Abstract
Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα) autophosphorylation at Thr286 and Thr305/Thr306 regulates kinase activity, modulates subcellular targeting, and is critical for normal synaptic plasticity and learning and memory. Here, a mass spectrometry-based approach was used to identify Ca2+-dependent and -independent in vitro autophosphorylation sites in recombinant CaMKIIα and CaMKIIβ. CaMKII holoenzymes were then immunoprecipitated from subcellular fractions of forebrains isolated from either wildtype (WT) mice or mice with a Thr286 to Ala knock-in mutation of CaMKIIα (T286A-KI mice) and analyzed using the same approach in order to characterize in vivo phosphorylation sites in both CaMKII isoforms and identify CaMKII associated proteins (CaMKAPs). A total of 6 and 7 autophosphorylation sites in CaMKIIα and CaMKIIβ, respectively, were detected in WT mice. Thr286-phosphorylated CaMKIIα and Thr287-phosphorylated CaMKIIβ were selectively enriched in WT Triton-insoluble (synaptic) fractions compared to Triton-soluble (membrane) and cytosolic fractions. In contrast, Thr306-phosphorylated CaMKIIα and Ser315- and Thr320/Thr321-phosphorylated CaMKIIβ were selectively enriched in WT cytosolic fractions. The T286A-KI mutation significantly reduced levels of phosphorylation of CaMKIIα at Ser275 across all subcellular fractions, and of cytosolic CaMKIIβ at Ser315 and Thr320/Thr321. Significantly more CaMKAPs co-precipitated with WT CaMKII holoenzymes in the synaptic fraction compared to the membrane fraction, with functions including scaffolding, microtubule organization, actin organization, ribosomal function, vesicle trafficking, and others. The T286A-KI mutation altered the interactions of multiple CaMKAPs with CaMKII, including several proteins linked to autism spectrum disorders. These data identify CaMKII isoform phosphorylation sites and a network of synaptic protein interactions that are sensitive to the abrogation of Thr286 autophosphorylation of CaMKIIα, likely contributing to the diverse synaptic and behavioral deficits of T286A-KI mice.
Keywords: Synaptic plasticity, postsynaptic density, autophosphorylation, proteomics, mass spectrometry, subcellular fractionation, protein-protein interactions
Introduction
The α and β isoforms of Ca2+/calmodulin-dependent protein kinase II (CaMKII) account for up to 1–2% of total protein in several regions of mammalian forebrain.1 Studies of genetically manipulated mice have revealed critical roles for both CaMKII isoforms in regulating synaptic plasticity and behavior.2–8 Both isoforms can be autophosphorylated at multiple sites in vitro9–17 with distinct effects on kinase activity and/or on interactions with other neuronal proteins. The best understood example is the Ca2+/calmodulin (CaM)-dependent autophosphorylation of CaMKIIα at Thr286, which generates a Ca2+/CaM-independent (autonomous) form of the kinase9–11 and enhances CaMKII interactions with both Ca2+/CaM and GluN2B subunits of the NMDA-type glutamate receptor.18–20 In contrast, CaMKIIα autophosphorylation at Thr305 or Thr306 blocks binding of Ca2+/CaM and α-actinin, thereby interfering with kinase activation.13–15, 21 Studies of transgenic mouse lines with knock-in mutations at Thr286 or Thr305/6 have demonstrated important roles for these two phosphorylation sites in the synaptic targeting of CaMKII, synaptic plasticity, and several neurobehaviors.22–26
While the “classical” view is that CaMKII activation is critical for long-term potentiation (LTP) of synaptic transmission, more recent studies suggest a more complex picture. For example, CaMKIIα is required for long-term depression (LTD) in cerebellar Purkinje neurons, whereas CaMKIIβ is required for LTP.2, 7 Moreover, recent studies show that CaMKIIα (and Thr286 autophosphorylation) is required for different forms of long-term depression in the hippocampus, in addition to the classical role in LTP27, 28. These findings emphasize the lack of detailed understanding of CaMKII interactions, targeting, and function in neurons, and of the inter-relationships and roles of CaMKIIα and CaMKIIβ autophosphorylation sites in vivo.
Here, we analyze mouse forebrain CaMKII holoenzymes using unbiased mass spectrometry-based approaches to identify CaMKIIα/β phosphorylation sites and CaMKII-associated proteins (CaMKAPs). We found that CaMKII holoenzymes isolated from different subcellular fractions of wild-type (WT) mice are differentially phosphorylated and interact with distinct networks of over 100 known and novel CaMKAPs, many having established roles in modulating synaptic structure and function. Parallel analyses in mice with a Thr286 to Ala knock-in mutation of CaMKIIα (T286A-KI mice) revealed changes in levels of phosphorylation at other sites in CaMKIIα and CaMKIIβ, and in the relative levels of co-precipitating CaMKAPs, which presumably contribute to the synaptic plasticity deficits and multiple behavioral phenotypes of these mice.22, 26 In combination, these data provide novel unbiased insights into the regulation, function and subcellular targeting of CaMKII and into the deficits in specific downstream molecular signaling pathways.
Results and Discussion
Analysis of in vitro autophosphorylation sites in CaMKIIα and CaMKIIβ
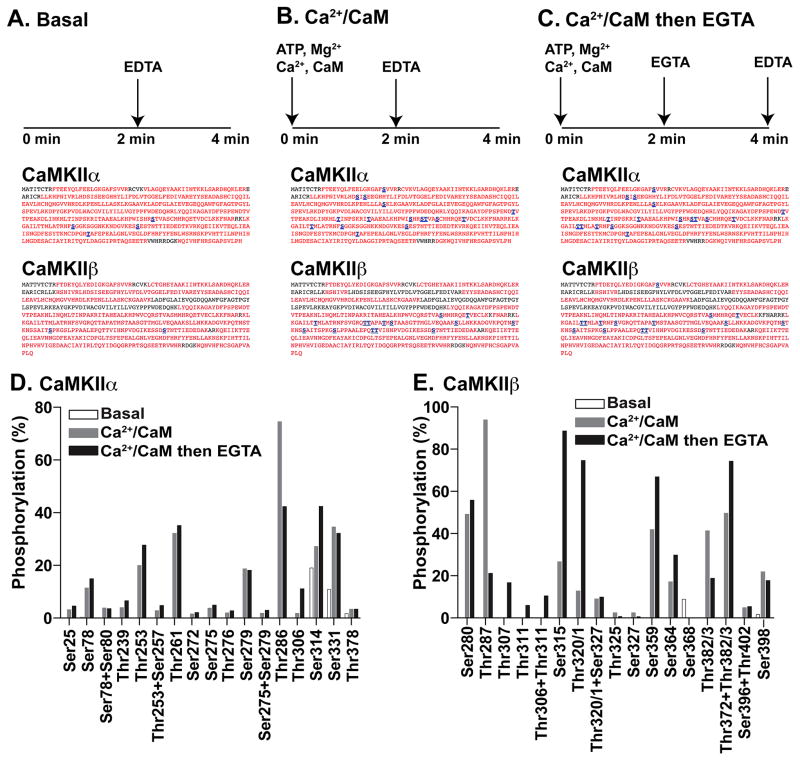
Recombinant CaMKIIα was purified from insect cells and then incubated under three conditions. 1) control conditions in the absence of ATP (basal); 2) to allow autophosphorylation in the presence of Ca2+/calmodulin alone (Ca2+/CaM); 3) to allow autophosphorylation first in the presence of Ca2+/calmodulin and then following Ca2+ chelation with EGTA to dissociate calmodulin (Ca2+/CaM then EGTA; sequential) (see Materials and Methods for details). Similar conditions were used to identify the preferential autophosphorylation of CaMKIIα at Thr286 in the presence of Ca2+/calmodulin, and at Thr305 or Thr306 and Ser314 in the presence of EGTA.9–15 However, the full repertoire of sites phosphorylated under these conditions is poorly understood. Our MS-based analyses recovered tryptic peptides covering >93% of the entire amino acid sequence of CaMKIIα under each condition (Fig 1A–1C) and detected a total of 19 residues that were phosphorylated in at least one sample, 16 of which were detected only following in vitro autophosphorylation (Fig 1D and Table S1). MS/MS spectra for each non-phosphorylated and phosphorylated tryptic peptide were confirmed and annotated (Fig S1). The chromatographic retention, percent phosphorylation, and PPM mass error of each peptide is shown in Table S1.
Figure 1. Identification of in vitro CaMKIIα and CaMKIIβ phosphorylation sites.
A–C. Sequence coverage (red) and phosphorylation-site detection (blue, underlined) in purified CaMKIIα or CaMKIIβ following either: A, a control incubation (Basal), B, phosphorylation in the presence of Ca2+/calmodulin alone (Ca2+/CaM), or C, sequential phosphorylation in the presence of Ca2+/calmodulin and then EGTA (Ca2+/CaM then EGTA) (see Methods). D, E. The AUCs of XICs were used to compare relative levels of phosphorylation of CaMKIIα at 16 different phosphorylation sites (D) or CaMKIIβ at 15 different phosphorylation sites (E) in each sample.
Since CaMKIIβ autophosphorylation has not been characterized extensively, we used the same approach to identify phosphorylation sites in purified recombinant CaMKIIβ (Fig 1A–C, 1E), yielding ~80% amino acid sequence coverage across the three incubation conditions. A total of 15 phosphorylation sites on CaMKIIβ were phosphorylated in at least one sample, with 13 sites detected only following in vitro autophosphorylation (Fig 1E, Table S2, Fig S2).
Comparison of in vitro autophosphorylated CaMKIIα and CaMKIIβ under different conditions
In order to compare relative levels of phosphorylation at each site under each condition, we estimated relative phosphorylation stoichiometries from the areas under the curve (AUC) of extracted ion chromatograms (XICs) for each phosphorylated and non-phosphorylated peptide pair. This approach provides a relatively crude estimate of the absolute levels of phosphorylation at each site (see Methods), so our interpretations are focused on relative differences between the preincubation conditions. As expected, the homologous Thr286 and Thr287 sites in CaMKIIα and CaMKIIβ, respectively, appeared to be substantially autophosphorylated in the presence of Ca2+/CaM. Additional Ca2+-independent incubation (plus EGTA) appeared to decrease the levels of Thr286/Thr287 phosphorylation, perhaps reflecting a lack of precision of this type of analysis. However, this decrease may also be due to dephosphorylation in the presence of EGTA due to a previously reported auto-catalytic event,29 or a contaminating phosphatase. The Ca2+-independent reaction also selectively increased phosphorylation of CaMKIIα at Thr306 and Ser314, confirming previous reports.14 Prior studies using site-directed mutagenesis indicated that both Thr305 and Thr306 in CaMKIIα could be phosphorylated in the Ca2+-independent phase,14, 15 but we detected only low levels of Thr305 phosphorylation, and then only when this tryptic fragment was also phosphorylated at Thr310 (Table S1). The Ca2+-independent phase of CaMKIIβ autophosphorylation (plus EGTA) selectively enhanced modifications at Thr306, Thr307, Thr311, and Ser315 (Fig 1 and Table S1, S2). Similar to the homologous CaMKIIα Thr305 site, CaMKIIβ phosphorylation at Thr306 was only detected in the simultaneous presence of Thr311 phosphorylation (Fig 1E and Table S2). However, these data cannot exclude the possibility that Thr305(α) or Thr306(β) can be phosphorylated alone. Interestingly, substantial CaMKIIβ phosphorylation at Ser315 was detected only following Ca2+-independent autophosphorylation, whereas phosphorylation of CaMKIIα at the homologous Ser314 residue was detected in all three samples, presumably due to basal phosphorylation of this site in the insect cell expression system prior to purification.
In addition to several previously identified in vitro autophosphoryation sites in both CaMKIIα (Thr253, Ser275, Ser279) and CaMKIIβ1 (Ser280, Ser343, Thr382/Thr383),9, 13, 16, 17 we detected autophosphorylation of CaMKIIα at several novel sites in the catalytic domain (e.g., Ser78 and Thr261), as well as at Ser331 and Thr378 in the C-terminal association domain. We did not detect CaMKIIβ autophosphorylation in the catalytic domain, but the association domain was prominently autophosphorylated at 8 previously unidentified sites. Most of the novel CaMKIIβ phosphorylation sites (Thr320/1, Thr325, Ser327, Ser359, Ser368, Thr372, and Thr382/3) are located within two domains implicated in F-actin binding (residues 317–342 and 354–393),8 and are not conserved in CaMKIIα (Fig S3). Notably, Thr320/1 in CaMKIIβ appeared to be phosphorylated predominantly in the Ca2+-independent reaction (plus EGTA). However, even though the recombinant CaMKII isoforms used for these studies were highly purified (see Methods), we cannot completely exclude contributions of contaminating kinases to total CaMKIIα/ β phosphorylation under these conditions.
In combination, these in vitro data confirm that Thr286 in CaMKIIα and Thr287 in CaMKIIβ are among several sites phosphorylated in the presence of Ca2+/CaM and also indicate that the predominant sites of Ca2+-independent autophosphorylation are Thr306 and Ser314 in CaMKIIα and Thr307, Ser315 and Th320/1 in CaMKIIβ. However, they also identify several additional sites that can be autophosphorylated under these in vitro conditions.
Phosphorylation of CaMKII in subcellular fractions from WT mouse brain
Having established methods to detect phosphorylation sites in purified CaMKIIα and CaMKIIβ, we analyzed mouse forebrain CaMKII holoenzymes isolated by immunoprecipitation in the presence of protease and protein phosphatase inhibitors from cytosolic (S1), Triton-soluble (membrane; S2), and Triton-insoluble (synaptic; S3) fractions that were rapidly prepared using progressively harsher detergents at approximately physiological ionic strength.26 Following SDS-PAGE, bands containing CaMKIIα or CaMKIIβ (~50 and ~60 kDa, respectively) were excised (Fig 2A), separately digested with trypsin, and then analyzed by LC-MS/MS. Relative levels of phosphorylation at each site between subcellular fractions were compared based on the ratio of AUCs for the XICs of phosphorylated and non-phosphorylated tryptic peptides (see Methods).
Figure 2. Identification of phosphorylation sites in mouse forebrain CaMKIIα and CaMKIIβ.
A. Cytosolic (S1), membrane (S2), and synaptic (S3) fractions were immunoprecipitated using control or anti-CaMKII IgG, and immune complexes were analyzed by SDS-PAGE followed by staining with Sypro Ruby. B. Semi-quantitative analysis of relative levels of CaMKIIα phosphorylation at 6 different sites in each fraction normalized to the highest level for each site. The synaptic fraction was selectively enriched for Thr286 phosphorylation, whereas the cytosolic fraction was selectively enriched for Thr306 phosphorylation. C. Immunoblot analysis of subcellular fractions confirms that the synaptic (S3) fraction is significantly enriched in Thr286 phosphorylated CaMKIIα. D. Semi-quantitative analysis of relative levels of CaMKIIβ phosphorylation at 6 different sites in each fraction normalized to the highest level for each site. The cytosolic fraction was selectively enriched for phosphorylation at Ser315 and Thr320/1. *p<0.05; ***p<0.001; ****p<0.0001; in comparison to the highest level at that site. E. Summary of CaMKII phosphorylation sites. Horizontal bars indicate aligned domain structures of the canonical CaMKIIα and CaMKIIβ isoforms based on a sequence alignment (accession numbers: P11798 and P28652, respectively). Sequence alignments of selected regions are indicated above the bars, whereas CaMKIIβ-specific sequences of the actin-binding domain (ABD) are indicated below. All phosphorylation sites detected in this study (in vitro or in vivo) are indicated by red dots adjacent to amino acid sequences or by residues in red font adjacent to the domain bars. Black dots and fonts indicate additional residues detected in prior global phospho-proteomics studies.36–41 Yellow highlighted labels indicate phosphorylation sites identified in only one study.
Coverage of CaMKIIα varied from 45–93% across the three fractions in four biological replicates (projects A–D; Supplementary Table S4), detecting a total of six phosphorylation sites with estimated ratios ranging from 0.1–11.4% (Ser275, Thr286, Thr306, Ser314, Ser331, and Thr337). Ser275, Ser314, and Ser331 were phosphorylated at similar levels in all three subcellular fractions (Fig 2B). However, the level of Thr286 phosphorylation in synaptic fractions was 5-fold or 2.4-fold higher than detected in the cytosolic or membrane fractions, respectively (Fig 2B). Differential Thr286 phosphorylation of CaMKIIα between subcellular fractions was confirmed by immunoblotting using a phospho-Thr286 specific antibody (Fig 2C). Notably, the selective enrichment of Thr286-autophosphorylated CaMKIIα in synaptic fractions is generally consistent with prior studies showing that Thr286 autophosphorylation enhances synaptic targeting of CaMKIIα26, 30, 31 In contrast, levels of Thr306 phosphorylation were >6-fold higher in the cytosolic kinase relative to membrane or synaptic pools of kinase (Fig 2B). These data are consistent with previous observations that Thr305/306 phosphorylation destabilizes synaptic targeting of CaMKIIα.23, 30 It is interesting that CaMKIIα was predominantly phosphorylated at Thr306 rather than Thr305, because phosphorylation at Thr306, but not at Thr305, blocks the CaMKII interaction with α-actinin-2,21 a major synaptic F-actin-binding protein. Taken together, these data show that distinct subcellular pools of CaMKIIα are phosphorylated in a site-specific manner.
Mouse forebrain CaMKIIβ was phosphorylated at 7 sites (Thr287, Ser315, Thr320/Thr321, Ser367, Thr381/Thr382, Ser397, and Thr401), with estimated ratios varying from 0.4–54.0%. Alternative mRNA splicing, mostly in the C-terminal domain, generates several CaMKIIβ variants, and our data cannot address whether these variants are differentially phosphorylated at sites in the conserved domains. Therefore, all CaMKIIβ residues are numbered according to the major, canonical, CaMKIIβ isoform (Accession # P28652). Somewhat surprisingly, we did not detect significant phosphorylation of CaMKIIβ at Thr306 or Thr307 in any subcellular fraction. Levels of phosphorylation at Ser367, Ser397, and Thr401 were not significantly different between subcellular fractions (Fig 2D), but there was a trend for enrichment of Thr287 phosphorylation in synaptic CaMKIIβ, as noted for Thr286 phosphorylation of CaMKIIα (see above). However, phosphorylation at multiple sites was selectively enriched in cytosolic CaMKIIβ. Specifically, levels of Ser315 and Thr320/Thr321 phosphorylation in cytosolic CaMKIIβ were >5-fold and >9-fold higher, respectively, than in the membrane or synaptic kinases (Fig 2B). Thr381/Thr382 phosphorylation was detected in only one replicate, but levels were >7-fold higher in the cytosolic fraction compared with membrane and synaptic fractions. Although we could not unambiguously distinguish modifications at Thr320/Thr321 or Thr381/Thr382, these sites lie within previously defined F-actin binding domains (Fig. 2E).8 Thus, it seems reasonable to suggest that phosphorylation of these sites modulates CaMKIIβ binding to F-actin, and thereby F-actin assembly and dendritic spine morphology/function.3, 32–35 In combination, these data show that distinct subcellular pools of CaMKIIβ are differentially phosphorylated at specific sites.
All of the phosphorylation sites that we detected in mouse brain CaMKII isoforms were also identified as in vitro autophosphorylation sites. However, we cannot exclude potential contributions of other protein kinases to CaMKII phosphorylation in vivo. Fig. 2E integrates data from our in vitro and in vivo studies with the repertoire of CaMKIIα and CaMKIIβ phosphorylation sites that have been identified in previous global mouse brain proteomics studies.36–41 Even though our total coverage of both CaMKII isoforms was high (32–93% depending on fraction, genotype and replicate), we detected a more limited number of in vivo sites than detected in the prior studies or in vitro. Notably, whereas prior studies detected phosphorylation of several additional sites in some tryptic fragments, we identified a more limited number of sites in the same tryptic fragments (Fig. 2E). The lower sensitivity of our in vivo studies relative to the in vitro studies may reflect a lower efficiency of digestion/extraction of the kinase from polyacrylamide gels versus TCA precipitates, or ion suppression from peptides derived from co-immunoprecipitated proteins. Moreover, the greater sensitivity of prior in vivo studies likely reflects the use of metal affinity-based methods to enrich for phosphopeptides. Nevertheless, the present study provides new insights into CaMKII biology by demonstrated that specific sites are differentially phosphorylated between subcellular fractions.
Surprisingly, we failed to detect some previously well-characterized autophosphorylation sites in vivo. For example, CaMKIIα phosphorylation at Thr253 in vivo was detected in prior proteomics studies,36–39, 42, 43 and using phospho-site specific antibodies.17, 44 However, we failed to detect phosphorylation of CaMKIIα at Thr253 or of CaMKIIβ at Thr254 in samples from mouse brain, even though in vitro Thr253 phosphorylation was readily detected (Fig. 1D). Perhaps the phosphorylation stoichiometry was relatively low in our in vivo samples because Thr253 phosphorylation is favored under physiological or pathophysiological conditions that were not prevalent prior to, or during, tissue isolation. Alternatively, the phospho-Thr253 peptide may contain an additional unknown covalent modification that affects its mass, so that it could not be identified. Similarly, even though commercially available antibodies raised to phospho-Thr305 in CaMKIIα can detect CaMKIIα phosphorylation in the brain, we detected substantial phosphorylation at Thr306, but not at Thr305. Thus, data obtained using phospho-Thr305 antibodies should be cautiously interpreted because it is unclear whether these antibodies consistently detect phospho-Thr306 in CaMKIIα.
Effect of T286A-KI mutation on CaMKII phosphorylation in mouse brain
CaMKII holoenzymes isolated from T286A-KI mouse forebrain subcellular fractions were analyzed in parallel with WT samples discussed above. There was a robust effect of the T286A-KI genotype (p<0.0001) to reduce Ser275 phosphorylation by >90% in all three subcellular fractions (Fig 3A). These data suggest that Ser275 phosphorylation may require prior phosphorylation at Thr286, although it is unclear whether Ser275 phosphorylation results from autophosphorylation (Fig. 1D), or from PKC phosphorylation.45 We also found that the T286A-KI mutation decreased Ser314 phosphorylation by 27–47% across the three subcellular fractions (p=0.0003) (Fig 3C), perhaps because the lack of Thr286 phosphorylation reduces autonomous CaMKII activity. However, there was no effect of genotype on CaMKIIα phosphorylation at Thr306, with similar enrichment in cytosolic CaMKII holoenzymes in both T286A-KI and WT mice (p<0.0001) (Fig 3B). In addition, genotype or fractionation effects on CaMKIIα phosphorylation at Ser331 (Fig 3D) or Thr337 were not detected (Fig 3E).
Figure 3. Effect of T286A-KI mutation on CaMKIIα and CaMKIIβ phosphorylation at other sites.
Levels of phosphorylation of CaMKIIα (A–E) and CaMKIIβ (F–K) at the indicated sites were compared across subcellular fractions isolated in parallel from WT and T286A-KI mice. Data are the mean from 4 (A–E) or 3 (F–K) biological replicates after normalization to the estimated level in the WT S1 fraction within each replicate. A 2-way ANOVA revealed a significant genotype effect on phosphorylation of CaMKIIα at Ser275 (A; F(1,15) = 157.8; p<0.0001) and Ser314 (C; F(1,15) = 22.42; p=0.0003) and a significant fractionation effect on Thr306 phosphorylation (B; F(2,15) = 35.41; p<0.0001). For CaMKIIβ there were significant fractionation and genotype effects and an interaction effect on the phosphorylation at Ser315 (E; Fractionation effect (F(2,9) = 153.8, p<0.0001), Genotype effect (F(1,9) = 49.4, p<0.0001), Interaction (F(2,9) = 33.55, p<0.0001) and Thr320/Thr321(F; Fractionation effect (F(2,9) = 58.31, p<0.0001), Genotype effect (F(1,9) = 9.115, p=0.0145), Interaction (F(2,9) = 6.159, p=0.0206). Significant differences revealed by post-hoc analyses are coded as follows: *, compared to S1 WT. $, compared to S2 WT. @, compared to S3 WT. #, compared to S1 KI. Single, double, triple and quadruple symbols indicate p<0.05, p<0.01, p<0.001, and p<0.0001, respectively.
The T286A-KI genotype had a significant effect on, and a significant interaction with subcellular fraction, on CaMKIIβ phosphorylation at Ser315 (fraction, p<0.0001; genotype, p<0.0001; interaction, p<0.0001; Fig 3G) and Thr320/Thr321 (fraction, p<0.0001; genotype, p<0.0145; interaction, p<0.0206; Fig 3H), with decreased levels in T286A-KI cytosolic fractions compared to WT cytosolic fractions (by 59.5% and 49.0%, respectively). Moreover, in the one biological replicate that detected phosphorylation at Thr381/Thr382, the levels were 3.2-fold higher in the WT cytosolic fraction that in the T286A-KI fraction. However, there was no statistically significant effect of genotype or fractionation on CaMKIIβ phosphorylation at Thr287 (Fig 3F), Ser367 (Fig 3I), Ser397 (Fig 3J), or Thr401 (Fig 3K).
In combination, these data show that abrogation of Thr286 phosphorylation in CaMKIIα by the T286A-KI mutation affects the levels of phosphorylation at other sites in CaMKIIα and in CaMKIIβ in different subcellular fractions, perhaps representing some form of compensatory adaptation to the disruption of synaptic CaMKII targeting in T286A-KI mice and also suggesting previously unrecognized functional linkages between sites. Further studies are required to investigate the contributions of these sites to CaMKII regulation, and perhaps to the phenotypes of T286A-KI mice.
Characterization of CaMKII associated proteins (CaMKAPs)
We also performed an unbiased proteomic screen for CaMKAPs associated with cytosolic, membrane, and synaptic CaMKII holoenzymes. Although the precise macromolecular nature of the solubilized synaptic fraction is unclear, we previously showed that it is highly enriched in PSD marker proteins, such as PSD95, NMDAR subunits, and other cytoskeletal proteins.26 CaMKII holoenzymes were immunoprecipitated from each subcellular fraction and then separated by SDS-PAGE. Control samples were isolated from each fraction in parallel using a non-specific IgG. CaMKII (α or β) or CaMKAP (non-CaMKII, non-IgG regions) regions of the gel lanes (Fig 2A) were excised and separately analyzed using a shotgun LC-MS/MS approach.
The total numbers of mass spectra (spectral counts) matching all proteins in analyses of WT CaMKII or IgG control samples isolated from cytosolic, membrane, and synaptic fractions were used to calculate WT/IgG ratios of 1.2, 4.3, and 5.0, respectively, for the first biological replicate (project A) (Table S3). Thus, CaMKII immunoprecipitation from cytosolic fractions lacks specificity, perhaps in part due to the lack of detergent in these samples. Consequently, subsequent analyses focused on membrane and synaptic fractions. CaMKII-derived spectral counts were 2.9-fold higher in membrane compared to synaptic fractions (Fig 4A), consistent with immunoblotting and protein staining data showing higher total levels of CaMKII in membrane, compared to synaptic, fractions (Fig 2A, 2C). In contrast, the total number of CaMKAP-derived spectral counts was 3.2-fold higher in the synaptic fraction compared to the membrane fraction (Fig 4B), even though the harsher solubilization conditions might destabilize some protein-protein interactions. Thus, it appears that CaMKAPs are preferentially associated with synaptic CaMKII holoenzymes. Unsurprisingly, most of the non-CaMKII derived spectral counts were detected in analyses of the two CaMKAP gel regions (above CaMKIIβ and below CaMKIIα), although a few co-precipitating proteins were detected in CaMKIIα or CaMKIIβ gel regions. Similar trends in the spectral count data were observed in two independent biological replicates (Projects B and D) (Fig S4; Table S3).
Figure 4. Comparative levels of CaMKII and CaMKAPs in WT S2 and S3 fractions.

A. Fewer total spectral counts derived from all CaMKII isoforms were detected in the S3 fraction compared to S2 fraction. B. The total number of spectral counts derived from proteins other than CaMKII was larger in the S3 fraction that in the S2 fraction. C. More proteins other than CaMKII were detected in S3 complexes compared to S2 complexes. These data are derived from project A; relative differences between fractions are representative of two other independent biological replicates (see Figure S4).
All proteins detected in CaMKII complexes for all three projects are shown in Table S3. This list was then filtered to remove three types of “non-specific” proteins. 1. Proteins also detected in the cytosolic fraction (due to the lack of specificity; see above). 2. Proteins detected with <7 spectral counts in membrane and synaptic fractions combined. 3. Proteins with WT/IgG spectral count ratios of <4 (since a WT/IgG ratio of 3.8 was calculated for CaMKIIα-derived spectral counts in the synaptic fraction). Remaining proteins from the three independently analyzed gel regions were then combined to form a final filtered list of 138 CaMKAPs (Table 1). Scaffold peptide probability46 and XCorr values for all peptides matching to any of the 138 CaMKAPs are shown in Table S4. The membrane and synaptic fractions contained 47 and 110 CaMKAPs, respectively, with ≥7 spectral counts (Fig 4C), and 34 CaMKAPs were detected with <7 spectral counts in either fraction, individually, but ≥ 7 total spectral counts in both fractions combined. Based on the number of spectral counts, a total of 15 CaMKAPs were selectively enriched in the membrane CaMKII complex, whereas 93 CaMKAPs were selectively enriched in the synaptic CaMKII complex, with 30 showing little selectivity (≤ 2-fold difference in number of spectral counts between the two fractions). About half of the CaMKAPs identified in Project A were also detected in at least one of the two independent replicates (Projects B and D) (Table S3). Note that our definition of CaMKAPs as proteins that specifically co-precipitate with CaMKII is not intended to imply direct binding to CaMKII, because CaMKII holoenzymes may be components of large multiprotein complexes. However, CaMKAPs may be important downstream targets whether or not they directly interact with CaMKII.
Table 1.
Spectral counts for CaMKII and CaMKAPs identified in membrane-associated (S2), and synaptic (S3) fractions prepared from WT mouse forebrain.
| Protein Name | Gene | Alternate Name | Uniprot ID | S2 WT | S3/P3 WT | WT/ IgG | Reps? | % coverage |
|---|---|---|---|---|---|---|---|---|
| Calcium/calmodulin-dependent protein kinase type II subunit alpha | Camk2a | CaMKIIα | P11798 | 2106 | 663 | 8.2 | Yes | 93% |
| Calcium/calmodulin-dependent protein kinase type II subunit beta | Camk2b | CaMKIIβ | P28652 | 893 | 450 | 9.7 | Yes | 82% |
| Calcium/calmodulin-dependent protein kinase type II subunit gamma | Camk2g | CaMKIIγ | Q6PHZ2 | 141 | 67 | 26.0 | Yes | 69% |
| Calcium/calmodulin-dependent protein kinase type II subunit delta | Camk2d | CaMKIIδ | Q923T9 | 253 | 105 | 71.6 | Yes | 61% |
| Brain-specific angiogenesis inhibitor 1-associated protein 2 | Baiap2 | IRSp53 | Q8BKX1 | 0 | 364 | ∞ | Yes | 65% |
| Ankyrin repeat and sterile alpha motif domain-containing protein 1B | Anks1b | AIDA-1 | Q8BIZ1 | 8 | 195 | ∞ | Yes | 13% |
| SH3 and multiple ankyrin repeat domains protein 3 | Shank3 | Shank3 | Q4ACU6 | 3 | 88 | ∞ | Yes | 46% |
| TNF receptor-associated factor 3 | Traf3 | Craf1 | Q60803 | 0 | 88 | ∞ | Yes | 35% |
| IQ motif and SEC7 domain-containing protein 2 | Iqsec2 | Iqsec2 | Q5DU25 | 3 | 54 | ∞ | Yes | 35% |
| Disks large homolog 2 | Dlg2 | PSD-93/Chapsyn-110 | Q91XM9 | 8 | 49 | ∞ | Yes | 54% |
| SH3 and multiple ankyrin repeat domains protein 1 | Shank1 | Shank1 | D3YZU1 | 5 | 39 | ∞ | Yes | 26% |
| Tubulin beta-4A chain | Tubb4a | β-4A tubulin | Q9D6F9 | 3 | 34 | ∞ | No | 43% |
| Glutamate [NMDA] receptor subunit zeta-1 | Grin1 | NMDAR NR1 (GluN1) | P35438 | 8 | 31 | ∞ | Yes | 29% |
| SH3 and multiple ankyrin repeat domains protein 2 | Shank2 | Shank2 | Q80Z38 | 4 | 28 | ∞ | Yes | 29% |
| Dynamin-1-like protein | Dnm1l | Dymple | Q8K1M6 | 3 | 28 | ∞ | Yes | 43% |
| Glutamate [NMDA] receptor subunit epsilon-2 | Grin2b | NMDAR NR2B (GluN2B) | Q01097 | 11 | 27 | ∞ | Yes | 23% |
| Tubulin beta-3 chain | Tubb3 | β-3 tubulin | Q9ERD7 | 3 | 25 | ∞ | Yes | 41% |
| Glutamate [NMDA] receptor subunit epsilon-1 | Grin2a | NMDAR NR2A (GluN2A) | P35436 | 1 | 25 | ∞ | No | 22% |
| Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | Ppp2r1a | PP2A subunit A isoform R1-α | Q76MZ3 | 0 | 24 | ∞ | No | 15% |
| Disks large-associated protein 3 | Dlgap3 | SAPAP3 | Q6PFD5 | 2 | 22 | ∞ | Yes | 30% |
| H/ACA ribonucleoprotein complex subunit 4 | Dkc1 | Dyskerin | Q9ESX5 | 0 | 22 | ∞ | No | 16% |
| IQ motif and SEC7 domain-containing protein 1 | Iqsec1 | Iqsec1 | Q8R0S2 | 6 | 21 | ∞ | Yes | 27% |
| Limbic system-associated membrane protein | Lsamp | Lsamp | Q8BLK3 | 6 | 21 | ∞ | No | 11% |
| Ataxin-10 | Ataxin10 | Brain protein E46 | P28658 | 0 | 21 | ∞ | Yes | 17% |
| Calcium/calmodulin-dependent 3′,5′-cyclic nucleotide phosphodiesterase 1B | Pde1b | Cam-PDE 1B | Q01065 | 0 | 21 | ∞ | No | 19% |
| Disks large-associated protein 1 | Dlgap1 | SAPAP1 | Q9D415 | 3 | 20 | ∞ | Yes | 30% |
| Elongation factor 1-alpha 2 | Eef1a2 | EF-1-α-2 | P62631 | 0 | 20 | ∞ | Yes | 18% |
| Leucine-rich repeat-containing protein 7 | Lrrc7 | Densin | Q80TE7 | 24 | 19 | ∞ | No | 23% |
| Disks large-associated protein 4 | Dlgap4 | SAPAP4 | B1AZP2 | 3 | 18 | ∞ | No | 25% |
| Synapsin-2 | Syn2 | Synapsin-2 | Q64332 | 2 | 18 | ∞ | Yes | 18% |
| Disks large-associated protein 2 | Dlgap2 | SAPAP2 | Q8BJ42 | 1 | 18 | ∞ | Yes | 24% |
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 6 | Enpp6 | E-NPP 6 | Q8BGN3 | 0 | 16 | ∞ | No | 16% |
| Actin-binding LIM protein 1 | Ablim1 | abLim-1 | Q8K4G5 | 0 | 15 | ∞ | No | 18% |
| NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial | Ndufv1 | CI-51kD | Q91YT0 | 0 | 15 | ∞ | No | 12% |
| Elongation factor 1-gamma | Eef1g | EF-1-γ | Q9D8N0 | 1 | 14 | ∞ | Yes | 15% |
| Calcium/calmodulin-dependent 3′, 5′-cyclic nucleotide phosphodiesterase 1A | Pde1a | Cam-PDE 1A | Q61481 | 0 | 14 | ∞ | No | 13% |
| Eukaryotic initiation factor 4A-II | Eif4a2 | eIF-4A-II | P10630 | 0 | 14 | ∞ | Yes | 28% |
| Myosin-14 | Myh14 | Non-muscle myosin heavy chain IIc | Q6URW 6 | 3 | 13 | ∞ | No | 18% |
| Signal-induced proliferation-associated 1-like protein 1 | Sipa1l1 | SIPA1-like protein 1 | Q8C0T5 | 0 | 13 | ∞ | No | 12% |
| Citron Rho-interacting kinase | Cit | CRIK | P49025 | 0 | 12 | ∞ | No | 7% |
| 40S ribosomal protein S6 | Rps6 | 40S ribosomal protein 6 | P62754 | 6 | 11 | ∞ | No | 21% |
| PH and SEC7 domain-containing protein 1 | Psd | Exchange factor for ARF6 | Q5DTT2 | 1 | 11 | ∞ | No | 6% |
| Brain-enriched guanylate kinase-associated protein | Begain | Begain | Q68EF6 | 0 | 11 | ∞ | No | 21% |
| Tight junction protein ZO-1 | Tjp1 | Zo-1 | P39447 | 0 | 11 | ∞ | No | 8% |
| Connector enhancer of kinase suppressor of ras 2 | Cnksr2 | Connector enhancer of KSR 2 | Q80YA9 | 4 | 10 | ∞ | No | 9% |
| T-complex protein 1 subunit alpha | Tcp1 | TCP-1-α | P11983 | 2 | 10 | ∞ | Yes | 10% |
| Neurochondrin | Ncdn | Norbin | Q9Z0E0 | 1 | 10 | ∞ | Yes | 21% |
| Contactin-associated protein 1 | Cntnap1 | Cell recognition molecule Caspr1 | O54991 | 0 | 10 | ∞ | No | 9% |
| Protein prune homolog | Prune | PRUNEM1 | Q8BIW1 | 0 | 10 | ∞ | No | 6% |
| Unconvential myosin-XVIIIa | Myo18a | Myosin-18a | Q9JMH9 | 9 | 9 | ∞ | Yes | 11% |
| F-actin-capping protein subunit alpha-2 | Capza2 | CapZ α-2 | P47754 | 3 | 9 | ∞ | No | 43% |
| Synaptosomal-associated protein 47 | Snap47 | SNAP-47 | Q8R570 | 0 | 9 | ∞ | No | 26% |
| Band 4.1-like protein 3 | Epb41l3 | 4.1B | Q9WV92 | 3 | 8 | ∞ | No | 13% |
| Protein piccolo | Pclo | Piccolo | Q9QYX7 | 3 | 8 | ∞ | Yes | 3% |
| Choline transporter-like protein 1 | Slc44a1 | Solute carrier family 44 member 1 | Q6X893 | 0 | 8 | ∞ | No | 5% |
| Heat shock protein HSP 90-alpha | Hsp90aa1 | HSP 90-α | P07901 | 4 | 7 | ∞ | No | 3% |
| Dynamin-3 | Dnm3 | Dynamin-3 | Q8BZ98 | 0 | 7 | ∞ | No | 16% |
| Serine/threonine-protein kinase OSR1 | Oxsr1 | Oxidative stress-responsive 1 protein | Q6P9R2 | 0 | 7 | ∞ | No | 6% |
| Protein shisa-7 | Shisa7 | Shisa-7 | Q8C3Q5 | 4 | 6 | ∞ | No | 5% |
| Uncharacterized protein C9orf172 homolog | Gm996 | Uncharacterized protein C9orf172 homolog | A2AJA9 | 3 | 6 | ∞ | No | 10% |
| Protein unc-13 homolog A | Unc13a | Munc13-1 | Q4KUS2 | 2 | 6 | ∞ | Yes | 5% |
| T-complex protein 1 subunit zeta | Cct6a | TCP-1-ζ | P80317 | 2 | 6 | ∞ | No | 4% |
| 60S ribosomal protein L7a | Rpl7a | Surfeit locus protein 3 | P12970 | 9 | 4 | ∞ | No | 26% |
| Diacylglycerol kinase epsilon | Dgke | DAG kinase ε | Q9R1C6 | 7 | 4 | ∞ | No | 5% |
| Sideroflexin-3 | Sfxn3 | Sideroflexin-3 | Q91V61 | 6 | 4 | ∞ | Yes | 28% |
| Ankyrin-2 | Ank2 | Ankyrin-2 | Q8C8R3 | 3 | 4 | ∞ | No | 2% |
| Regulating synaptic membrane exocytosis protein 2 | Rims2 | Rab-3-interacting molecule 2 | Q9EQZ7 | 3 | 4 | ∞ | No | 4% |
| Mitochondrial glutamate carrier 1 | Slc25a22 | GC-1 | Q9D6M3 | 11 | 3 | ∞ | Yes | 4% |
| Fibrinogen beta chain | Fgb | Fibrinogen beta chain | Q8K0E8 | 8 | 3 | ∞ | Yes | 7% |
| Neurabin-2 | Ppp1r9b | Spinophilin | Q6R891 | 6 | 1 | ∞ | No | 20% |
| Phosphorylase b kinase regulatory subunit beta | Phkb | Phosphorylase kinase subunit β | Q7TSH2 | 21 | 0 | ∞ | Yes | 21% |
| 6-phosphofructokinase, muscle type | Pfkm | PFK-M | P47857 | 10 | 0 | ∞ | Yes | 14% |
| Homer protein homolog 1 | Homer1 | Homer-1 | Q9Z2Y3 | 45 | 277 | 46.0 | Yes | 81% |
| Dynamin-1 | Dnm1 | Dynamin-1 | P39053 | 5 | 35 | 40.0 | Yes | 37% |
| Leucine-rich glioma-inactivated protein 1 | Lgi1 | Epitempin-1 | Q9JIA1 | 12 | 23 | 35.0 | Yes | 17% |
| Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 2 | Agap2 | PIKE | Q3UHD9 | 20 | 15 | 35.0 | Yes | 27% |
| Tubulin beta-4B chain | Tubb4b | β-4B tubulin | P68372 | 49 | 299 | 26.8 | No | 55% |
| Vimentin | Vim | Vimentin | P20152 | 1 | 71 | 24.0 | Yes | 50% |
| Claudin-11 | Cldn11 | Oligodendrocyte -specific protein | Q60771 | 0 | 92 | 23.0 | No | 23% |
| T-complex protein 1 subunit theta | Cct8 | TCP-1-θ | P42932 | 0 | 20 | 20.0 | No | 18% |
| Tubulin beta-2A chain | Tubb2a | β-2A tubulin | Q7TMM 9 | 81 | 305 | 19.3 | Yes | 73% |
| Tubulin beta-5 chain | Tubb5 | β-5 tubulin | P99024 | 11 | 64 | 18.8 | Yes | 65% |
| Disks large homolog 4 | Dlg4 | PSD-95 | Q62108 | 10 | 82 | 18.4 | Yes | 62% |
| Microtubule-associated protein 6 | Map6 | MAP-6 | Q7TSJ2 | 20 | 16 | 18.0 | Yes | 36% |
| Actin-related protein 3 | Actr3 | Arp-3 | Q99JY9 | 2 | 16 | 18.0 | No | 19% |
| Heat shock protein HSP 90-beta | Hsp90ab1 | HSP 90-β | P11499 | 5 | 13 | 18.0 | Yes | 23% |
| 60S ribosomal protein L3 | Rpl3 | J1 protein | P27659 | 7 | 10 | 17.0 | No | 23% |
| Heat shock 70 kDa protein 12A | Hspa12a | Heat shock 70 kDa protein 12A | Q8K0U4 | 4 | 12 | 16.0 | Yes | 22% |
| Vesicle-fusing ATPase | Nsf | NEM-sensitive fusion protein | P46460 | 5 | 11 | 16.0 | Yes | 16% |
| Ras GTPase-activating protein SynGAP | Syngap1 | Synaptic Ras-GAP 1 | F6SEU4 | 20 | 135 | 15.5 | No | 65% |
| Unconventional myosin-Id | Myo1d | Myosin-Id | Q5SYD0 | 0 | 27 | 13.5 | Yes | 29% |
| AP-2 complex subunit alpha-1 | Ap2a1 | Adaptor protein complex AP-2 subunit α-1 | P17426 | 10 | 17 | 13.5 | Yes | 23% |
| Tubulin alpha-1C chain | Tuba1c | α-1C tubulin | P68373 | 171 | 381 | 13.5 | Yes | 49% |
| CLIP-associating protein 2 | Clasp2 | Cytoplasmic linker-associated protein 2 | Q8BRT1 | 2 | 11 | 13.0 | No | 12% |
| 6-phosphofructokinase type C | Pfkp | PFK-C | Q9WUA 3 | 10 | 3 | 13.0 | Yes | 20% |
| Cytoplasmic dynein 1 heavy chain 1 | Dync1h1 | Cytoplasmic dynein heavy chain 1 | Q9JHU4 | 5 | 31 | 12.0 | Yes | 10% |
| Transcriptional activator protein Pur-beta | Purb | Purine-rich element-binding protein B | O35295 | 10 | 2 | 12.0 | No | 60% |
| Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform | Phka1 | Phosphorylase kinase α M subunit | P18826 | 12 | 0 | 12.0 | Yes | 18% |
| 60S ribosomal protein L4 | Rpl4 | 60S ribosomal protein L4 | Q9D8E6 | 27 | 73 | 11.1 | No | 40% |
| Neurotrimin | Ntm | Hnt | Q99PJ0 | 1 | 10 | 11.0 | No | 9% |
| V-type proton ATPase catalytic subunit A | Atp6v1a | V-ATPase subunit A | P50516 | 4 | 7 | 11.0 | No | 14% |
| Flotillin-1 | Flot1 | Flotillin-1 | O08917 | 0 | 53 | 10.6 | No | 46% |
| Neural cell adhesion molecule 1 | Ncam1 | NCAM-1 | P13595 | 3 | 7 | 10.0 | No | 7% |
| Probable ATP-dependent RNA helicase DDX5 | Ddx5 | DEAD box RNA helicase DEAD1 | Q61656 | 4 | 6 | 10.0 | Yes | 14% |
| Plectin | Plec | Plectin-1 | Q9QXS1 | 8 | 91 | 9.0 | Yes | 23% |
| Regulating synaptic membrane exocytosis protein 1 | Rims1 | Rab-3-interacting molecule 1 | Q99NE5 | 1 | 8 | 9.0 | No | 13% |
| 60S ribosomal protein L8 | Rpl8 | 60S ribosomal protein L8 | P62918 | 6 | 3 | 9.0 | No | 22% |
| Synaptic vesicle glycoprotein 2A | Sv2a | Synaptic vesicle protein 2 | Q9JIS5 | 9 | 0 | 9.0 | Yes | 16% |
| Spectrin beta chain, brain 1 | Sptbn1 | β-II spectrin | Q62261 | 29 | 58 | 8.7 | Yes | 29% |
| Tropomodulin-2 | Tmod2 | Neuronal tropomodulin | Q9JKK7 | 8 | 9 | 8.5 | Yes | 30% |
| 60S ribosomal protein L6 | Rpl6 | 60S ribosomal protein L6 | P47911 | 10 | 7 | 8.5 | No | 30% |
| Myelin proteolipid protein | Plp1 | Lipophilin | P60202 | 3 | 47 | 8.3 | No | 19% |
| Contactin-1 | Cntn1 | Neural cell surface protein F3 | P12960 | 9 | 15 | 8.0 | No | 20% |
| Nesprin-1 | Syne1 | Enaptin | Q6ZWR 6 | 1 | 7 | 8.0 | Yes | 1% |
| Gamma-aminobutyric acid receptor subunit alpha-1 | Gabra1 | GABA(A) receptor subunit α-1 | P62812 | 3 | 5 | 8.0 | Yes | 8% |
| Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | Ppp1cc | Protein phosphatase 1 γ | P63087 | 6 | 2 | 8.0 | No | 17% |
| Putative oxidoreductase GLYR1 | Glyr1 | Glyoxylate reductase 1 homolog | Q922P9 | 0 | 7 | 7.0 | No | 9% |
| Phosphate carrier protein, mitochondrial slc25a3 | Slc25a3 | Phosphate transport protein | Q8VEM 8 | 6 | 1 | 7.0 | Yes | 22% |
| Spectrin alpha chain, brain | Sptan1 | α-II spectrin | P16546 | 45 | 93 | 6.9 | No | 43% |
| Myosin-10 | Myh10 | Non-muscle myosin heavy chain IIb | Q61879 | 74 | 68 | 6.5 | Yes | 50% |
| Neurofilament heavy polypeptide | Nfh | 200 kDa neurofilament protein | P19246 | 2 | 17 | 6.3 | Yes | 18% |
| ATPase family AAA domain-containing protein 3 | Atad3 | AAA-ATPase TOB3 | Q925I1 | 8 | 10 | 6.0 | Yes | 18% |
| Serine/threonine-protein kinase DCLK1 | Dclk1 | Doublecortin-like and CAM kinase-like 1 | Q9JLM8 | 3 | 9 | 6.0 | No | 13% |
| Mitochondrial 2-oxoglutarate/malate carrier protein | Slc25a11 | Solute carrier family 25 member 11 | Q9CR62 | 7 | 5 | 6.0 | Yes | 21% |
| Neuronal growth regulator 1 | Negr1 | Kindred of IgLON | Q80Z24 | 4 | 13 | 5.7 | No | 9% |
| Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Ppp3ca | Calmodulin-dependent calcineurin A subunit α isoform | P63328 | 0 | 16 | 5.3 | No | 13% |
| ERC protein 2 | Erc2 | CAZ-associated structural protein 1 | Q6PH08 | 4 | 17 | 5.3 | Yes | 21% |
| Unconventional Myosin-Va | Myo5a | Dilute myosin heavy chain, non-muscle | Q99104 | 47 | 75 | 5.1 | Yes | 37% |
| Synapsin I | Syn1 | Synapsin I | O88935 | 1 | 14 | 5.0 | No | 34% |
| Heterochromatin protein 1-binding protein 3 | Hp1bp3 | Heterochromatin protein 1-binding protein 3 | Q3TEA8 | 2 | 8 | 5.0 | No | 22% |
| Calcium-binding mitochondrial carrier protein Aralar1 | Slc25a12 | Mitochondrial aspartate glutamate carrier 1 | Q8BH59 | 33 | 30 | 4.5 | Yes | 49% |
| Microtubule-associated protein 1B | Map1b | MAP1(X) | P14873 | 10 | 8 | 4.5 | Yes | 10% |
| Elongation factor 1-alpha 1 | Eef1a1 | Elongation factor Tu | P10126 | 12 | 65 | 4.3 | No | 22% |
| 40S ribosomal protein S3a | Rps3a | Protein TU-11 | P97351 | 11 | 6 | 4.3 | No | 35% |
| Neurofilament light polypeptide | Nfl | 68 kDa neurofilament protein | P08551 | 13 | 135 | 4.2 | Yes | 62% |
| Tubulin alpha-4A chain | Tuba4a | α-4A tubulin | P68368 | 12 | 30 | 4.2 | Yes | 53% |
| Sodium/potassium-transporting ATPase subunit beta-1 | Atp1b1 | Sodium/potassiu m-dependent ATPase subunit β-1 | P14094 | 20 | 13 | 4.1 | Yes | 16% |
| AP-2 complex subunit beta | Ap2b1 | Adaptor protein complex AP-2 subunit β | Q9DBG3 | 18 | 19 | 4.1 | Yes | 17% |
| Microtubule-associated protein 1A | Map1a | Microtubule-associated protein 1A | Q9QYR6 | 41 | 4 | 4.1 | No | 18% |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | Ndufa10 | NADH-ubiquinone oxidoreductase 42 kDa subunit | Q99LC3 | 0 | 8 | 4.0 | No | 27% |
| Unconventional myosin-VI | Myo6 | Unconventional myosin-6 | Q64331 | 2 | 6 | 4.0 | Yes | 9% |
| Guanine nucleotide-binding protein G(z) subunit alpha | Gnao1 | Guanine nucleotide-binding protein G(z) subunit alpha | P18872 | 6 | 2 | 4.0 | No | 41% |
Gene names, Alternate Protein Names, and Uniprot IDs are shown, along with the number of spectral counts detected in CaMKII complexes isolated from S2 and S3 fractions of WT CaMKII. The WT/IgG ratio was calculated by dividing the total number of spectra detected in CaMKII immunoprecipitates of the S2 AND S3 fractions by the total number of spectra detected in the corresponding IgG controls. Only CaMKAPs with ≥7 spectral counts in the S2 and S3 fractions combined, and a WT/IgG ratio of ≥4 are listed. The “Replicated” column indicates whether the protein was detected in independent biological sample. The final column indicates the highest percentage coverage of the entire CaMKAP in the analysis of a single gel region from a single subcellular fraction of either a WT or T286A-KI mouse brain.
Several synaptic CaMKAPs identified here were previously shown to directly interact with CaMKII, validating our approach. For example, NMDA receptor subunits (GluN1, GluN2B, and GluN2A), densin (LRRC7), spinophilin (PPP1R9b; neurabin-2), and myosin Va can directly bind CaMKII and modulate kinase activity and/or subcellular localization.19–21, 47–51 CaMKII interaction with GluN2B was recently shown to be important for normal synaptic plasticity.52–54 However, some previously characterized CaMKAPs were either undetected (e.g., diacylglycerol lipase-α55) or did not meet our rigorous cutoffs (e.g., α-actinin21). This may be due to dissociation of the interacting protein from CaMKII during subcellular fractionation, weakness of the interaction in the tissue analyzed here, or low abundance. Moreover, some weaker interactions may be easier to detect when the CaMKAP, rather than CaMKII, is immunoprecipitated (e.g., diacylglycerol lipase-α55). However, most CaMKAPs detected here have not been previously reported to associate with CaMKII, including several synaptic scaffolding proteins (e.g., SAPAP1/2/3/4, Shank1/2/3, and Brain-enriched guanylate kinase-associated protein) and proteins with other functions (e.g., brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2)); TNF receptor-associated factor 3; multiple subunits of phosphorylase b kinase; Limbic system-associated membrane protein; Eukaryotic initiation factor 4A (eIF4A); and phosphodiesterase 1. Additional studies will be required to ascertain whether these novel CaMKAPs directly bind to CaMKII, or are components of larger macromolecular complexes.
To provide initial insight into functionally related groupings and roles of the CaMKAPs, the STRING database of mouse proteins, containing all but one CaMKAP (SynGAP), was used to assign CaMKAPs into 14 groups based on known associations and functions (CaMKII; NMDARs; synaptic scaffolds; myosins; tubulin and microtubules; actin cytoskeleton; ribosome and translation; ATPase/GTPase; metabolism and mitochondria; junction and myelin; general signaling, intermediate filaments; vesicle trafficking; and other) (Fig. 5). The top Kyoto Encyclopedia of Genes and Genomes (KEGG; www.kegg.jp56) pathways in this network were then identified using WebGestalt57, 58 (Table S5), including Ca2+ signaling, long-term potentiation, and the ribosome. Taken together, these analyses emphasize that CaMKII is involved in diverse pathways in multiple cellular compartments. However, additional studies will be required to identify specific functional roles of the interactions.
Figure 5. A CaMKAP interaction map.
The STRING-db identifies interactions utilizing genomic context, high-throughput experiments, co-expression, and previous knowledge. Default parameters were used to generate this map. Associations from multiple sources and/or contexts have thicker lines. Proteins were arbitrarily assigned to 14 different groups based on protein function and interaction data.
Effect of the T286A-KI mutation on CaMKII interactions
In order to provide additional perspective about the biological relevance of CaMKAPs, we isolated and analyzed in parallel synaptic CaMKII complexes from WT and T286A-KI mice. Since T286A-KI mice display robust changes in synaptic plasticity and diverse behavioral deficits,22, 26 we hypothesized that changes in the CaMKII interactome would provide insights into molecular mechanisms underlying behavioral and synaptic phenotypes. Consistent with a prior observation that synaptic levels of CaMKIIα are reduced in T286A-KI mice relative to WT,26 the number of CaMKII spectral counts in synaptic T286A-KI complexes were reduced by ~33% relative to WT complexes. Therefore, to provide an initial semi-quantitative comparison of the relative levels of CaMKAPs associated with CaMKII holoenzymes in WT and T286A-KI synaptic fractions, we normalized the number of CaMKAP spectral counts to the number of CaMKII spectral counts (all isoforms), and then expressed normalized values as a KI/WT ratio. KI/WT ratios of between 0.6 and 1.4 for the majority (10 out of 18) of CaMKAPs, indicated minimal changes in relative association. However, average KI/WT ratios across two biological replicates were ≥1.40 for eIF4A, GluN1, GluN2B, PSD-95, SAPAP1, SAPAP2, and Shank3, but less than 0.6 for only one CaMKAP, BAIAP2 (Table 2).
Table 2.
Comparison of CaMKII and CaMKAPs in synaptic S3 fractions isolated in parallel from forebrains of WT and T286A-KI mice.
| Spectral Counts | Normalized Spectral Counts | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Project A | Project B | Project A | Project B | ||||||||
| WT | KI | WT | KI | WT | KI | Ratio KI/WT | WT | KI | Ratio KI/WT | Avg ratio | |
| CaMKIIα | 663 | 443 | 399 | 319 | 0.67 | 0.80 | 0.73 | ||||
| CaMKII all isoforms | 1285 | 1052 | 419 | 344 | 0.82 | 0.82 | 0.82 | ||||
| NMDARs | |||||||||||
| GluN1 | 31 | 44 | 9 | 10 | 0.024 | 0.042 | 1.73 | 0.021 | 0.029 | 1.35 | 1.54 |
| GluN2B | 27 | 34 | 8 | 10 | 0.021 | 0.032 | 1.54 | 0.019 | 0.029 | 1.52 | 1.53 |
| GluN2A | 25 | 18 | ND | ND | 0.019 | 0.017 | 0.88 | NA | NA | NA | 0.88 |
| Synaptic Scaffolding Proteins | |||||||||||
| Homer 1 | 277 | 250 | 73 | 69 | 0.216 | 0.238 | 1.10 | 0.174 | 0.201 | 1.15 | 1.13 |
| PSD-93 | 49 | 46 | 8 | 8 | 0.038 | 0.044 | 1.15 | 0.019 | 0.023 | 1.22 | 1.18 |
| PSD-95 | 82 | 105 | 20 | 27 | 0.064 | 0.100 | 1.56 | 0.048 | 0.078 | 1.64 | 1.60 |
| SAPAP1 | 20 | 27 | 6 | 7 | 0.016 | 0.026 | 1.65 | 0.014 | 0.020 | 1.42 | 1.54 |
| SAPAP2 | 18 | 21 | 3 | 5 | 0.014 | 0.020 | 1.43 | 0.007 | 0.015 | 2.03 | 1.73 |
| SAPAP3 | 22 | 25 | 8 | 7 | 0.017 | 0.024 | 1.39 | 0.019 | 0.020 | 1.07 | 1.23 |
| Shank1 | 39 | 43 | 7 | 3 | 0.030 | 0.041 | 1.35 | 0.017 | 0.009 | 0.52 | 0.93 |
| Shank2 | 28 | 25 | 5 | 1 | 0.022 | 0.024 | 1.09 | 0.012 | 0.003 | 0.24 | 0.67 |
| Shank3 | 88 | 82 | 11 | 15 | 0.068 | 0.078 | 1.14 | 0.026 | 0.044 | 1.66 | 1.40 |
| Cytoskeletal Proteins | |||||||||||
| BAIAP2 | 364 | 147 | 38 | 0 | 0.283 | 0.140 | 0.49 | 0.091 | 0.000 | 0.00 | 0.25 |
| Myosin Va | 75 | 63 | 38 | 29 | 0.058 | 0.060 | 1.03 | 0.091 | 0.084 | 0.93 | 0.98 |
| Myosin-Id | 27 | 20 | 6 | 2 | 0.021 | 0.019 | 0.90 | 0.014 | 0.006 | 0.41 | 0.66 |
| Spectrin beta chain | 58 | 46 | 3 | 1 | 0.045 | 0.044 | 0.97 | 0.007 | 0.003 | 0.41 | 0.69 |
| Tubulin all chains | 1167 | 600 | 140 | 85 | 0.908 | 0.570 | 0.63 | 0.334 | 0.247 | 0.74 | 0.68 |
| Translation Factor | |||||||||||
| EIF4A (all isoforms) | 17 | 89 | 1 | 7 | 0.013 | 0.085 | 6.39 | 0.002 | 0.020 | 8.53 | 7.46 |
The table shows the total number of spectral counts detected in all excised gel regions in projects A and B identified using either an Orbitrap Velos or Orbitrap XL. Spectral counts for each CaMKAP were normalized to the number of spectral counts derived from all isoforms of CaMKII in the corresponding project. The average normalized KI/WT ratio and the range of ratios across the biological replicates are also shown. Proteins highlighted in pink have an average KI/WT ratio greater than 1.4. The protein highlighted in green has an average KI/WT ratio less than 0.5.
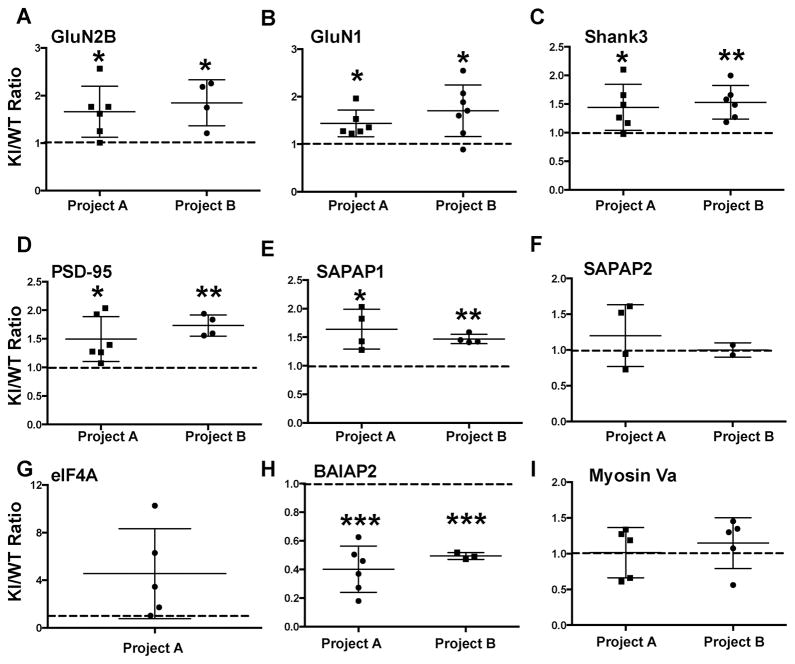
We also compared relative levels of CaMKII and CaMKAPs in WT and T286A-KI synaptic complexes by measuring AUCs of XICs for several peptides. In project A, this analysis indicated that levels of CaMKIIα and CaMKIIβ in T286A-KI complexes were 46.4±5.1% (9 peptides) and 57.3±13.8% (7 peptides) of the levels in WT complexes, respectively. In project B, levels of CaMKIIα in T286A-KI complexes were 49.5±4.7% (9 peptides) of the levels in WT complexes (CaMKIIβ was not analyzed in this project). Raw AUCs of XICs for 2–9 peptides matching each CaMKAP were then normalized to the mean CaMKIIα AUC of XIC in the same project, before calculating a KI/WT ratio. KI/WT ratios were significantly greater than 1.0 for GluN2B, GluN1, Shank3, PSD-95, and SAPAP1 in both projects A and B (Fig. 6A–E), generally similar to KI/WT ratios calculated from spectral counts (see above), with the exception of SAPAP2 (Fig 6F; Table 2). As a control, KI/WT ratios for myosin Va were not significantly different from 1.0 by AUC analysis in either run (Fig. 6I), consistent with the KI/WT ratio based on spectral counts (Table 2). A KI/WT ratio for eIF4A could be calculated in only one biological replicate because relevant MS/MS spectra could not be validated in the other analysis. The 4.5-fold increase in mean KI/WT ratio for eIF4A based on AUCs was consistent with the 7.5-fold increase based on spectral counts, although the increased AUC-based ratio was not statistically significant (Fig 6G). In contrast, mean AUC-based KI/WT ratios for BAIAP2 were significantly decreased in both projects (Fig 6H), consistent with KI/WT ratios based on spectral counts.
Figure 6. Effect of T286A-KI mutation of CaMKIIα on the association of selected CaMKAPs with CaMKII holoenzymes in the S3 fraction.
Relative levels of each CaMKAP in WT and T286A-KI samples were estimated from the AUC of XICs for multiple peptides, normalized to the relative levels of CaMKIIα (similarly estimated from AUCs for 9 peptides) and then expressed as a ratio (KI/WT). Data from 1 or 2 independent biological replicates (“projects”) are shown. Each data point is the ratio calculated for a single CaMKAP derived peptide, with the mean and S.E.M indicated. A. GluN2B, B. GluN1, C. Shank3, D. PSD-95, E. SAPAP1, F. SAPAP2, G. EIF4A, H. BAIAP2, I. Myosin Va. Data for each project were compared to a theoretical value of 1 (no difference; dashed line) using a one-column t-test. *p<0.05, **p<0.01, ***p<0.001. ****p<0.0001.
These independent analytical approaches indicate that T286A-KI mutation of CaMKIIα significantly affects an array of protein-protein interactions in S3 synaptic fractions. Since Thr286 autophosphorylation stabilizes direct interactions of CaMKII with GluN2B in vitro,19, 20 it is perhaps surprising that we detected a significant increase in KI/WT ratio for GluN2B. However, this observation is consistent with a previous analyses of NMDAR subunits association with synaptic CaMKII holoenzymes in T286A-KI mice by Western blotting.26 We also detected significant increases in KI/WT ratios for several other CaMKAPs (e.g., GluN1, PSD-95, SAPAP1, SAPAP2, Shank3, eIF4A) using both methods (Table 2; Fig 6A, B). However, KI/WT ratios for related CaMKAPs (e.g., homer, SAPAP3, shank1, shank2 and myosin Va) were essentially unchanged, suggesting that this is not a technical artifact (Table 2; Fig. 6I). Although changes in KI/WT ratios may not reflect direct effects of a lack of Thr286 autophosphorylation on an interaction with CaMKII, these data indicate a broad but selective impact of T286A-KI mutation on synaptic protein-protein interactions.
Insights into novel roles of CaMKII
The classical model of a critical role for CaMKII during LTP has been extended by recent observations that CaMKII is also involved in different forms of LTD.27, 28 The identification of 138 CaMKAPs in the present study suggests novel mechanisms of postsynaptic CaMKII signaling that may contribute to these different functions. For example, direct or indirect association of Homer and Shank proteins with CaMKII might be important for mGluR1/5-dependent signaling pathways that are critical for some forms of LTD, especially because some of these interactions are sensitive to Thr286 mutation (Fig. 6; Table 2). Thus, the current observations provide novel insights into the physiological targeting of CaMKII to distinct pathways that mediate diverse synaptic outcomes.
Several of the novel CaMKAPs detected here have been linked to neurological and/or psychiatric disorders. For example, BAIAP2 (also known as IRSp53) is highly abundant in CaMKII complexes, based on the number of spectral counts detected, and was the only CaMKAP predominantly detected in the CaMKIIα gel segment. Moreover, it was the only CaMKAP with a significantly decreased KI/WT ratio, estimated by both methods. The simplest interpretation of this observation is that BAIAP2 association with CaMKII is decreased in T286A-KI mice, although it is also possible that changes in posttranslational modifications of BAIAP2 (e.g. phosphorylation) in T286A-KI synaptic fractions affect the electrophoretic mobility of BAIAP2 to decrease the amount of BAIAP2 that essentially co-migrates with CaMKIIα. Nevertheless, BAIAP2 merits validation as a CaMKAP followed by functional analyses because of its known roles in binding PSD95, in regulating actin dynamics, filopodia formation, and excitatory synaptic transmission and in modulating learning and memory.59–62
Several novel CaMKAPs are involved in mRNA translation, and T286A-KI mutation increased relative levels of eIF4A associated with CaMKII (Table 2; Fig 6G). As part of the eIF4 translation initiation complex, eIF4A is a D-E-A-D-box RNA helicase.63, 64 Interestingly, CaMKII regulates the recruitment of eIF4GII to the eIF4 initiation complex,65 and CaMKII phosphorylation of cytoplasmic polyadenylation element-binding protein enhances protein synthesis during LTP.66 Since abnormal protein synthesis is being increasingly implicated in neurological and psychiatric disorders,67, 68 our data support additional investigation of interactions between CaMKII and the protein synthesis machinery.
Synaptic scaffolding proteins implicated in neurological and/or psychiatric disorders were also identified as novel synaptic CaMKAPs. For example, the Shank3, Dlgap2, and Syngap1 genes have been linked to Autism Spectrum Disorder (ASD),69 and specific haplotypes in the Dlgap2 gene (SAPAP2) correlate with increased risk for schizophrenia.70 Moreover, Shank3 knockout mice exhibit ASD-like behaviors,71 and some ASD-related disorders have been linked to abnormal CaMKII signaling (e.g. Angelman syndrome25, 72). Notably, we detected increased KI/WT ratios for PSD95, Shank3, and SAPAP1, indicating that these interactions are sensitive to Thr286 autophosphorylation (Table 2; Fig 6C–E). Thus, regardless of whether CaMKII interacts directly or indirectly with Shank3, SAPAPs, or Syngap1, our data suggest that T286A-KI mutation causes significant changes in postsynaptic protein architecture. The potential involvement of CaMKII in the physiological regulation of these proteins, and in associated pathologies, should be further investigated.
Summary and Conclusions
Prior studies have established that CaMKIIα and CaMKIIβ are critical for many aspects of synaptic regulation and behavior, and that CaMKIIα autophosphorylation is a key regulator of kinase activity, interactions with other proteins, and subcellular location. However, understanding specific molecular mechanisms underlying the diverse synaptic roles of CaMKII requires comprehensive information about CaMKII phosphorylation and the CaMKII interactome in situ. Here, we used an unbiased semi-quantitative proteomics approach to identify biologically relevant phosphorylation sites on both CaMKIIα and CaMKIIβ. Our data are consistent with known roles for Thr286/287 and Thr306 phosphorylation in regulating CaMKII localization. Additional novel sites are differentially phosphorylated in the subcellular fractions, and analyses of T286A-KI mice suggest an inter-dependence of some phosphorylation events and/or compensatory changes in response to mutation. These data suggest a complex physiological interdependence between distinct subcellular pools of CaMKII. Using the same unbiased approach, we also identified 138 CaMKAPs linked to diverse cellular functions, the majority of which (~100) are preferentially associated with synaptic CaMKII holoenzymes. Notably, all the CaMKAPs appear to be present at sub-stoichiometric levels relative to CaMKII itself (Fig. 2A), consistent with a model in which subpopulations of CaMKII holoenzymes are associated with different CaMKAPs to subserve distinct roles. Moreover, interactions with CaMKAPs appear to be differentially affected by T286A-KI mutation of CaMKIIα, providing insight into molecular changes that may contribute to well-established synaptic and behavioral phenotypes of these mice. In combination, these findings should promote future studies to identify novel CaMKII-regulated pathways involved in the normal and pathological modulation of synaptic physiology, and various forms of learning and memory.
Materials and Methods
DNA constructs and CaMKIIα/β expression
Murine CaMKIIα (Uniprot #P11798) was expressed in Sf9 insect cells.73 Rat CaMKIIβ, with an Ala inserted at position 341 in the canonical sequence (Uniprot #P08413), was PCR amplified from a construct provided by Dr. L. Redmond Hardy33 and inserted into pcDNA3.1 for transfection into HEK293 cells growing in suspension. Thus, residue numbering of rat CaMKIIβ used for in vitro studies is increased by one relative to the canonical sequence after the Ala insertion site. Both isoforms were purified to ≥95% purity, essentially as described.73,74.
In vitro autophosphorylation of CaMKII
Purified recombinant CaMKIIα and CaMKIIβ (5.5 and 2.5 μM, respectively) were incubated for 2 min at 30°C in 50 mM HEPES pH 7.5, 2 mM DTT in the absence or presence of 10 mM Mg(CH3COO)2, 1.5 mM CaCl2, 10 μM calmodulin, 500 μM ATP (basal or Ca2+/CaM conditions, respectively). A third sample was first autophosphorylated in the presence of Ca2+/calmodulin prior to the addition of EGTA (4 mM final) to allow continued Ca2+-independent autophosphorylation for 2 min at 30°C (Ca2+/CaM then EGTA; sequential). All reactions were terminated by addition of EDTA (25 mM final) and then mixed with trichloracetic acid (10% (w/v) final). After incubation on ice for 30 minutes and centrifugation (10,000 x g for 15 min), protein pellets were washed by resuspension in cold acetone and then re-centrifuged.
Mice
T286A-KI or WT littermates (male: 3–6 months of age) bred from heterozygous breeding pairs on a C57BL/6J background (as described previously26) were used for separate comparative proteomics analysis. All animal protocols were approved by the Vanderbilt Institutional Care and Use Committee.
Immunoprecipitations from whole forebrain fractions
Cytosolic (S1), Triton-soluble membrane (S2), and Triton/deoxycholate soluble synaptic (S3) fractions of mouse forebrain75 (~3 mg total protein each) were immunoprecipitated using a polyclonal goat CaMKII antibody (5.4 μg) or a goat IgG control (purified from pre-immune serum by ammonium sulfate precipitation).51, 73 The CaMKII antibody exhibits approximately equivalent sensitivity for purified α, β, γ and δ isoforms of CaMKII in immunoblots. Since CaMKII constitutes ~1% of total forebrain protein, each input contains ~30 μg of CaMKII, likely saturating the antibody during immunoprecipitation, consistent with the similar SYPRO Ruby staining intensity for IgG and CaMKII bands in the immunoprecipitation lanes in Fig. 2A.
Immunoblotting
Mouse forebrain subcellular fractions were immunoblotted using phospho-Thr286-specific (Santa Cruz, 12886-R; 1:1000-1:2000 dilution) and total (Thermo-Pierce, MA1-048; 1:4000-1:10000 dilution) CaMKIIα antibodies.75
Mass spectrometric analysis
CaMKII and control immunoprecipitates were resolved by SDS-PAGE and stained with either SYPRO Ruby (Life Technologies) or colloidal blue stain (Life Technologies). Each gel lane was excised in segments, corresponding to the CaMKIIα or CaMKIIβ regions and two CaMKAP regions below CaMKIIα and above CaMKIIβ (Fig. 2A). Thus, the gel segment containing the highly abundant IgG heavy chain was excluded from all subsequent analyses in order to minimize the suppression of MS signals from less abundant proteins. In some replicates, only the CaMKIIα and/or CaMKIIβ segments were analyzed for phosphorylation sites, whereas other replicates analyzed all four segments to detect co-precipitating proteins. Gel segments were incubated with 100 mM ammonium bicarbonate, pH 8, reduced with 4 mM DTT or TCEP, alkylated with 8 mM iodoacetamide and digested overnight with trypsin (10 ng/μl; 37 °C). Extracted peptides were dissolved in 0.1% formic acid and resolved using a reverse-phase C18 capillary column (360 μm o.d and 100 μm i.d.) packed with Jupiter beads (3-μm, 300 Å; Phenomenex) and equipped with a laser-pulled emitter tip using an Eksigent Ultra LC and autosampler. Mobile phases were 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% acetonitrile (solvent B). The elution gradient (500 nl/min) was: 0–10 min, 2% B; 10–50 min, 2–35% B; 50–60 min, 35–90% B; 60–65 min, 90% B, 65–70 min, 90–2% B, 70–90 min, 2% B. Eluted peptides were analyzed on LTQ Orbitrap XL or LTQ Orbitrap Velos mass spectrometers (Thermo Scientific) using a data-dependent method with dynamic exclusion enabled. Full scan (m/z 300–2,000) spectra were acquired with the Orbitrap as the mass analyzer, and the most abundant ions (5 on the LTQ Orbitrap XL or 12 on the LTQ Orbitrap Velos) in each MS scan were selected for fragmentation via collision-induced dissociation (CID). All tandem mass spectra were converted into DTA files using Scansifter and matched to a mouse subset of the UniProtKB protein database (also containing reversed (decoy) protein sequences) using a custom version of SEQUEST76 on the Vanderbilt ACCRE Linux cluster. The results were assembled in Scaffold 3 (Proteome Software) with minimum filtering criteria of 95% peptide probability.46 Searches were configured to use variable modifications of cysteine carbamidomethylation, methionine oxidation, and serine, threonine, and tyrosine phosphorylation. Modifications were validated by manual inspection of raw tandem mass spectra using QualBrowser (Xcalibur 2.1.0, Thermo Scientific) (Fig. S1, S2). Peptides originating from CaMKIIα and CaMKIIβ were matched to the canonical mRNA splice variants in the UniProt database (P11798 and P28652, respectively).
Spectral Counting from Assembled Scaffold Files
Relative protein abundance in samples from WT and T286A-KI mice was estimated from the total number of spectral counts (in all regions of the gel analyzed in a given biological replicate) matching a specific protein based on Scaffold files, using both unmodified and modified (i.e. cysteine alkylation, methionine oxidation, STY phosphorylation) spectra. The global list of ranked proteins, with common contaminants (e.g., keratin) deleted, separated by the segment of the parent gel in which they were identified is available online as Table S3, which also lists the number of spectral counts detected for each protein in each segment and project. For normalizing CaMKAPs from WT and KI synaptic fractions, the total number of spectral counts matching a specific CaMKAP was divided by the total number of spectral counts matching all of the CaMKII isoforms isolated from the synaptic fraction of the same biological replicate.
Extracted Ion Chromatogram (XIC) Analysis
Accurate mass measurements were used to generate extracted ion chromatograms (XICs) with a 10 PPM tolerance for non-phosphorylated and phosphorylated peptide pairs, and were validated from the MS/MS fragmentation pattern. Monoisotopic m/z values of observed precursor ions (across different charge states) were used to generate XICs. The abundance of each phosphorylated and non-phosphorylated peptide pair was estimated from areas under the curve (AUC) of each XIC. An estimated “percentage phosphorylation” was calculated as the ratio of the AUC for the phosphorylated tryptic peptide divided by the sum of AUCs for the phosphorylated and non-phosphorylated tryptic peptides (AUCPhospho / (AUCPhospho + AUCnon-phospho). AUC based methods can be used to compare relative stoichiometries of phosphorylation between different experimental groups,77 although effects of covalent modifications on peptide ionization efficiencies and peptide “flyability” suggest caution when interpreting absolute values.78 However, even with this caveat, the patterns of absolute percentages of CaMKIIα phosphorylation under the in vitro conditions (Fig. 1D, E) are consistent with previous data showing that Thr286 is a preferred site of Ca2+/CaM-dependent autophosphorylation,9–11 whereas Ser314 is a preferred site for Ca2+/CaM-independent autophosphorylation (Ca2+/CaM followed by EGTA conditions).13, 14 In cases where tryptic fragments contains multiple modifications (e.g., oxidation, phosphorylation at other sites in the same peptide), or a missed cleavage resulted in the separation of multiple tryptic peptides containing the same site-specific modification, the percentage phosphorylation was calculated based on the sum of AUCs for all relevant XICs. The percentage phosphorylation at a given site in each subcellular fraction from WT or T286A-KI mice within each biological replicate (processed and analyzed in parallel) was normalized to the highest level of phosphorylation detected in a WT subcellular fraction. Normalized ratios were then averaged across biological replicates.
AUCs of XICs for precursor ions of a specific tryptic fragment of a CaMKAP were also used to compare the abundance of specific CaMKAPs in WT and T286A-KI CaMKII holoenzyme samples prepared and analyzed in parallel. For these AUC analyses, we carefully selected tryptic fragments lacking any modified (e.g., phosphorylated) versions in the Scaffold files. If an XIC was detected and validated by MS/MS in either the WT or KI and a corresponding peak of the correct precursor mass and retention time window was detected in the other genotype without an associated MS/MS scan, this peak was used for quantification. AUCs for each CaMKAP peptide were expressed as a ratio (KI/WT) and normalized to the average KI/WT AUC ratio calculated from 9 unique XICs matching CaMKIIα. Thus, the normalized ratio represents the relative abundance of specific tryptic precursor ions in samples derived from the two genotypes.
Network Analyses
CaMKAPs (Uniprot IDs) were analyzed using WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/). Analyses for Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichments were performed and are provided either as tables or interactive webpages. To identify known or predicted interactions between CaMKAPs, Uniprot IDs were input to the STRING database (http://www.string-db.org), which identifies interactions based on direct and indirect evidence, including genomic context, high-throughput experiments, co-expression, and previous knowledge.
Statistics
Comparisons were made using a one-way or two-way ANOVA, as appropriate, followed by Tukey’s post-hoc test to compare specific groups. A one-column t-test was used to compare KI/WT ratios to a theoretical value of 1 (Fig 6).
Supplementary Material
Acknowledgments
We would like to thank the expert technical assistance of Ms. Salisha Hill (Vanderbilt University School of Medicine). The authors would also like to thank Drs. Yasunori Hayashi (Brain Science Institute, RIKEN, Wako, Japan), Andy Hudmon (Indiana University School of Medicine), John Lisman (Brandeis University), and Kevin Schey (Vanderbilt University School of Medicine) for critical evaluation of drafts of the manuscript. Funding for these studies was provided by the NIH (K01-NS073700 to AJB; R01-MH063232 to RJC; S10RR027714 to Vanderbilt Mass Spectrometry Research Center Proteomics Core), and Indiana University-Purdue University, Indianapolis (AJB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The version of rat CaMKIIβ expressed for these studies has an alanine inserted at position 341 compared to the canonical rat CaMKIIβ sequence (accession number: P08413), increasing residue numbering by 1 after this site.
Author contributions. Created reagents (AJB, BCS, RJC). Performed experiments (AJB, BCS, KLR). Analyzed data (AJB, KLR, RJC). Wrote the paper (AJB, RJC); all authors edited drafts and approved the final version.
Supporting Information Available. This manuscript is accompanied by four supplementary figures (Figs. S1–S4) plus five supplementary tables (Excel data files) (Tables S1–S5), which are each referenced in the text. This information is available free of charge via the Internet at http://pubs.acs.org/.
Bibliography
- 1.Erondu NE, Kennedy MB. Regional distribution of type II Ca2+/calmodulin-dependent protein kinase in rat brain. J Neurosci. 1985;5:3270–3277. doi: 10.1523/JNEUROSCI.05-12-03270.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Woerden GM, Hoebeek FE, Gao Z, Nagaraja RY, Hoogenraad CC, Kushner SA, Hansel C, De Zeeuw CI, Elgersma Y. betaCaMKII controls the direction of plasticity at parallel fiber-Purkinje cell synapses. Nat Neurosci. 2009;12:823–825. doi: 10.1038/nn.2329. [DOI] [PubMed] [Google Scholar]
- 3.Fink CC, Bayer KU, Myers JW, Ferrell JE, Jr, Schulman H, Meyer T. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 4.Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 6.Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 7.Hansel C, de Jeu M, Belmeguenai A, Houtman SH, Buitendijk GH, Andreev D, De Zeeuw CI, Elgersma Y. alphaCaMKII Is essential for cerebellar LTD and motor learning. Neuron. 2006;51:835–843. doi: 10.1016/j.neuron.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 8.O’Leary H, Lasda E, Bayer KU. CaMKIIbeta association with the actin cytoskeleton is regulated by alternative splicing. Mol Biol Cell. 2006;17:4656–4665. doi: 10.1091/mbc.E06-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller SG, Patton BL, Kennedy MB. Sequences of autophosphorylation sites in neuronal type II CaM kinase that control Ca2(+)-independent activity. Neuron. 1988;1:593–604. doi: 10.1016/0896-6273(88)90109-2. [DOI] [PubMed] [Google Scholar]
- 10.Schworer CM, Colbran RJ, Keefer JR, Soderling TR. Ca2+/calmodulin-dependent protein kinase II. Identification of a regulatory autophosphorylation site adjacent to the inhibitory and calmodulin-binding domains. J Biol Chem. 1988;263:13486–13489. [PubMed] [Google Scholar]
- 11.Thiel G, Czernik AJ, Gorelick F, Nairn AC, Greengard P. Ca2+/calmodulin-dependent protein kinase II: identification of threonine-286 as the autophosphorylation site in the alpha subunit associated with the generation of Ca2+-independent activity. Proc Natl Acad Sci U S A. 1988;85:6337–6341. doi: 10.1073/pnas.85.17.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson PI, Kapiloff MS, Lou LL, Rosenfeld MG, Schulman H. Expression of a multifunctional Ca2+/calmodulin-dependent protein kinase and mutational analysis of its autoregulation. Neuron. 1989;3:59–70. doi: 10.1016/0896-6273(89)90115-3. [DOI] [PubMed] [Google Scholar]
- 13.Patton BL, Miller SG, Kennedy MB. Activation of type II calcium/calmodulin-dependent protein kinase by Ca2+/calmodulin is inhibited by autophosphorylation of threonine within the calmodulin-binding domain. J Biol Chem. 1990;265:11204–11212. [PubMed] [Google Scholar]
- 14.Hanson PI, Schulman H. Inhibitory autophosphorylation of multifunctional Ca2+/calmodulin-dependent protein kinase analyzed by site-directed mutagenesis. J Biol Chem. 1992;267:17216–17224. [PubMed] [Google Scholar]
- 15.Colbran RJ. Inactivation of Ca2+/calmodulin-dependent protein kinase II by basal autophosphorylation. J Biol Chem. 1993;268:7163–7170. [PubMed] [Google Scholar]
- 16.Dosemeci A, Gollop N, Jaffe H. Identification of a major autophosphorylation site on postsynaptic density-associated Ca2+/calmodulin-dependent protein kinase. J Biol Chem. 1994;269:31330–31333. [PubMed] [Google Scholar]
- 17.Migues PV, Lehmann IT, Fluechter L, Cammarota M, Gurd JW, Sim AT, Dickson PW, Rostas JA. Phosphorylation of CaMKII at Thr253 occurs in vivo and enhances binding to isolated postsynaptic densities. J Neurochem. 2006;98:289–299. doi: 10.1111/j.1471-4159.2006.03876.x. [DOI] [PubMed] [Google Scholar]
- 18.Meyer T, Hanson PI, Stryer L, Schulman H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science. 1992;256:1199–1202. doi: 10.1126/science.256.5060.1199. [DOI] [PubMed] [Google Scholar]
- 19.Strack S, Colbran RJ. Autophosphorylation-dependent targeting of calcium/calmodulin-dependent protein kinase II by the NR2B subunit of the N-methyl-D-aspartate receptor. J Biol Chem. 1998;273:20689–20692. doi: 10.1074/jbc.273.33.20689. [DOI] [PubMed] [Google Scholar]
- 20.Bayer KU, De Koninck P, Leonard AS, Hell JW, Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411:801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- 21.Jalan-Sakrikar N, Bartlett RK, Baucum AJ, Colbran RJ. Substrate-selective and calcium-independent activation of CaMKII by alpha-actinin. J Biol Chem. 2012;287:15275–15283. doi: 10.1074/jbc.M112.351817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giese KP, Fedorov NB, Filipkowski RK, Silva AJ. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science. 1998;279:870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- 23.Elgersma Y, Fedorov NB, Ikonen S, Choi ES, Elgersma M, Carvalho OM, Giese KP, Silva AJ. Inhibitory autophosphorylation of CaMKII controls PSD association, plasticity, and learning. Neuron. 2002;36:493–505. doi: 10.1016/s0896-6273(02)01007-3. [DOI] [PubMed] [Google Scholar]
- 24.Kimura R, Silva AJ, Ohno M. Autophosphorylation of alphaCaMKII is differentially involved in new learning and unlearning mechanisms of memory extinction. Learn Mem. 2008;15:837–843. doi: 10.1101/lm.1049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, de Avila Freire R, Jiang YH, Elgersma Y, Weeber EJ. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of alphaCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10:280–282. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- 26.Gustin RM, Shonesy BC, Robinson SL, Rentz TJ, Baucum AJ, 2nd, Jalan-Sakrikar N, Winder DG, Stanwood GD, Colbran RJ. Loss of Thr286 phosphorylation disrupts synaptic CaMKIIalpha targeting, NMDAR activity and behavior in pre-adolescent mice. Mol Cell Neurosci. 2011;47:286–292. doi: 10.1016/j.mcn.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernard PB, Castano AM, Bayer KU, Benke TA. Necessary, but not sufficient: insights into the mechanisms of mGluR mediated long-term depression from a rat model of early life seizures. Neuropharmacology. 2014;84:1–12. doi: 10.1016/j.neuropharm.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coultrap SJ, Freund RK, O’Leary H, Sanderson JL, Roche KW, Dell’Acqua ML, Bayer KU. Autonomous CaMKII mediates both LTP and LTD using a mechanism for differential substrate site selection. Cell Rep. 2014;6:431–437. doi: 10.1016/j.celrep.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SA, Hudmon A, Volmer A, Waxham MN. CaM-kinase II dephosphorylates Thr(286) by a reversal of the autophosphorylation reaction. Biochem Biophys Res Commun. 2001;282:773–780. doi: 10.1006/bbrc.2001.4651. [DOI] [PubMed] [Google Scholar]
- 30.Shen K, Teruel MN, Connor JH, Shenolikar S, Meyer T. Molecular memory by reversible translocation of calcium/calmodulin-dependent protein kinase II. Nat Neurosci. 2000;3:881–886. doi: 10.1038/78783. [DOI] [PubMed] [Google Scholar]
- 31.Strack S, Choi S, Lovinger DM, Colbran RJ. Translocation of autophosphorylated calcium/calmodulin-dependent protein kinase II to the postsynaptic density. J Biol Chem. 1997;272:13467–13470. doi: 10.1074/jbc.272.21.13467. [DOI] [PubMed] [Google Scholar]
- 32.Shen K, Teruel MN, Subramanian K, Meyer T. CaMKIIbeta functions as an F-actin targeting module that localizes CaMKIIalpha/beta heterooligomers to dendritic spines. Neuron. 1998;21:593–606. doi: 10.1016/s0896-6273(00)80569-3. [DOI] [PubMed] [Google Scholar]
- 33.Lin YC, Redmond L. CaMKIIbeta binding to stable F-actin in vivo regulates F-actin filament stability. Proc Natl Acad Sci U S A. 2008;105:15791–15796. doi: 10.1073/pnas.0804399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto K, Narayanan R, Lee SH, Murata K, Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci U S A. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanabria H, Swulius MT, Kolodziej SJ, Liu J, Waxham MN. {beta}CaMKII regulates actin assembly and structure. J Biol Chem. 2009;284:9770–9780. doi: 10.1074/jbc.M809518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wisniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J Proteome Res. 2010;9:3280–3289. doi: 10.1021/pr1002214. [DOI] [PubMed] [Google Scholar]
- 37.Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, Burlingame AL. Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics. 2008;7:684–696. doi: 10.1074/mcp.M700170-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballif BA, Carey GR, Sunyaev SR, Gygi SP. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J Proteome Res. 2008;7:311–318. doi: 10.1021/pr0701254. [DOI] [PubMed] [Google Scholar]
- 41.Tweedie-Cullen RY, Reck JM, Mansuy IM. Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J Proteome Res. 2009;8:4966–4982. doi: 10.1021/pr9003739. [DOI] [PubMed] [Google Scholar]
- 42.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stokes MP, Farnsworth CL, Moritz A, Silva JC, Jia X, Lee KA, Guo A, Polakiewicz RD, Comb MJ. PTMScan direct: identification and quantification of peptides from critical signaling proteins by immunoaffinity enrichment coupled with LC-MS/MS. Mol Cell Proteomics. 2012;11:187–201. doi: 10.1074/mcp.M111.015883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skelding KA, Suzuki T, Gordon S, Xue J, Verrills NM, Dickson PW, Rostas JA. Regulation of CaMKII by phospho-Thr253 or phospho-Thr286 sensitive targeting alters cellular function. Cell Signal. 2010;22:759–769. doi: 10.1016/j.cellsig.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Hvalby O, Hemmings HC, Jr, Paulsen O, Czernik AJ, Nairn AC, Godfraind JM, Jensen V, Raastad M, Storm JF, Andersen P, et al. Specificity of protein kinase inhibitor peptides and induction of long-term potentiation. Proc Natl Acad Sci U S A. 1994;91:4761–4765. doi: 10.1073/pnas.91.11.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 47.Costa MC, Mani F, Santoro W, Jr, Espreafico EM, Larson RE. Brain myosin-V, a calmodulin-carrying myosin, binds to calmodulin-dependent protein kinase II and activates its kinase activity. J Biol Chem. 1999;274:15811–15819. doi: 10.1074/jbc.274.22.15811. [DOI] [PubMed] [Google Scholar]
- 48.Leonard AS, Bayer KU, Merrill MA, Lim IA, Shea MA, Schulman H, Hell JW. Regulation of calcium/calmodulin-dependent protein kinase II docking to N-methyl-D-aspartate receptors by calcium/calmodulin and alpha-actinin. J Biol Chem. 2002;277:48441–48448. doi: 10.1074/jbc.M205164200. [DOI] [PubMed] [Google Scholar]
- 49.Robison AJ, Bartlett RK, Bass MA, Colbran RJ. Differential modulation of Ca2+/calmodulin-dependent protein kinase II activity by regulated interactions with N-methyl-D-aspartate receptor NR2B subunits and alpha-actinin. J Biol Chem. 2005;280:39316–39323. doi: 10.1074/jbc.M508189200. [DOI] [PubMed] [Google Scholar]
- 50.Jiao Y, Jalan-Sakrikar N, Robison AJ, Baucum AJ, 2nd, Bass MA, Colbran RJ. Characterization of a central Ca2+/calmodulin-dependent protein kinase IIalpha/beta binding domain in densin that selectively modulates glutamate receptor subunit phosphorylation. J Biol Chem. 2011;286:24806–24818. doi: 10.1074/jbc.M110.216010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baucum AJ, 2nd, Strack S, Colbran RJ. Age-dependent targeting of protein phosphatase 1 to Ca2+/calmodulin-dependent protein kinase II by spinophilin in mouse striatum. PLoS One. 2012;7:e31554. doi: 10.1371/journal.pone.0031554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. doi: 10.1016/j.neuron.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 53.Halt AR, Dallapiazza RF, Zhou Y, Stein IS, Qian H, Juntti S, Wojcik S, Brose N, Silva AJ, Hell JW. CaMKII binding to GluN2B is critical during memory consolidation. EMBO J. 2012;31:1203–1216. doi: 10.1038/emboj.2011.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanhueza M, Fernandez-Villalobos G, Stein IS, Kasumova G, Zhang P, Bayer KU, Otmakhov N, Hell JW, Lisman J. Role of the CaMKII/NMDA receptor complex in the maintenance of synaptic strength. J Neurosci. 2011;31:9170–9178. doi: 10.1523/JNEUROSCI.1250-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shonesy BC, Wang X, Rose KL, Ramikie TS, Cavener VS, Rentz T, Baucum AJ, 2nd, Jalan-Sakrikar N, Mackie K, Winder DG, Patel S, Colbran RJ. CaMKII regulates diacylglycerol lipase-alpha and striatal endocannabinoid signaling. Nat Neurosci. 2013;16:456–463. doi: 10.1038/nn.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robens JM, Yeow-Fong L, Ng E, Hall C, Manser E. Regulation of IRSp53-dependent filopodial dynamics by antagonism between 14-3-3 binding and SH3-mediated localization. Mol Cell Biol. 2010;30:829–844. doi: 10.1128/MCB.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim MH, Choi J, Yang J, Chung W, Kim JH, Paik SK, Kim K, Han S, Won H, Bae YS, Cho SH, Seo J, Bae YC, Choi SY, Kim E. Enhanced NMDA receptor-mediated synaptic transmission, enhanced long-term potentiation, and impaired learning and memory in mice lacking IRSp53. J Neurosci. 2009;29:1586–1595. doi: 10.1523/JNEUROSCI.4306-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen CJ, Shih CH, Chang YJ, Hong SJ, Li TN, Wang LH, Chen L. SH2B1 and IRSp53 Promotes the Formation of Dendrites and Dendritic Branches. J Biol Chem. 2015 doi: 10.1074/jbc.M114.603795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung W, Choi SY, Lee E, Park H, Kang J, Park H, Choi Y, Lee D, Park SG, Kim R, Cho YS, Choi J, Kim MH, Lee JW, Lee S, Rhim I, Jung MW, Kim D, Bae YC, Kim E. Social deficits in IRSp53 mutant mice improved by NMDAR and mGluR5 suppression. Nat Neurosci. 2015 doi: 10.1038/nn.3927. [DOI] [PubMed] [Google Scholar]
- 63.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 64.Schmid SR, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 65.Srivastava T, Fortin DA, Nygaard S, Kaech S, Sonenberg N, Edelman AM, Soderling TR. Regulation of neuronal mRNA translation by CaM-kinase I phosphorylation of eIF4GII. J Neurosci. 2012;32:5620–5630. doi: 10.1523/JNEUROSCI.0030-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atkins CM, Davare MA, Oh MC, Derkach V, Soderling TR. Bidirectional regulation of cytoplasmic polyadenylation element-binding protein phosphorylation by Ca2+/calmodulin-dependent protein kinase II and protein phosphatase 1 during hippocampal long-term potentiation. J Neurosci. 2005;25:5604–5610. doi: 10.1523/JNEUROSCI.5051-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 68.Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 69.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, Piven J, Ponting CP, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Sequeira AF, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stein O, Sykes N, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Webber C, Weksberg R, Wing K, Wittemeyer K, Wood S, Wu J, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Devlin B, Ennis S, Gallagher L, Geschwind DH, Gill M, Haines JL, Hallmayer J, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Scherer SW, Sutcliffe JS, Betancur C. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li JM, Lu CL, Cheng MC, Luu SU, Hsu SH, Hu TM, Tsai HY, Chen CH. Role of the DLGAP2 gene encoding the SAP90/PSD-95-associated protein 2 in schizophrenia. PLoS One. 2014;9:e85373. doi: 10.1371/journal.pone.0085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, Brown SE, Christian JM, Mirnikjoo B, Silva A, Beaudet AL, Sweatt JD. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23:2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McNeill RB, Colbran RJ. Interaction of autophosphorylated Ca2+/calmodulin-dependent protein kinase II with neuronal cytoskeletal proteins. Characterization of binding to a 190-kDa postsynaptic density protein. J Biol Chem. 1995;270:10043–10049. doi: 10.1074/jbc.270.17.10043. [DOI] [PubMed] [Google Scholar]
- 74.Brickey DA, Colbran RJ, Fong YL, Soderling TR. Expression and characterization of the alpha-subunit of Ca2+/calmodulin-dependent protein kinase II using the baculovirus expression system. Biochem Biophys Res Commun. 1990;173:578–584. doi: 10.1016/s0006-291x(05)80074-9. [DOI] [PubMed] [Google Scholar]
- 75.Baucum AJ, 2nd, Brown AM, Colbran RJ. Differential association of postsynaptic signaling protein complexes in striatum and hippocampus. J Neurochem. 2013;124:490–501. doi: 10.1111/jnc.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 77.Ratnikov B, Ptak C, Han J, Shabanowitz J, Hunt DF, Ginsberg MH. Talin phosphorylation sites mapped by mass spectrometry. J Cell Sci. 2005;118:4921–4923. doi: 10.1242/jcs.02682. [DOI] [PubMed] [Google Scholar]
- 78.Steen H, Jebanathirajah JA, Springer M, Kirschner MW. Stable isotope-free relative and absolute quantitation of protein phosphorylation stoichiometry by MS. Proc Natl Acad Sci U S A. 2005;102:3948–3953. doi: 10.1073/pnas.0409536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.