of these reactions, three are major sources of NADPH. One is catalyzed by malic enzyme (ME, EC: 1.1.1.40). The other two are in the pentose phosphate pathway and are catalyzed by glucose-6-phosphate dehydrogenase (G6PD, EC: 1.1.1.49) and phosphogluconate dehydrogenase (PGD, EC: 1.1.1.44), respectively. Their corresponding fluxes of NADPH were 1.41 mmol/gDW/h, 1.49 mmol/gDW/h, and 1.49 mmol/gDW/h. In addition to NADPH, acetyl-CoA, which is the precursor for fatty acid de novo biosynthesis, also plays an important role. The flux of acetyl-CoA used for fatty acid synthesis was 1.88 mmol/gDW/h.
Biochemical engineering strategies for improving DHA production by in silico simulation
Malate plays an important role in the TCA cycle. Catalyzed by ME, malate can be converted into pyruvate, accompanied by NADPH. Based on the YE medium, when the maximum uptake rate of malate was set at 1 mmol/gDW/h, DHA production increased 24.5 %. This was in agreement with Ren’s report that, after adding 4 g/L malate, the DHA content of the total fatty acids increased from 35 % to 60 % [14]. It was also proved that the addition of malate lead to an increase in DHA production of 40.02 % [32]. Flux balance analysis (FBA) results showed that, by adding malate, the flux of NADPH supplied by the reaction catalyzed by ME increased from 1.4 mmol/gDW/h to 1.8 mmol/gDW/h, an increase of 28.6 %. Additionally, flux distribution showed that the pentose phosphate pathway was enhanced by 23.5 %, which means that, by adding malate, more NADPH was supplied for DHA biosynthesis. In addition to NADPH, citrate lyase (ACL, EC: 2.3.3.8), which can catalyze the cleavage of citrate, is the source of acetyl-CoA for fatty acid biosynthesis [33]. Adding citrate in silico led to a 37.1 % increase in DHA production. The corresponding acetyl-CoA flux provided by this pathway was also increased by 23.2 %. This also agreed with Wang’s report that a 47.17 % improvement in DHA production was gained by adding citrate [32].
DCW, lipid content, and DHA percentage of total fatty acids are three important parameters for evaluating the fermentation process. And amino acids can be utilized by microorganisms both as a carbon source and a nitrogen source for cell growth. In YE medium, we first calculated the uptake rate of 20 amino acids by FBA. For nine amino acids (Ala, Gly, Asn, Asp, Cys, Glu, Gln, Ser, and Thr), the uptake rate could reach the set maximum values (Fig. 6a). For the others, the uptake rates were lower than 0.01 mmol/gDW/h. This means these nine amino acids could promote the growth of S. limacinum SR21, so the effect of each amino acid on DHA production was simulated by adding them individually to the MG medium. The maximum uptake rate of each amino acid was fixed at 1 mmol/gDW/h, and only the nine amino acids could promote both growth and DHA production. DHA production was increased by 32.7 %, 16.3 %, 32.7 %, 32.7 %, 50.3 %, 50.3 %, 52.3 %, 27.5 %, and 45.8 %, respectively (Fig. 6b).
Fig. 6.
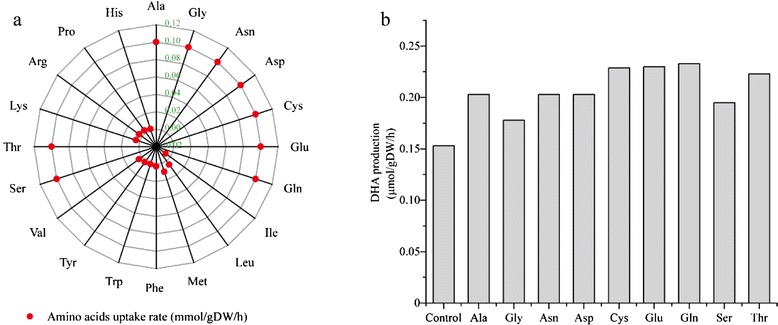
Simulation results of amino acids uptake rates and their effects on DHA production. a In silico result of 20 different amino acids uptake rates in YE medium. b Effect on DHA production by adding different amino acids to MG medium
Furthermore, these simulation results were verified by experiment. The experimental results showed that addition of eight of the nine amino acids (except cysteine) could promote cell growth (Fig. 7). The addition of asparagine could improve the dry cell weight from 26.0 g/L to 37.4 g/L, an increase of 43.8 %, whereas adding cysteine to the culture medium inhibited the growth of SR21. The reason may be that only cysteine contains sulfur, and excess sulfur may inhibit cell growth. The robustness analysis results also showed that the optimized sulfate uptake rate for growth was 0.2 mmol/gDW/h (Additional file 6A). Besides, the experimental results also proved that the addition of Ala, Gly, Asn, Glu, Ser, and Thr (66.7 % of the selected amino acids) could improve DHA production (Fig. 7). Compared to the control group, the addition of Asn led to a 50.6 % increase (9.56 g/L) in DHA production, bringing the DHA content up to 35.0 % of total lipids (Additional file 6B).
Fig. 7.
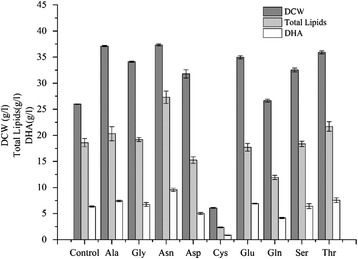
Effects on dry cell weight, total lipids, and DHA production of adding nine different amino acids to S. limacinum cultures
Combined with the FBA results, after adding Asn, SR21 could uptake Asn directly from the in silico medium, instead of using the pathway to generate Asn starting from Asp. Via adenylosuccinate synthase (EC: 6.3.4.4) and adenylosuccinate lyase (EC: 4.3.2.2), extra Asp was converted into fumarate, enhancing the TCA cycle. As a result, fluxes of two reactions involving acetyl-CoA were also enhanced significantly. For example, the flux of acetyl-CoA supplied by pyruvate dehydrogenase complex (PDC, EC: 1.2.4.1, 2.3.1.12, 1.8.1.4) was increased 28.1 %, and acetyl-CoA flux used for the first step of fatty acid biosynthesis via acetyl-CoA carboxylase (ACC, EC: 6.4.1.2) was also increased 20.9 %. In addition to enhancing acetyl-CoA generation, the addition of Asn also increased NADPH flux for producing DHA. After adding Asn, two fluxes in the pentose phosphate pathway catalyzed by G6PD and PGD both increased 122.6 %. This means adding these amino acids could improve DHA production through enhancing the supply of both acetyl-CoA and NADPH.
Genetic engineering strategies for improving DHA production by in silico overexpression
MOMA was used to re-calculate the fluxes for the overexpression algorithm. This simulation was carried out based on YE medium. The lower bound of the DHA exchange reaction was set as 0.01 mmol/gDW/h. Then each reaction that had non-zero flux value in the FBA simulation was overexpressed computationally. After finishing the cycle for over-expression, according to equation 1, 32 reactions catalysed by 30 genes were identified as potential targets (Fig. 8, Additional file 7).
Fig. 8.
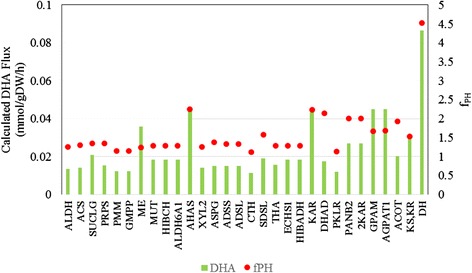
The effect of single gene overexpression on the specific DHA production and fPH
These potential targets could be classified into two groups, one of which is directly involved in DHA synthesis while the other is involved in cell growth (Additional file 7). During the biosynthesis of DHA, acetyl-CoA synthetase (ACS, EC: 6.2.1.1) could promote acetyl-CoA generation [34]. Overexpressing ACS led to an increase in DHA production from 0.01 mmol/gDW/h to 0.0143 mmol/gDW/h (an increase of 43.0 %). ME was assumed to be the major supplier of NADPH for fatty acid biosynthesis [35, 36]. When ME was overexpressed in silico, DHA production rose to 0.0307 mmol/gDW/h. As a dehydrase/isomerase (DH) involved in the last step of DHA synthesis in the PKS system, 0.0863 mmol/gDW/h DHA was accumulated by overexpressing this gene. Genes involved in cell growth were distributed across many metabolic subsystems, such as carbohydrate metabolism, amino acid metabolism and nucleotide metabolism. For example, phosphomannomutase (PMM, EC: 5.4.2.8) and guanosine diphosphomannose phosphorylase (GMPP, EC: 2.7.7.22) are two genes involved in the synthesis of L-galactose, which is a precursor of the biomass function. The growth rate decreased by 5.7 %, and DHA production increased 22.0 % by overexpressing these two genes. After overexpressing PRPP synthetase (PRPS, EC: 2.7.6.1), fluxes used for the synthesis of AMP increased from 0 to 0.0264 mmol/gDW/h, which led to a 52.0 % improvement in DHA production and an 11.5 % decrease in growth rate.
Conclusions
A GSMM of S. limacinum SR21 for DHA production, named iCY1170_DHA, which contained 1769 reactions, 1659 metabolites, and 1170 genes, was successfully reconstructed. Based on glucose and glycerol constraint conditions, the simulated results for growth rate were only 8.5 % and 1.1 % lower than experimental results, respectively. Use of 15 carbon sources and 3 nitrogen sources by SR21 also agreed well with literature reports. Moreover, after the addition of malate and citrate, DHA production increased 24.5 % and 37.1 %, respectively. Furthermore, 9 of 20 amino acids (Ala, Gly, Asn, Asp, Cys, Glu, Gln, Ser, and Thr) were predicted to increase DHA production. According to experimental results, of these 9 amino acids, 6 have been proved to improve DHA production. The addition of Asn could lead a 50.55 % increase in DHA production. Based on MOMA, 30 overexpressed genes, such as those encoding acetyl-CoA synthetase and malic enzyme, were identified as having a positive effect on DHA production.
Methods
Reconstruction of the genome-scale metabolic model
The metabolic model of S. limacinum was initially reconstructed based on genome annotation information and the metabolic pathway database. First, the genome sequence of S. limacinum SR21, which contains 181 scaffolds, was downloaded from the JGI database (http://genome.jgi-psf.org/Aurli1/Aurli1.home.html). Three existing models for Mortierella alpina ATCC 32,222 [37], cyanobacterium Cyanothece sp. ATCC 51,142 [38], and Arabidopsis thaliana [39] were used as reference models, based on protein homology (identity ≥ 40 %, e-value ≤ 1E-30) [40]. KEGG Ontology (KO) and Gene Ontology (GO) identifiers were used to additionally infer reactions that could not be found in the reference strains. To refine the draft model, CELLO [41] and WoLFPSORT [42] were used to determine subcellular compartmentalization. The MetaCyc [43] and BioPath [44] databases were used to judge reaction direction and reversibility. Transport information was obtained by cross-referencing BLATSp searches and the Transporter Classification Database (TCDB) [45].
Biomass composition
Knowledge of the cellular biomass composition is an important prerequisite for the in silico flux analysis, especially during the exponential growth phase, where the primary cellular objective is to maximize growth. The cellular composition of S. limacinum SR21 consists of lipids, proteins, carbohydrates, ash, and nucleic acids [46]. The lipid composition was obtained from previous publications on S. limacinum [47]. Amino acid and metal ion compositions were determined according to Pyle’s report [46]. The overall DNA and RNA compositions were assumed to be the same as cyanobacterium Cyanothece sp. ATCC 51,142 [38] since no data were available on S. limacinum. The individual weights of nucleotides in the DNA and RNA were calculated based on the genome sequence of S. limacinum, in which the G + C content accounts for 45.17 %. The cell wall composition of S. limacinum was assumed to be the same as S. aggregatum [48]. Detailed information about biomass composition can found in Additional file 8.
Constraints-based flux analysis
In order to perform in silico simulations, and to predict the metabolic characteristics of S. limacinum, constraints-based flux analysis, including flux balance analysis (FBA) [49], was carried out under the assumption of a pseudo-steady state. For growth simulation, the biomass equation was set as the objective function. A complex medium, named yeast extract (YE) medium, which contains the basic elements and 20 amino acids, was used for simulation. The glucose uptake rate was 1.4 mmol/gDW/h according to experimental results [24], and all amino acid maximum uptake rates were set as 0.1 mmol/gDW/h [23]. When glycerol was used as the carbon source, its uptake rate was set as 1.6 mmol/gDW/h [25]. In addition to the YE medium, a minimal glucose (MG) medium that did not contain any amino acids was used for different carbon or nitrogen simulations.
The overexpression algorithm involves five steps [23]. (1) We imposed a DHA production flux of 0.01 mmol/gDW/h, which was higher than the wild-type model production (0.00021 mmol/gDW/h). (2) Flux for each reaction was calculated based on the YE medium. (3) A two-fold flux amplification was imposed for biochemical reactions with non-zero flux (to simulate the effect of gene overexpression). (4) Minimization of metabolic adjustment (MOMA) [50] was used to solve the overexpression problem. (5) An overexpressed target that delivers higher DHA production (over 0.01 mmol/gDW/h) and an fPH value > 1 were identified (Eq. 1), where fPH is the product of the specific biomass overexpression rate and the specific DHA overexpression rate. Steps 3 and 4 were iterated for every reaction within the model.
| 1 |
In this study, implementation of constraint-based analysis was performed using Cobra Toolbox 2.05 [51] with MATLAB 2012b and Gurobi 5.6.0 optimizer [52].
Strain, medium, and culture conditions
Schizochytrium limacinum SR21 was gifted by Prof Xiaobin Yu. The fermentation medium contained 120 g/L glucose, 10 g/L peptone, 5 g/L yeast extract, and 20 g/L crystal sea salt [32]. Cells were grown in 500-mL Erlenmeyer flasks each containing 50 mL of medium and incubated at 25 °C in an orbital shaker set at 200 rpm.
Cell density, dry cell weight, and fatty acid analysis
Cell density was calculated from the optical density measured at 660 nm. DCW was determined by transferring a 5-mL cell suspension to a pre-weighed centrifuge tube and then centrifuging at 5000 r/min for 5 min. The cell pellet was then washed twice with distilled water, and dried at 70 °C to constant weight. Fatty acid analysis was accomplished by first harvesting freshly produced cells and freeze drying overnight. The subsequent methods for fatty acid methyl ester (FAME) preparation and gas chromatographic (GC) analysis are the same as reported by Wang [32].
Ethics
This manuscript does not involve Ethics.
Acknowledgements
We thank Prof. Jackie L. Collier for providing access to Aurantiochytrium limacinum ATCC MYA-1381 genome data produced by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This work was supported by the National High-Tech R & D Program of China (No. 2014AA021701), the National Natural Science Foundation of China (21422602), the Program for Innovative Research Team in University (IRT1249).
Abbreviation
- DHA
Docosahexaenoic acid
- GSMM
Genome-scale metabolic model, PKS, polyketide synthase
- PUFA
Polyunsaturated fatty acid
- DCW
Dry cell weight
- ARA
Arachidonic acid
- EPA
Eicosapentaenoic acid
- ORFs
Open reading frames
- MG
Minimal glucose
- YE
Yeast extract
- FAS
Fatty acid synthesis
- ACP
Acyl carrier protein
- KS
3-ketoacyl synthase
- KR
3-ketoacyl-ACP reductase
- ER
Enoyl reducatase
- DH
Dehydrase/isomerase
- ME
Malic enzyme
- G6PD
Glucose-6-phosphate dehydrogenase
- PGD
Phosphogluconate dehydrogenase
- ACL
Citrate lyase
- ACC
Acetyl-CoA carboxylase
- MOMA
Minimization of Metabolic Adjustment algorithm
- ACS
Acetyl-CoA synthetase
- PMM
Phosphomannomutase
- GMPP
Guanosine diphosphomannose phosphorylase
- PRPS
PRPP synthetase
- KO
KEGG Ontology
- GO
Gene Ontology
- TCDB
Transporter Classification Database
- FAME
Fatty acid methyl ester
- GC
Gas chromatographic
- FBA
Flux balance analysis.
Additional files
Detailed information about model i CY1170_DHA. (XML 3178 kb)
Essential genes identified by different in silico media. (XLSX 38 kb)
Comparison among three existing models of S. limacinum, M. alpina, and Y. lipolytica.(XLSX 89 kb)
The annotation results about PKS proteins. (XLSX 9 kb)
Reactions involving NADPH which have fluxes in YE medium. (XLSX 12 kb)
A: RobustnessAnalysis result of the relationship between sulfate uptake rate and growth rate. B: The gas chromatography analysis result of adding Asn to the medium. (DOCX 25 kb)
Potential targets identified by MOMA which could enhance DHA production. (XLSX 10 kb)
Detailed information about the biomass composition of S. limacinum. (XLSX 173 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Chao Ye carried out the construction of iCY1170_DHA, performed the statistical analysis, and drafted the manuscript. Weihua Qiao carried out literature mining for S. limacinum, provided help for the reconstruction of model. Jackie L. Collier provided the genome sequence of SR21 and helped to draft the manuscript. Xiaobin Yu provided the strain S. limacinum SR21 for experimental verification. Xiaojun Ji and He Huang carried out the genome annotation of S. limacinum. Liming Liu participated in the design of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Chao Ye, Email: ziye0911@gmail.com.
Weihua Qiao, Email: qiaoweihua2012@163.com.
Xiaobin Yu, Email: xbyu@jiangnan.edu.cn.
Xiaojun Ji, Email: xiaojunji@njtech.edu.cn.
He Huang, Email: huangh@njtech.edu.cn.
Jackie L. Collier, Email: jackie.collier@stonybrook.edu
Liming Liu, Email: mingll@jiangnan.edu.cn.
References
- 1.Horrocks LA, Yeo YK. Health benefits of docosahexaenoic acid (DHA) Pharmacol Res. 1999;40(3):211–225. doi: 10.1006/phrs.1999.0495. [DOI] [PubMed] [Google Scholar]
- 2.Innis SM. Dietary omega 3 fatty acids and the developing brain. Brain Res. 2008;1237:35–43. doi: 10.1016/j.brainres.2008.08.078. [DOI] [PubMed] [Google Scholar]
- 3.Graham IA, Larson T, Napier JA. Rational metabolic engineering of transgenic plants for biosynthesis of omega-3 polyunsaturates. Curr Opin Biotech. 2007;18(2):142–147. doi: 10.1016/j.copbio.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Song X, Tan Y, Liu Y, Zhang J, Liu G, Feng Y, et al. Different Impacts of Short-Chain Fatty Acids on Saturated and Polyunsaturated Fatty Acid Biosynthesis in Aurantiochytrium sp. SD116. J Agr Food Chem. 2013;61(41):9876–81. [DOI] [PubMed]
- 5.Morita E, Kumon Y, Nakahara T, Kagiwada S, Noguchi T. Docosahexaenoic acid production and lipid-body formation in Schizochytrium limacinum SR21. Mar Biotechnol. 2006;8(3):319–327. doi: 10.1007/s10126-005-5060-y. [DOI] [PubMed] [Google Scholar]
- 6.Yokochi T, Honda D, Higashihara T, Nakahara T. Optimization of docosahexaenoic acid production by Schizochytrium limacinum SR21. Appl Microbiol Biot. 1998;49(1):72–76. doi: 10.1007/s002530051139. [DOI] [Google Scholar]
- 7.Kroes R, Schaefer EJ, Squire RA, Williams GM. A review of the safety of DHA45-oil. Food Chem Toxicol. 2003;41(11):1433–1446. doi: 10.1016/S0278-6915(03)00163-7. [DOI] [PubMed] [Google Scholar]
- 8.Fedorova-Dahms I, Marone PA, Bailey-Hall E, Ryan AS. Safety evaluation of Algal Oil from Schizochytrium sp. Food Chem Toxicol. 2011;49(1):70–77. doi: 10.1016/j.fct.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 9.Sijtsma L, De Swaaf M. Biotechnological production and applications of the ω-3 polyunsaturated fatty acid docosahexaenoic acid. Appl Microbiol Biotechnol. 2004;64(2):146–153. doi: 10.1007/s00253-003-1525-y. [DOI] [PubMed] [Google Scholar]
- 10.Chi Z, Liu Y, Frear C, Chen S. Study of a two-stage growth of DHA-producing marine algae Schizochytrium limacinum SR21 with shifting dissolved oxygen level. Appl Microbiol Biotechnol. 2009;81(6):1141–1148. doi: 10.1007/s00253-008-1740-7. [DOI] [PubMed] [Google Scholar]
- 11.Ren LJ, Sun LN, Zhuang XY, Qu L, Ji XJ, Huang H. Regulation of docosahexaenoic acid production by Schizochytrium sp.: effect of nitrogen addition. Bioprocess Biosyst Eng. 2014;37(5):865–872. doi: 10.1007/s00449-013-1057-5. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Liu R, Chang G, Li X, Chang M, Liu Y, et al. A strategy for the highly efficient production of docosahexaenoic acid by Aurantiochytrium limacinum SR21 using glucose and glycerol as the mixed carbon sources. Bioresource Technol. 2015;177:51–7. [DOI] [PubMed]
- 13.Ganuza E, Anderson AJ, Ratledge C. High-cell-density cultivation of Schizochytrium sp in an ammonium/pH-auxostat fed-batch system. Biotechnol Lett. 2008;30(9):1559–1564. doi: 10.1007/s10529-008-9723-4. [DOI] [PubMed] [Google Scholar]
- 14.Ren LJ, Huang H, Xiao AH, Lian M, Jin LJ, Ji XJ. Enhanced docosahexaenoic acid production by reinforcing acetyl-CoA and NADPH supply in Schizochytrium sp HX-308. Bioprocess Biosyst Eng. 2009;32(6):837–843. doi: 10.1007/s00449-009-0310-4. [DOI] [PubMed] [Google Scholar]
- 15.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7(2):129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu LM, Agren R, Bordel S, Nielsen J. Use of genome-scale metabolic models for understanding microbial physiology. FEBS Lett. 2010;584(12):2556–2564. doi: 10.1016/j.febslet.2010.04.052. [DOI] [PubMed] [Google Scholar]
- 17.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15(2):107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 18.Oberhardt MA, Palsson BØ, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5. [DOI] [PMC free article] [PubMed]
- 19.Durot M, Bourguignon PY, Schachter V. Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS Microbiol Rev. 2009;33(1):164–190. doi: 10.1111/j.1574-6976.2008.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan P, Hua Q. Reconstruction and In Silico Analysis of Metabolic Network for an Oleaginous Yeast, Yarrowia lipolytica. PloS One. 2012;7(12):e51535. doi: 10.1371/journal.pone.0051535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda D, Yokochi T, Nakahara T, Erata M, Higashihara T. Schizochytrium limacinum sp. nov., a new thraustochytrid from a mangrove area in the west Pacific Ocean. Mycol Res. 1998;102(4):439–448. doi: 10.1017/S0953756297005170. [DOI] [Google Scholar]
- 22.Nagano N, Taoka Y, Honda D, Hayashi M. Optimization of culture conditions for growth and docosahexaenoic acid production by a Marine Thraustochytrid, Aurantiochytrium limacinum mh0186. J Oleo Sci. 2009;58(12):623–628. doi: 10.5650/jos.58.623. [DOI] [PubMed] [Google Scholar]
- 23.Boghigian BA, Armando J, Salas D, Pfeifer BA. Computational identification of gene over-expression targets for metabolic engineering of taxadiene production. Appl Microbiol Biotechnol. 2012;93(5):2063–2073. doi: 10.1007/s00253-011-3725-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song XJ, Zhang XC, Kuang CH, Zhu LY, Zhao XB. Batch kinetics and modeling of DHA production by S. limacinum OUC88. Food Bioprod Process. 2010;88(C1):26–30. doi: 10.1016/j.fbp.2009.12.004. [DOI] [Google Scholar]
- 25.Liang Y, Sarkany N, Cui Y, Blackburn JW. Batch stage study of lipid production from crude glycerol derived from yellow grease or animal fats through microalgal fermentation. Bioresource Technol. 2010;101(17):6745–6750. doi: 10.1016/j.biortech.2010.03.087. [DOI] [PubMed] [Google Scholar]
- 26.Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 2001;293(5528):290–3. [DOI] [PubMed]
- 27.Ratledge C. Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie. 2004;86(11):807–815. doi: 10.1016/j.biochi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Wallis JG, Watts JL, Browse J. Polyunsaturated fatty acid synthesis: what will they think of next? Trends Biochem Sci. 2002;27(9):467–473. doi: 10.1016/S0968-0004(02)02168-0. [DOI] [PubMed] [Google Scholar]
- 29.Qiu X. Biosynthesis of docosahexaenoic acid (DHA, 22:6–4, 7,10,13,16,19): two distinct pathways. Prostaglandins Leukot Essent Fatty Acids. 2003;68(2):181–186. doi: 10.1016/S0952-3278(02)00268-5. [DOI] [PubMed] [Google Scholar]
- 30.Qu L, Ji XJ, Ren LJ, Nie ZK, Feng Y, Wu WJ, et al. Enhancement of docosahexaenoic acid production by Schizochytrium sp. using a two-stage oxygen supply control strategy based on oxygen transfer coefficient. Lett Appl Microbiol. 2011;52(1):22–7. [DOI] [PubMed]
- 31.Zhang L, Zhao H, Lai Y, Wu J, Chen H. Improving docosahexaenoic acid productivity of Schizochytrium sp. by a two-stage AEMR/shake mixed culture mode. Bioresour Technol. 2013;142:719–22. [DOI] [PubMed]
- 32.Wang S, Luo W, Jiang Y, Yu X. Enhancing DHA synthesis in Schizochytrium limacinum by exogenous additives. Chin J Bioprocess Eng. 2013;11(5):21–25. [Google Scholar]
- 33.Feng Y, Ren L, Wei P, Tong Q, Ji X, Huang H. Progress in metabolic mechanism of docosahexenoic acid production by fermentation. Chin J Biotechnol. 2010;26(9):1225–1231. [PubMed] [Google Scholar]
- 34.Yan JF, Cheng RB, Lin XZ, You S, Li K, Rong H, et al. Overexpression of acetyl-CoA synthetase increased the biomass and fatty acid proportion in microalga Schizochytrium. Appl Microbiol Biotechnol. 2013;97(5):1933–9. [DOI] [PubMed]
- 35.Wynn JP, bin Abdul Hamid A, Ratledge C. The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi. Microbiology. 1999;145(8):1911–1917. doi: 10.1099/13500872-145-8-1911. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Zhang L, Chen H, Chen YQ, Ratledge C, Song Y, et al. Regulatory properties of malic enzyme in the oleaginous yeast, Yarrowia lipolytica, and its non-involvement in lipid accumulation. Biotechnol Lett. 2013;1–8. [DOI] [PubMed]
- 37.Ye C, Xu N, Chen H, Chen YQ, Chen W, Liu L. Reconstruction and analysis of a genome-scale metabolic model of the oleaginous fungus Mortierella alpina. BMC Syst Biol. 2015;9(1):1–11. doi: 10.1186/s12918-014-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vu TT, Stolyar SM, Pinchuk GE, Hill EA, Kucek LA, Brown RN, et al. Genome-scale modeling of light-driven reductant partitioning and carbon fluxes in diazotrophic unicellular cyanobacterium Cyanothece sp. ATCC 51142. PLoS Comput Biol. 2012;8(4):e1002460. [DOI] [PMC free article] [PubMed]
- 39.Dal’Molin CGD, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. AraGEM, a Genome-Scale Reconstruction of the Primary Metabolic Network in Arabidopsis. Plant Physiol. 2010;152(2):579–589. doi: 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol. 2003;333(4):863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 41.Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13(5):1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier C, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35 suppl 2:W585–7. [DOI] [PMC free article] [PubMed]
- 43.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2008;36 suppl 1:D623–31. [DOI] [PMC free article] [PubMed]
- 44.Reitz M, Sacher O, Tarkhov A, Trümbach D, Gasteiger J. Enabling the exploration of biochemical pathways. Org Biomol Chem. 2004;2(22):3226–3237. doi: 10.1039/b410949j. [DOI] [PubMed] [Google Scholar]
- 45.Saier MH, Tran CV, Barabote RD. TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34(suppl 1):D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pyle DJ, Garcia RA, Wen Z. Producing docosahexaenoic acid (DHA)-rich algae from blodiesel-derived crude glycerol: Effects of impurities on DHA production and algal biomass composition. J Agr Food Chem. 2008;56(11):3933–3939. doi: 10.1021/jf800602s. [DOI] [PubMed] [Google Scholar]
- 47.Wang G, Wang T. Characterization of Lipid Components in Two Microalgae for Biofuel Application. J Am Oil Chem Soc. 2011;89(1):135–143. doi: 10.1007/s11746-011-1879-8. [DOI] [Google Scholar]
- 48.Darley WM, Porter D, Fuller MS. Cell wall composition and synthesis via Golgi-directed scale formation in the marine eucaryote, Schizochytrium aggregatum, with a note on Thraustochytrium sp. Arch Mikrobiol. 1973;90(2):89–106. doi: 10.1007/BF00414512. [DOI] [PubMed] [Google Scholar]
- 49.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segre D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002;99(23):15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2. 0. Nat Protoc. 2011;6(9):1290–307. [DOI] [PMC free article] [PubMed]
- 52.Yin W. Gurobi Mex: a MATLAB interface for Gurobi. Online at http://wikimizatio/wikimization/indexphp/gurobi_mex 2009.


