Abstract
Many bacteria use small RNAs (sRNAs) and the RNA chaperone Hfq to regulate mRNA stability and translation. Hfq, a ring-shaped homohexamer, has multiple faces that can bind both sRNAs and their mRNA targets. We find that Hfq has at least two distinct ways in which it interacts with sRNAs; these different binding properties have strong effects on the stability of the sRNA in vivo and the sequence requirements of regulated mRNAs. Class I sRNAs depend on proximal and rim Hfq sites for stability and turn over rapidly. Class II sRNAs are more stable and depend on the proximal and distal Hfq sites for stabilization. Using deletions and chimeras, we find that while Class I sRNAs regulate mRNA targets with previously defined ARN repeats, Class II sRNAs regulate mRNAs carrying UA-rich rim-binding sites. We discuss how these different binding modes may correlate with different roles in the cell, with Class I sRNAs acting as emergency responders and Class II sRNAs acting as silencers.
Keywords: ChiX, Hfq, MgrR, RyhB
Introduction
Hfq was originally characterized as a host factor from Escherichia coli required for RNA bacteriophage Qβ replication (Franze de Fernandez et al, 1968). Since then, it has been implicated in many facets of RNA metabolism, including RNA decay and transcription termination (reviewed in Vogel & Luisi, 2011; Sobrero & Valverde, 2012). One of its most well-studied roles is as an RNA chaperone facilitating pairing between bacterial small RNAs (sRNAs) and their mRNA targets. This pairing typically leads to changes in the stability or translation of the target mRNA transcript (reviewed in Gottesman & Storz, 2011).
Hfq is a member of a larger family of Sm and Sm-like (LSm) RNA-binding proteins found in eukaryotes and archaea. This family of multimeric proteins is characterized by two conserved domains, Sm1 and Sm2, that consist of an N-terminal α-helix stacked on top of a closed barrel comprised of five anti-parallel β strands (reviewed in Sauer, 2013; Wilusz & Wilusz, 2013). Unlike Sm and LSm proteins, which commonly form heteroheptameric rings (reviewed in Wilusz & Wilusz, 2013), Hfq is a homohexamer (Møller et al, 2002; Schumacher et al, 2002; Zhang et al, 2002; Sauter et al, 2003).
In vitro and crystal structure studies of Hfq initially revealed two distinct surfaces for RNA interaction (reviewed in Brennan & Link, 2007). One surface, termed the proximal face, has an affinity for uridine-rich sequences, where it can bind up to six nucleotides (one per monomer) (Schumacher et al, 2002; Mikulecky et al, 2004; Link et al, 2009). This surface recognizes and binds to the U-rich stretch at the 3′ end of Rho-independent terminators, a common attribute of the 3′ end of sRNAs (Otaka et al, 2011; Sauer & Weichenrieder, 2011). A second surface, termed the distal face, has a strong affinity for adenine-rich sequences (de Haseth & Uhlenbeck, 1980; Mikulecky et al, 2004). Such a sequence motif was originally identified within the 5′ untranslated region (5′ UTR) of the rpoS mRNA, where a AAYAA sequence significantly enhanced Hfq binding and formation of a stable ternary complex between Hfq, rpoS, and DsrA (Soper & Woodson, 2008). Crystal structures of Hfq from E. coli in complex with poly(A) revealed that three nucleotides bind per subunit of Hfq on the distal face, and led to definition of the binding motif as (A-R-N)n sequences, the term used here, where A is an adenine nucleotide, R is any purine nucleotide, and N is any nucleotide (Link et al, 2009; Wang et al, 2013). Subsequently, ARN-like sequences were shown to be important for sRNA-dependent regulation of other sRNA/mRNA pairs (Salim & Feig, 2010; Beisel et al, 2012; Salim et al, 2012). A more recent study, which utilized tryptophan fluorescence quenching to more thoroughly map Hfq-RNA interactions, redefined the optimal binding motif for the distal face of the E. coli protein as (A-A-N)n (Robinson et al, 2014).
A third surface of Hfq, termed the rim or lateral face, has more recently been shown to interact with RNA. This face consists of a conserved arginine patch on the periphery of the E. coli Hfq ring. In vivo and in vitro studies have revealed that this basic patch plays a role in binding and stabilization of some sRNAs by making contact with UA-rich sequences within the sRNAs (Sauer et al, 2012; Zhang et al, 2013; Dimastrogiovanni et al, 2014). Beyond binding, this patch also has been implicated as an active site for the chaperone activity of Hfq, as mutations of these residues led to a disruption in the nucleation of model RNA helices (Panja et al, 2013). Thus, Hfq utilizes three distinct faces to facilitate sRNA regulation: the proximal face for binding U-rich sequences, the distal face for binding AAN motifs, and the rim for binding UA-rich sequences. Additionally, the unstructured C-terminus of Hfq may contribute to binding and/or exchange of sRNAs and mRNAs (Dimastrogiovanni et al, 2014).
Aside from having a Rho-independent terminator recognized by Hfq and a sequence for base pairing with target mRNAs termed the ‘seed’ region (Papenfort et al, 2010), the size, shape, and sequence of sRNAs can vary dramatically. For example, sRNAs may also contain additional uridine-rich stretches, which have also been shown to play a role in binding and regulation (Zhang et al, 2002; Balbontín et al, 2010; Ishikawa et al, 2012; Sauer et al, 2012). Despite the degenerate recognition sequences and variation in size and shape of sRNAs, Hfq preferentially binds base-pairing sRNAs and their mRNA partners (Zhang et al, 2003; Sittka et al, 2008).
To gain further insight into how Hfq binds sRNAs and mRNAs and facilitates their interaction, a recent study utilized a combination of in vivo approaches to gain a better perspective on RNA:Hfq protein interactions. Specifically, the effect of 14 different hfq mutations of residues on the proximal face, the distal face, and the rim on the in vivo activity and accumulation of a representative group of sRNAs was examined. Many of the results supported the basic in vitro conclusions that the proximal face of Hfq was necessary for sRNA stability and function. However, sRNAs differed in their patterns of accumulation in rim and distal mutants, suggesting that the sRNAs fall into two distinct groups (Zhang et al, 2013). Here, we further examine this difference in behavior, its basis, and implications for the function of Hfq and the sRNAs and mRNAs that bind to this protein.
Results
sRNAs fall into different classes based on accumulation in Hfq mutants
In a previous study, we examined the levels of five sRNAs (DsrA, RyhB, ArcZ, McaS, and ChiX) in 14 different hfq mutants in a MG1655 background (Zhang et al, 2013). Three of these sRNAs, DsrA, RyhB, and ArcZ (FigEV1) showed an overall decrease in accumulation in the rim mutants, but had high levels in the distal face mutants. Here, we define this group as Class I. The other two sRNAs, ChiX and McaS (FigEV1), had the inverse accumulation pattern with higher levels of accumulation in the rim mutants and reduced levels in the distal face mutants. We define this second group as Class II.
Figure EV1. Effects of hfq mutations on accumulation of sRNAs.
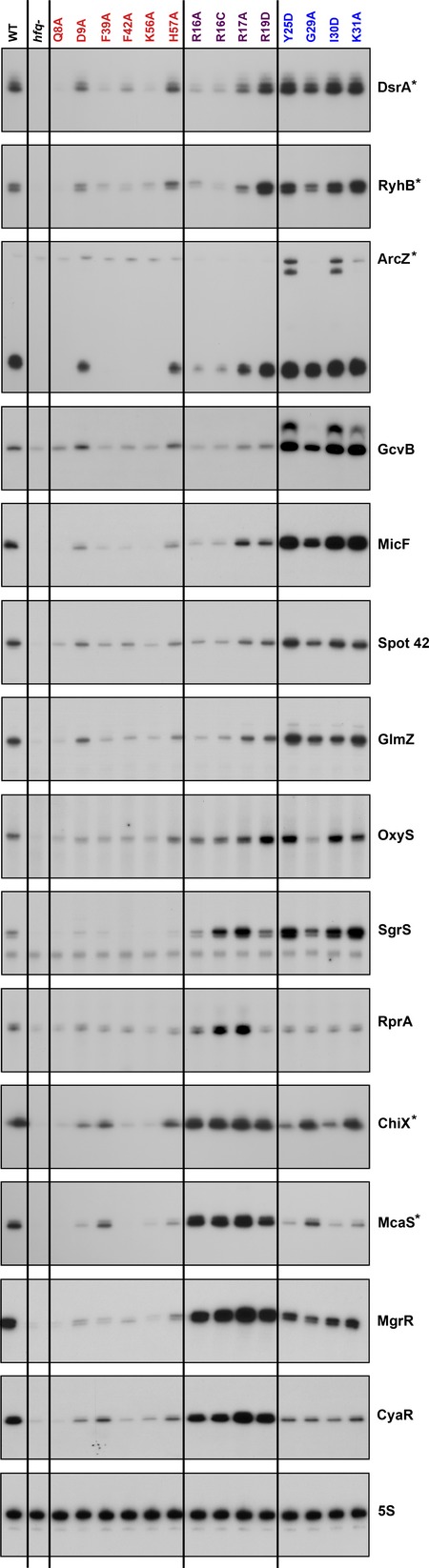
Primer extension and Northern assays shown in Fig1 together with those previously shown in Fig4 of Zhang et al (2013) (marked with asterisks). The color code for Hfq surfaces (here for the Hfq mutant allele labels) is red for the proximal surface, purple for the rim, and blue for distal surface.
The Class I and II definitions are based on accumulation levels of only five endogenously expressed sRNAs in multiple rim and distal site mutants. We were interested in determining whether the broader group of Hfq-binding sRNAs fell into these classes as well. We initially examined this by taking advantage of tiling arrays done in our previous study (Zhang et al, 2013). In those arrays, we examined the genome-wide accumulation of endogenous sRNAs as well as the levels of sRNAs that co-immunoprecipitated (IP) with Hfq in cells expressing wild-type (WT) Hfq or one of three mutants (proximal face Q8A, rim R16A, and distal face K31A). While both the total and IP RNA levels paralleled the accumulation data for DsrA, RyhB, ArcZ, ChiX, and McaS, the total expression levels of a few sRNAs were very low, making comparisons difficult. However, all of the sRNAs were enriched upon co-IP with Hfq, and thus, the Hfq IP numbers from table S3 in Zhang et al (2013) were used in this comparison (Table EV1). The ratio for the rim R16A mutant IP relative to the distal site K31A mutant IP was found to best distinguish between the original sRNAs classified as Class I and Class II. Based on the ratios for these mutants, additional sRNAs were initially assigned to Class I (low ratio, < 0.5, purple in Table EV1) or Class II (high ratio, close to or > 1, blue in Table EV1). We next tested these predicted classifications in the broader set of hfq mutants for sRNAs selected to cover a range of R16A/K31A ratios.
Of the 24 Hfq-dependent sRNAs included in this analysis, 14 (including DsrA, RyhB and ArcZ) had R16A/K31A ratios of < 0.5 for the co-IP samples in two independent data sets, the characteristics defined for Class I. Spot 42 and GlmZ had values just below the cutoff in one data set and just above in the other, but otherwise had other characteristics of Class I sRNAs. GcvB, MicF, Spot 42, and GlmZ were chosen from this set for further testing by examining accumulation in the full set of hfq alleles by primer extension analysis (Fig1). All four of these sRNAs had an accumulation pattern similar to that of the original Class I sRNAs: reduced levels in the rim mutants and higher levels in the distal mutants (Fig1). Therefore, including Spot 42 and GlmZ, 16 of 24 sRNAs can be categorized as Class I sRNAs on the basis of the co-IP data for R16A and K31A.
Figure 1. Effect of hfq mutations on accumulation of sRNAs.
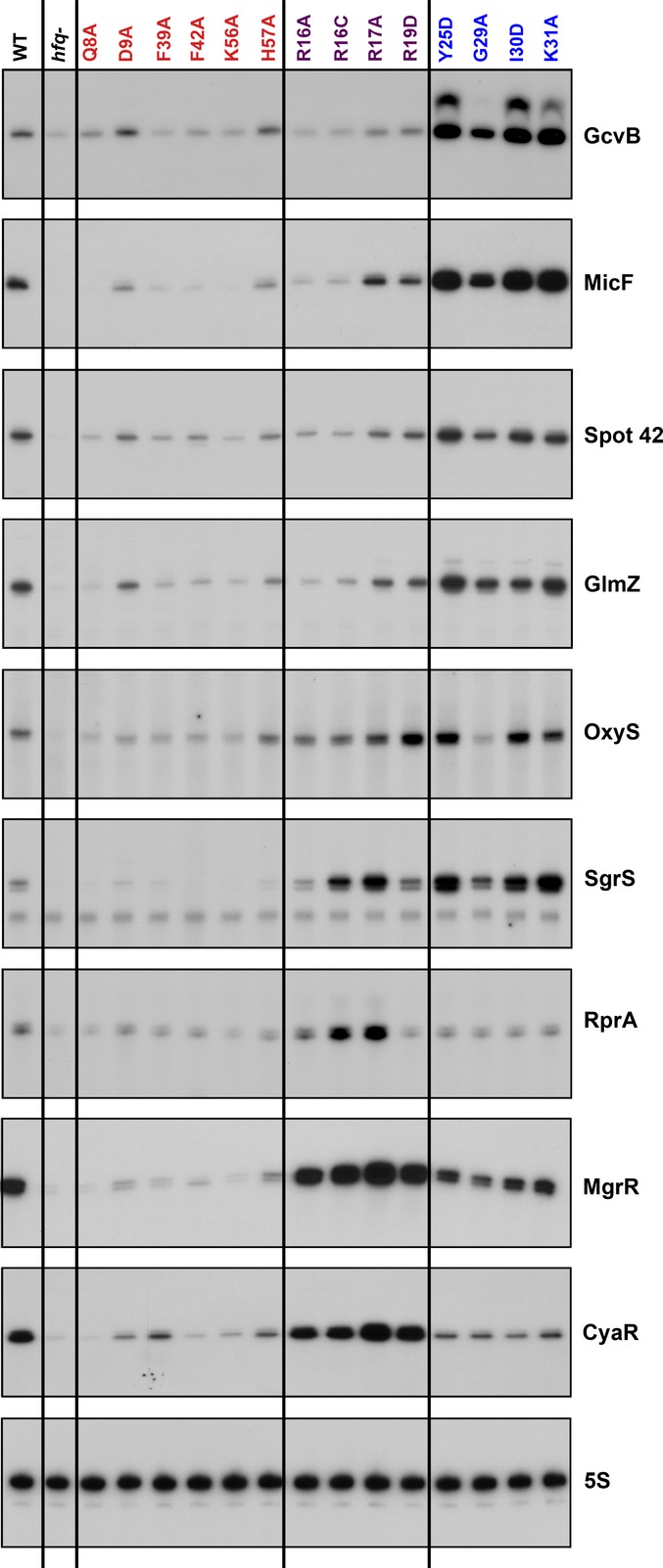
Extracts were prepared from wild-type (SG30214) and isogenic hfq mutants (SG30206 to SG30237A, listed in Appendix Table S1) grown in LB medium at 37°C to early stationary phase (OD600 ∼ 1.0). The levels of all the sRNAs were analyzed by primer extension analysis of 5 μg of total RNA. The same extracts tested in Fig4 of Zhang et al (2013) were used to analyze the sRNAs in this figure. The levels of 5S were determined by Northern analysis of the ArcZ blot in the same (Zhang et al, 2013) figure. The color code for Hfq surfaces, used throughout the paper (here for the Hfq mutant allele labels), is red for the proximal surface, purple for the rim, and blue for distal surface. We noted the presence of a second primer extension band (2–5 nt longer) for GcvB in the distal mutants. We do not know if it is due to modification or an additional transcription start site, but the band was also detected in WT cells when GcvB was expressed at high levels.
Four sRNAs had both total and IP R16A/K31A ratios close to or > 1, characteristic of Class II sRNAs. MgrR and CyaR were further examined and, like the defining Class II sRNAs ChiX and McaS, had higher levels in the rim mutants, and reduced levels in the distal mutants (Fig1).
The four remaining sRNAs, ArcZ, RprA, SgrS, and OxyS, had intermediate ratios for the co-IP samples (> 0.5 but < 1.0). ArcZ was previously tested; this sRNA is rapidly processed, and both the full length and processed forms accumulated with patterns much like that for DsrA and other Class I sRNAs (Zhang et al, 2013) (FigEV1). We assayed the other three sRNAs, RprA, SgrS, and OxyS, for accumulation in the larger group of hfq mutants by primer extension analysis (Fig1). These assays suggested that OxyS falls more within the category of a Class I sRNA, although the decrease in expression in the rim alleles was not as dramatic as seen for other Class I sRNAs. On the other hand, the accumulation profile for RprA was more similar to that of a Class II sRNA, with high levels of accumulation in some of the rim mutants and reduced levels in the distal face mutants. However, unlike the other Class II RNAs, the levels were low in two of the rim mutants, R16A and R19D. SgrS showed an accumulation pattern similar to both RprA and OxyS. We conclude that the sRNAs with an intermediate IP ratio (> 0.5 but < 1.0) may represent RNAs intermediate between Class I and Class II; these are color-coded light blue in Table EV1. Overall, ArcZ is more like a Class I sRNA, and RprA is more like a Class II sRNA, while OxyS and SgrS have characteristics of both classes. We focused here on characterizing sRNAs that were clearly Class I or Class II.
The majority of the sRNAs (20 of 24) could be assigned to Class I or Class II by their IP ratios (Table EV1), and the eleven that were further tested for the accumulation level in the full set of hfq alleles confirmed the prediction from the IP ratio. The four sRNAs that did not fit clearly into Class I or Class II by the original ratio were confirmed to have more complex behavior in the full set of alleles. Overall, these results strongly support the use of the R16A/K31A ratio as a robust assay for the assignment of sRNAs to Class I or II. We note that there are differences among the hfq alleles that we do not fully explore here. Some alleles, including H57A on the proximal face, K31A and G29A on the distal face, and R17A on the rim, are likely relatively leaky mutants, based on previous work (Zhang et al, 2013), and thus, the accumulation phenotypes are weaker (more like wild-type) than other alleles on the same face. R19, which is found on the distal face of the rim, may have both rim and distal face properties. For the rest of this work, we made use of the Q8A proximal mutant, the R16A rim mutant, and the Y25D distal mutant, all having consistently strong phenotypes for sRNA accumulation and repression of target genes (Zhang et al, 2013).
Accumulation of sRNAs in two classes is correlated with turnover
The differences in the pattern of accumulation of sRNAs in hfq mutants seen in Figs1 and EV1 are most easily explained by differences in turnover of these sRNAs, because transcriptional regulation of each of the sRNAs is different, and Hfq is known to affect the stability of sRNAs (reviewed in Vogel and Luisi, 2011). Since deletion of hfq destabilizes most of these sRNAs (see second lane in Figs1 and EV1), mutations in surfaces of Hfq needed for sRNA binding would be expected to destabilize the sRNA. In addition, we previously suggested that sRNA pairing to targets is accompanied by coupled degradation of the sRNA and mRNA (Massé et al, 2003), possibly due to their displacement from Hfq. Thus, mutations that interfere with mRNA binding and/or pairing might retain the sRNA on Hfq, also leading to increased stability.
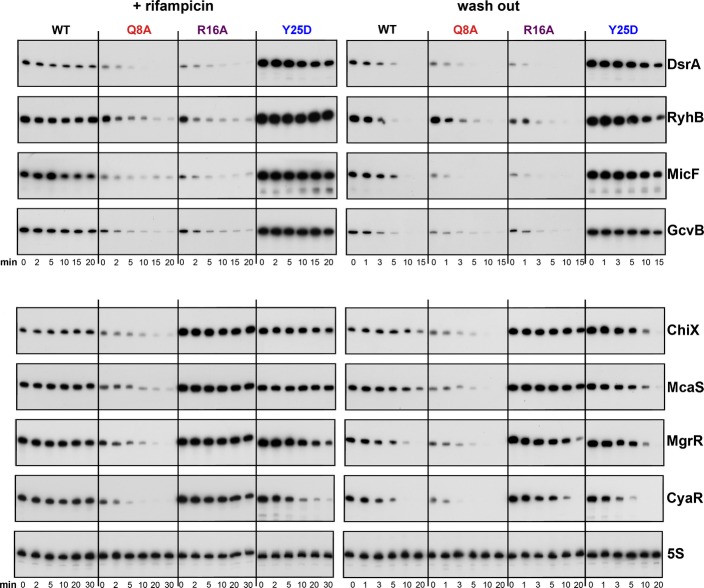
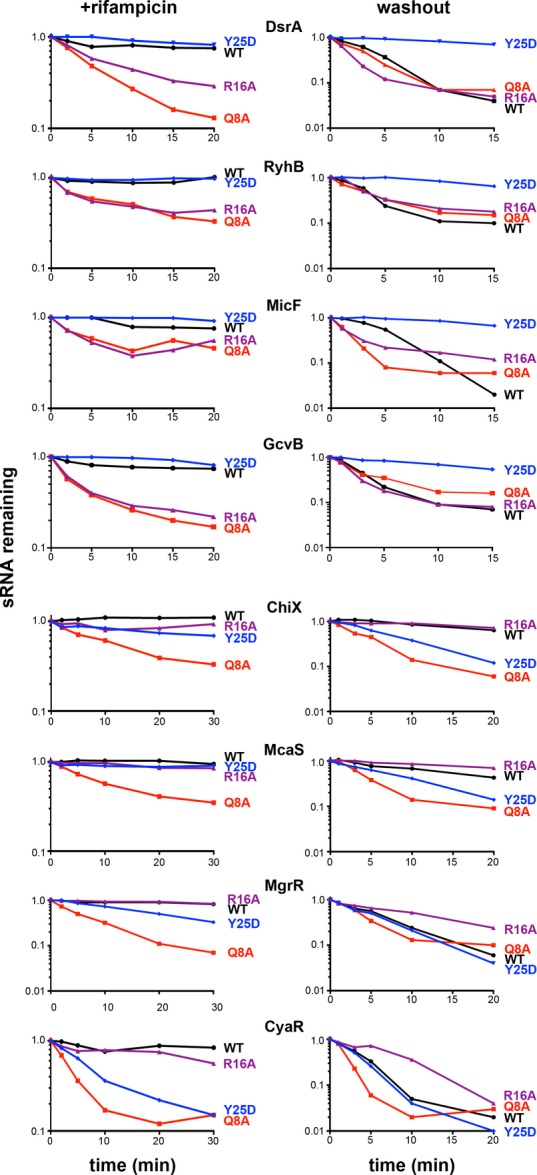
These predictions were tested by examining sRNA turnover in two ways. In both cases, the sRNA was expressed from a plasmid from a plac promoter induced for 15 min. In the first test, the intrinsic stability was measured by adding rifampicin to the culture (inhibiting all transcription) after induction and analyzing samples during a chase period by Northern analysis. In that assay, the four Class I sRNAs tested (DsrA, RyhB, MicF, and GcvB) and the four Class II sRNAs (ChiX, McaS, MgrR, and CyaR) were stable in an hfq+ host and were somewhat unstable in the hfqQ8A host (Figs2, left panels, and EV2). This is consistent with previous in vitro data supporting a role for Q8 in binding all sRNAs (Mikulecky et al, 2004; Updegrove & Wartell, 2011). In the rim mutant (R16A), Class I sRNAs also became unstable, while Class II sRNAs were stable, correlating with the reduced accumulation of Class I but not Class II sRNAs in rim alleles (Figs1 and EV1), and suggesting a role for the rim in binding and stabilizing Class I but not Class II sRNAs. We also observed some destabilization of ChiX, CyaR, and MgrR in the Y25D distal mutant (Fig EV2), suggesting that distal site binding might be involved in stabilizing Class II sRNAs.
Figure 2. Effect of Hfq mutations on stability of Class I and Class II sRNAs.
Wild-type and isogenic hfq mutants, deleted for the sRNA being examined and harboring a plasmid expressing the sRNA of interest under the control of Plac, were grown at 37°C and treated as described in Materials and Methods; all transcription was stopped by treatment with rifampicin (left), or transcription of just the sRNA was stopped by washing cells to remove IPTG (right). Total RNA (3 μg for RyhB, 3.5 μg for MicF and 4 μg for all other sRNAs) extracted from each sample was subject to Northern analysis using an oligonucleotide specific to the sRNA. The levels of 5S were determined by Northern analysis of the CyaR blot. Quantitation of the Northern blots is given in FigEV2. Strains were derived from hfq+ (SG30214), hfqQ8A (SG30206), hfqR16A (SG30207), and hfqY25D (SG30237A) by P1 transduction to delete a given sRNA gene, as listed in Appendix Table S1.
Figure EV2. Quantitation of Northern blot analysis in Fig2.
Phosphoimages of the Northern membranes were analyzed by using GE Typhoon imaging scanner and ImageJ software. Graphs were plotted with Prism software. Color code is as in Fig1.
Source Data are available online for this figure.
In the second set of experiments, we measured sRNA stability by washing out IPTG to stop further sRNA transcription, while general transcription of other RNAs, including target mRNAs, continued. We previously reported that under similar assay conditions, DsrA and RyhB were unstable in an hfq+ host, in contrast to their behavior upon rifampicin treatment (Massé et al, 2003); this degradation was interpreted as coupled degradation of the sRNA and mRNA. This pattern was confirmed for the four Class I RNAs (DsrA, RyhB, MicF, and GcvB) (Fig2, compare WT right and left panels; quantitated in Fig EV2). Both the Q8A and R16A washouts showed similar instability. However, strikingly, these Class I sRNAs were stable in the Y25D washout experiment (Figs2 and EV2, half-life increased from 3 to 5 min to greater than 15 min). If the rapid turnover of an sRNA after inducer washout in WT cells is due to coupled degradation of the sRNA and the mRNA, stabilization in Y25D may reflect failure of mRNA binding, leading to a defect in pairing. As a result, the Class I sRNAs would remain stably associated with Hfq for longer. This stabilization is consistent with higher levels of accumulation of endogenous Class I sRNAs in distal site mutants (Figs1 and EV1), and higher levels of the induced Class I sRNAs at 0 min in the Y25D mutant (Fig2).
Two of the four Class II RNAs (ChiX and McaS) were stable in the hfq+ washout experiment. Based on previous studies with ChiX (Overgaard et al, 2009b), Class II sRNAs may, in some cases, be stable regulators that can be reused in multiple rounds of pairing and turnover of their targets. Strikingly, however, each of the Class II sRNAs was more stable in the R16A rim mutant (FigEV2); in Y25D, degradation was either similar to or more rapid than in the WT host, a pattern that was the inverse of that seen for Class I.
Overall, these results agree with the pattern of accumulation of Class I and Class II sRNAs in the hfq mutants seen in Fig1 and confirm that the differences in accumulation are due to differences in stability. They also support roles for three distinct faces for interaction with RNA, consistent with work by others (de Haseth & Uhlenbeck, 1980; Schumacher et al, 2002; Mikulecky et al, 2004; Sauer et al, 2012), but suggest that Class I and Class II sRNAs use these interaction faces differently (Fig3). We assume here that the sRNAs are stable as long as they are bound to Hfq and thus that loss of stability reflects a reduction in Hfq binding. The second assumption is that, after pairing with target mRNAs, the sRNA is displaced from Hfq and therefore becomes sensitive to degradation. Based on the results in Fig2, Class I sRNAs use the proximal face and the rim for binding, and mutations in the distal face stabilize binding or prevent the sRNAs from being displaced from Hfq, while Class II sRNAs use the proximal and distal faces for binding, and mutations in the rim further stabilize binding (Fig3).
Figure 3. A model of alternative modes of RNA binding to Hfq.
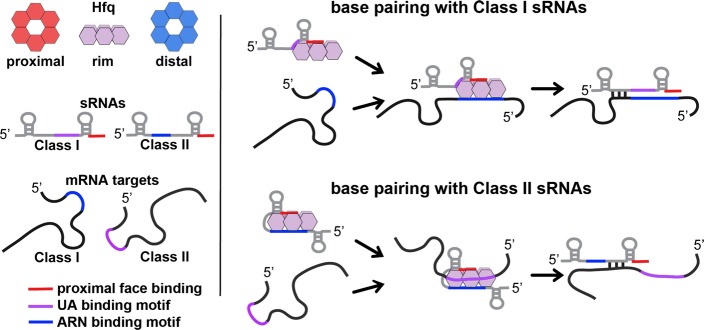
The cartoon model of the Hfq hexamer depicts the three RNA-binding surfaces of Hfq: proximal face (red), rim (purple), and distal face (blue). For sRNAs and mRNAs, elements in red represent sequences (rho-independent terminator) that bind to the proximal face, elements in purple represent UA sequences that bind the rim, and elements in blue represent ARN-binding motifs that bind to the distal face. The model depicts two alternative pathways for binding and regulation by sRNA:mRNA pairs. Class I sRNAs utilize a U-rich rho-independent terminator for binding the proximal face and a UA-binding motif for interaction with the rim. mRNA targets regulated by this class of sRNAs utilize ARN-binding motifs for interacting with the distal face of Hfq. Binding of the Class I sRNA and its corresponding mRNA to Hfq lead to base pairing and regulation and degradation of the sRNA. A second class of sRNAs (Class II) utilize the U-rich rho-independent terminator for binding the proximal face of Hfq and an ARN motif for binding to the distal face. mRNA targets regulated by this class of sRNAs contain UA-binding motifs that allow for binding to the rim of Hfq. Binding of the Class II sRNA and its corresponding mRNA to Hfq lead to base pairing and regulation, but not necessarily degradation of the sRNA.
ARN motifs in Class II sRNAs confer stability
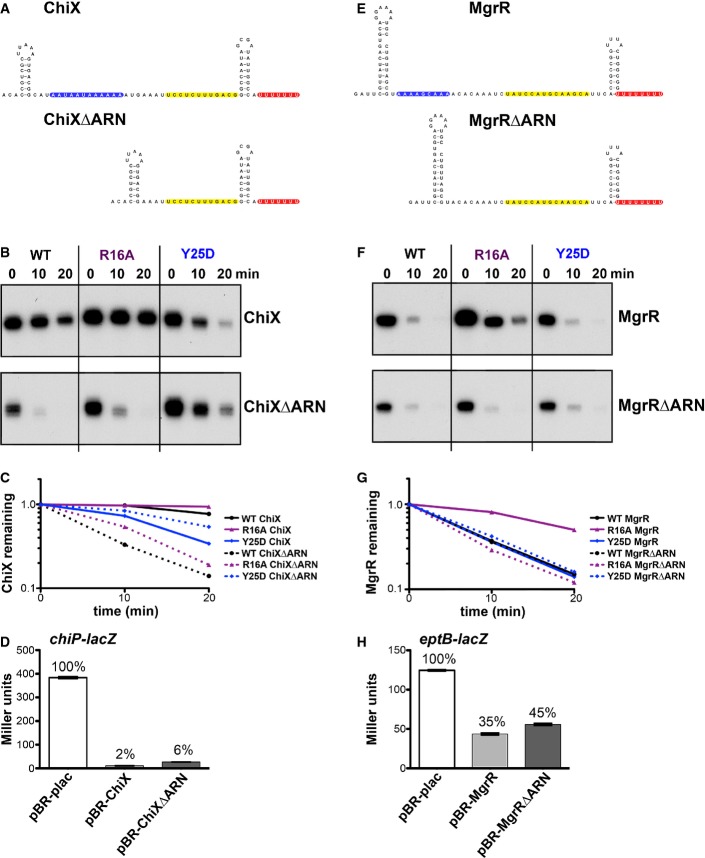
The results in Fig2 suggest a role for the distal face in Hfq binding to Class II sRNAs. We thus looked for elements present in Class II but not Class I sRNAs consistent with distal face binding. This analysis revealed, in Class II sRNAs, internal single-stranded adenosine-rich stretches similar to the ARN-binding motifs found to be required for mRNA binding to the distal face of Hfq (highlighted in blue in Fig4A and E and Appendix Fig S1). To examine the roles of these ARN sequence repeats, we deleted these sequences in the Class II sRNAs ChiX and MgrR. ChiX harbors four ARN repeats, and mutants were constructed in which one, two, three, or the entire ARN repeat region was removed (Appendix Fig S2A). The stability of the mutants compared to the WT was then measured in washout experiments similar to those in Fig2. The results of this analysis suggest that the individual ARN sequences have an additive effect on the stability of ChiX (Appendix Fig S2B and C). Deletion of one or two ARN sites decreased stability of the RNA relative to WT; deletion of three or all four ARN sequences made ChiX as unstable as RyhB (5 min half-life).
Figure 4.
- Reported secondary structure of the wild-type ChiX (Rasmussen et al, 2009) and predicted secondary structure of the corresponding ARN deletion mutant (ChiXΔARN). Predicted RNA structures in all figures were determined as described in Reuter and Mathews (2010). Color codes show the ARN motifs in blue, the base-pairing region to the chiP target (Rasmussen et al, 2009) in yellow, and the U-rich region of the rho-independent terminator in red.
- Northern blot analysis of washout experiments comparing the wild-type ChiX to the ChiXΔARN mutant. These experiments were carried out in wild-type and isogenic mutant derivatives of SG30200, each carrying ΔchiX::kan [WT hfq+ (DJS2784), hfqR16A (DJS2786), and hfqY25D (DJS2789)] and harboring a plasmid that expressed the wild-type ChiX (pBR-ChiX) or the ChiXΔARN mutant (pDJS2211). The washouts were carried out similarly to the experiments in Fig2, with the exception that 5 μg of total RNA was analyzed for each sample.
- Quantitation of the Northern blot in (B). Quantification was carried out as for Fig EV2. Wild-type ChiX results are shown with a solid line and ChiXΔARN with dotted lines and WT in black, R16A in purple, and Y25D in blue.
- β-Galactosidase activity measured in a PBAD-chiP-lacZ ΔchiX::kan strain (DJS2991) carrying a plasmid expressing wild-type ChiX (pBR-ChiX), ChiXΔARN (pDJS2211), or a vector control (pBR-plac). Strains were grown in LB medium containing 100 μg/ml ampicillin, 10 μM IPTG, and 0.002% arabinose at 37°C to early stationary phase (OD600 ∼ 1.0) and assayed for β-galactosidase activity. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
- Northern blot analysis of washout experiments comparing wild-type MgrR to the MgrRΔARN mutant. These experiments were carried out in wild-type and isogenic mutant derivatives of SG30200, each carrying ΔmgrR::kan [WT hfq (DJS2963), hfqR16A (DJS2965), and hfqY25D (DJS2966)] and harboring a plasmid that expressed the wild-type MgrR (pBR-MgrR) or the MgrRΔARN mutant (pDJS2225). The washouts were carried out as for Fig2, with the exception that 5 μg of total RNA was analyzed for each sample.
- Quantitation of the Northern membrane in (F) as for Fig EV2.
- β-galactosidase activity measured in a PBAD-eptB-lacZ ΔmgrR::kan strain (DJS3003) carrying a plasmid expressing wild-type MgrR (pBR-MgrR), MgrRΔARN (pDJS2225), or a vector control (pBR-plac). Samples were treated and analyzed as in (D), except that the LB contained 0.2% arabinose. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
Source data are available online for this figure.
The deletion of all four ARN motifs destabilized ChiX, as did the Y25D mutation (Fig4B and C), consistent with a role for the ARN motifs in binding to the distal site. If both Y25D and deletion of the ARN motifs block ChiX binding to the distal site, leading to instability, the effects should not be additive. To test this prediction, the stabilities of ChiX and ChiXΔARN in WT, R16A, and Y25D mutant cells were examined after treatment with rifampicin and after inducer washout (Fig4 and Appendix Fig S3). After inducer washout, full-length ChiX was stable in the WT and R16A mutant backgrounds (> 20 min half-life) and unstable (9 min half-life) in the Y25D mutant (Fig4B, solid lines in Fig4C), as seen in Fig2. In contrast, the ChiX-ΔARN mutant was rapidly turned over in both the WT and the R16A mutant backgrounds (5 min half-life). The observation that turnover in both the WT and rim mutant backgrounds was equally slow for ChiX and equally rapid for ChiXΔARN suggests that ChiX does not bind the rim, even in the absence of ARN motifs. In contrast, the rate of turnover of ChiX and the ChiXΔARN mutant were very similar in the Y25D mutant (Fig4B and C). This result suggests that mutations in the distal face of Hfq make ChiX turnover independent of the ARN motifs, and points to the distal face as the interaction surface with the ARN-binding motifs found in ChiX.
The abilities of ChiX and ChiXΔARN to repress a chiP-lacZ fusion were also compared (Fig4D). In these experiments, in which the ChiX derivatives were overproduced, both ChiX and ChiXΔARN effectively repressed expression of the fusion, although ChiX was slightly more effective. More striking differences between ChiX and ChiXΔARN-mediated regulation were observed when the sRNA was expressed from its native chromosomal locus rather than from a plasmid (Appendix Fig S4). In this experiment, the kinetics of chiP repression were analyzed after wild-type, chiXΔARN, and ΔchiX strains were treated with rifampicin. The chiP signal is barely detectable in cells expressing the wild-type ChiX, but is easily detected in either the absence of ChiX or in cells in which chromosomally expressed ChiX is deleted for its ARN sequences. By 10 min, there is some reduction in chiP levels in the chiXΔARN background, while there is no change in the ΔchiX strain. Thus, while ChiX is still partially functional in the absence of ARN sequences, this region is required for efficient repression.
We next compared the stabilities of MgrR and MgrRΔARN in WT, R16A, and Y25D mutant cells after washout (Fig4F and G) or after cells were treated with rifampicin (Fig2, Appendix Fig S3). In the washout experiments, MgrR was equally unstable with or without ARN motifs in WT cells or in a Y25D mutant. This suggests that for wild-type MgrR, binding to the distal face is not sufficiently strong to prevent displacement from Hfq after washout, and thus, stability is unchanged by either deleting the ARN motifs or mutating the distal binding site. However, while MgrR sRNA was stabilized in the R16A mutant (Fig4F and G), this was not observed for the MgrRΔARN mutant, suggesting that stabilization by the rim mutant requires interaction with the distal face. As for ChiX, deletion of the ARN motifs from MgrR did not significantly reduce the ability of this sRNA, when overexpressed, to repress expression of a target fusion, eptB-lacZ (Fig4H).
Given that the ARN motifs in MgrR are shorter and less A rich than those in ChiX (Fig4E), we also examined the effects of extending these sequences. Replacement of the AGC sequence with AAU and/or the addition of a fourth ARN (mutation of ACA to AAA) increased the stability of MgrR (Appendix Fig S2D–F), consistent with the role of these sequences in stabilizing ChiX.
Overall, for both ChiX and MgrR, the ARN motifs in these Class II sRNAs are not essential for function, at least when the sRNAs are expressed at high levels. However, the ARN motifs do improve the stability and efficacy of the Class II sRNAs (Fig4 and Appendix Figs S2 and S4).
The addition of ARN motifs stabilizes RyhB but inactivates the sRNA
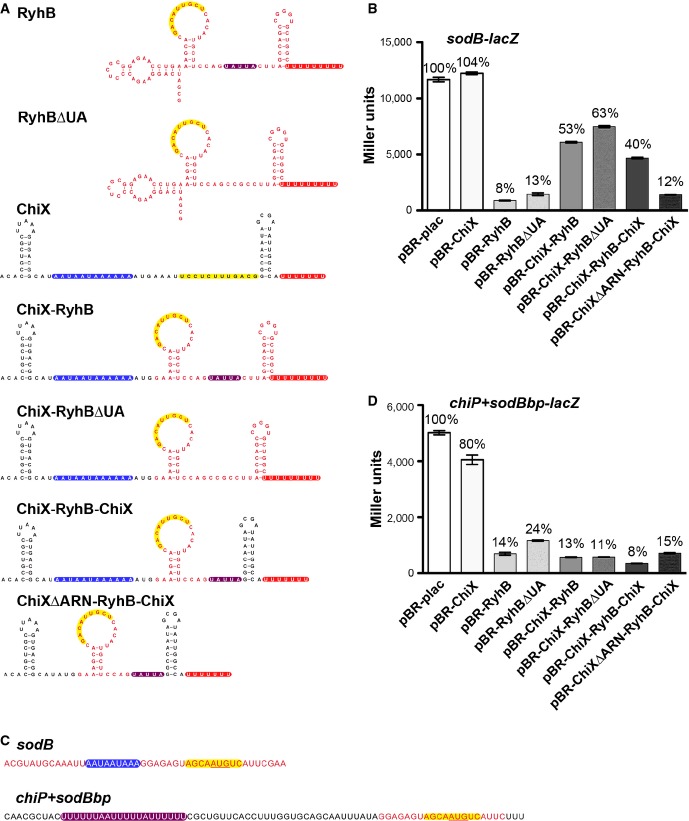
We observed that ARN motifs contribute to stabilization of the Class II sRNAs ChiX and MgrR. To ask whether these motifs could stabilize an unstable Class I sRNA, we fused the ARN-binding motifs from ChiX to the Class I sRNA RyhB. In the chimera considered here, the 5′ region of ChiX harboring an initial stem-loop followed by the ARN-binding motif was fused to RyhB truncated at the 5′ end, replacing the 5′ stem-loop usually found on RyhB (ChiX-RyhB) (Fig5A). Washout experiments were carried out with RyhB and the ChiX-RyhB chimera in WT, R16A, and Y25D Hfq backgrounds (Fig5B and C). The chimera harboring the ARN-binding motif demonstrated increased stability compared to WT RyhB (> 20 min half-life compared to 5 min for RyhB). This stabilization further supports the conclusion that the ARN-binding motif is responsible for the increased stability of Class II sRNAs. Most strikingly, the distal site mutant, Y25D, which stabilized WT RyhB, had the opposite effect in the chimera, modestly destabilizing the sRNA. In the Y25D strain, the stability of the chimera was quite similar to that of WT RyhB, consistent with lack of distal site binding similarly affecting both of these sRNAs.
Figure 5.
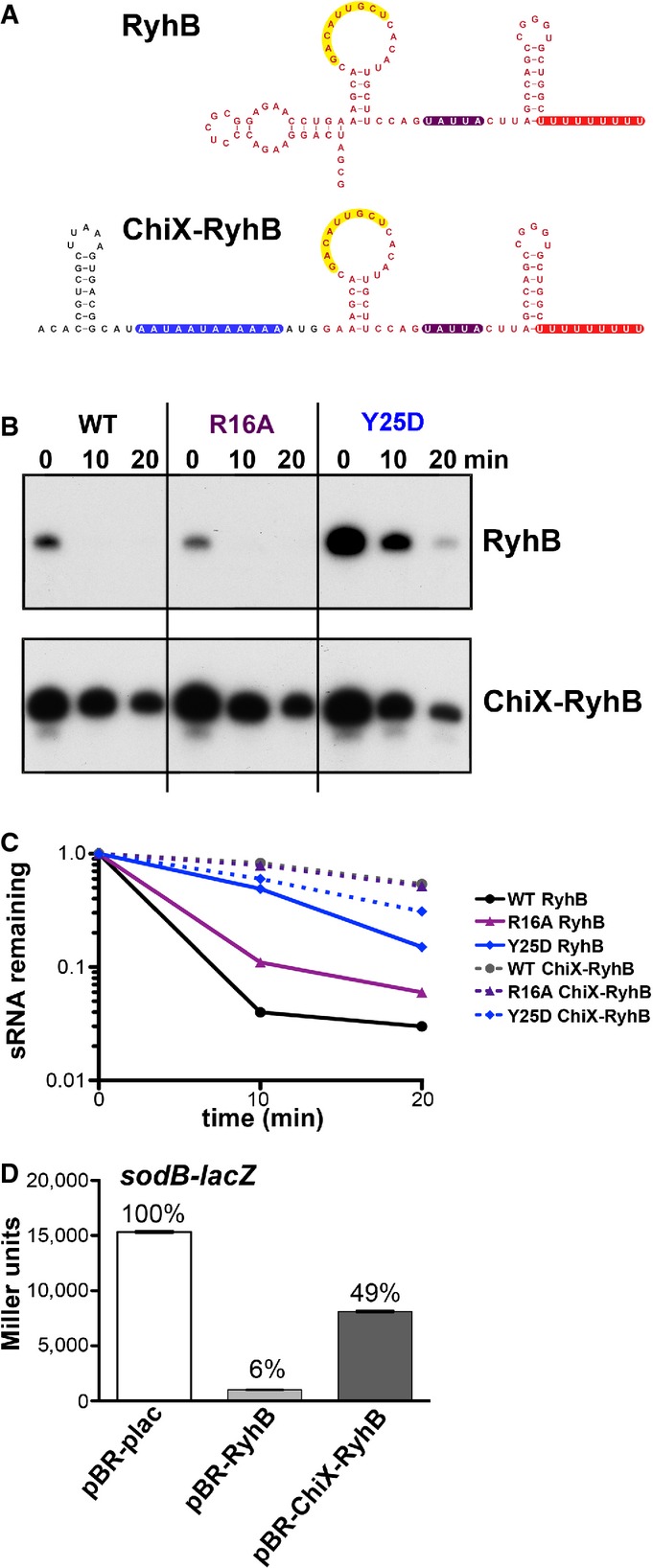
- Reported secondary structure of the wild-type RyhB (Geissmann & Touati, 2004) and the predicted secondary structure of the chimera in which the 5′ end of ChiX was fused to truncated RyhB (ChiX-RyhB). Color codes are as for Fig4A, with region of pairing to the sodB target (Geissmann & Touati, 2004) in yellow and the ARN motifs in blue. Red nucleotides correspond to bases from RyhB, and black nucleotides correspond to bases from ChiX.
- Northern blot analysis of washout experiments to compare the stability of the wild-type RyhB to the ChiX-RyhB chimera. These experiments were carried out in hfq+ ΔchiX::kan (DJS2784), hfqR16A ΔchiX::kan (DJS2786) and hfqY25D ΔchiX::kan (DJS2789), harboring a plasmid that expressed the wild-type RyhB (pBR-RyhB) or the ChiX-RyhB mutant (pDJS2219) under control of a plac promoter. The washouts were carried out as in Fig2, with the exception that 5 μg of total RNA was analyzed for each sample.
- Quantitation of the Northern membrane in (B) as in Fig EV2.
- β-Galactosidase activity measured in NRD537 (PBAD-sodB-lacZ) carrying a plasmid expressing wild-type RyhB (pBR-RyhB), ChiX-RyhB (pDJS2219), or a vector control (pBR-plac). Samples were treated and analyzed as in Fig4D. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
Source data are available online for this figure.
The stable ChiX-RyhB chimera was also tested for repression of a sodB-lacZ translational fusion (Fig5D). Repression was markedly reduced (from 16-fold for the WT RyhB to 2-fold for the chimera), even though the pairing region was preserved and the chimeric sRNA is very abundant (Appendix Fig S5A). Thus, although the addition of the ChiX ARN sequences conferred increased stability to a Class I sRNA, the motif interfered with the ability of RyhB to regulate its native target sodB.
The mRNA requirements for regulation by Class I and Class II sRNAs differ
To further understand why the RyhB RNA with the added ARN motifs was inactive for sodB regulation, we generated several additional chimeric and mutant sRNAs based on RyhB and ChiX; all were engineered to have the region for base pairing with sodB. Previous work has shown that RyhB contains several sequence elements that contribute to stability, including the rho-independent terminator and a UA-rich Hfq-binding site, a possible rim-binding site, upstream of the terminator (Ishikawa et al, 2012). Plasmid-expressed RyhB carrying a mutation of this UA-rich sequence, highlighted in purple, to CCGC (RyhBΔUA, Fig6A), regulated slightly less efficiently than WT RyhB, but better than the ChiX-RyhB chimera (Fig6B), even though the levels of these mutant sRNAs are similar (Appendix Fig S5A). The same mutation was introduced in the ChiX-RyhB chimera to create ChiX-RyhBΔUA. This mutant chimera, like the ChiX-RyhB sRNA, was also defective for sodB regulation (Fig6B).
Figure 6.
- Secondary structures of the wild-type RyhB and the corresponding predicted secondary structures of the various ChiX/RyhB chimeras as in Fig5A.
- β-Galactosidase activity measured in NRD537 (PBAD-sodB-lacZ) carrying a plasmid expressing a vector control (pBR-plac), wild-type RyhB (pBR-RyhB), wild-type ChiX (pBR-ChiX), ChiX-RyhB (pDJS2219), RyhBΔUA (pDJS2227), ChiX-RyhBΔUA (pDJS2226), ChiX-RyhB-ChiX (pDJS2229), or ChiXΔARN-RyhB-ChiX (pDJS2230). Samples were treated and analyzed as in Fig4D. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
- Sequences from the 5′ UTR of PBAD-sodB-lacZ and PBAD-chiP+sodBbp-lacZ fusions. Color codes show the sequence for the base-pairing region to the mRNA target sodB to the sRNA RyhB highlighted in yellow and the UA element from chiP in purple. Red text represents bases from sodB, and black text represents bases from chiP.
- β-Galactosidase activity measured in DJS2985 (PBAD-chiP+sodBbp-lacZ) carrying the same set of plasmids as in (B). Samples were treated and analyzed as in Fig4D. Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated.
Source data are available online for this figure.
In another derivative, the RyhB terminator stem-loop of ChiX-RyhB was replaced by the terminator stem-loop from ChiX (ChiX-RyhB-ChiX); this was nearly as defective for regulation of sodB as was the original ChiX-RyhB chimera (Fig6B). In a final construct, the ARN repeats (blue highlighted region in Fig6A) were deleted from the ChiX-RyhB-ChiX chimera to give ChiXΔARN-RyhB-ChiX. Interestingly, the deletion of the ARN repeats partially restored the ability of the chimera to regulate sodB (Fig6B), even though the stability of the ChiXΔARN-RyhB-ChiX construct was less than the ChiX-RyhB-ChiX chimera (Appendix Fig S5B). Therefore, the difference in activity is not consistent with a difference in stability.
From these results, we conclude that the presence of the ARN repeats on RyhB precludes regulation of sodB. Other sequences within the chimera (initial stem-loop, the UA-rich binding site as well as the specific RyhB terminator stem-loop) are not essential for regulation under these overproduction conditions.
We hypothesized that the ARN repeats in the RyhB chimeras prevent regulation of sodB because they prevent binding of Hfq to ARN sequences in the target mRNA. If so, and assuming mRNA binding to Hfq is a prerequisite for regulation, Class II targets may have sites other than ARNs recognized by Hfq. Rim-binding sites were a possible alternative, suggested by a number of observations. As noted above, Class I sRNAs were stabilized in the distal site Y25D mutant, while the Class II sRNAs MgrR and CyaR were stabilized in the R16A rim mutant (Figs2, 4F and EV2). If stabilization reflects lack of target mRNA binding to Hfq, preventing coupled turnover of the sRNA, this observation suggests that mRNA targets of Class II sRNAs require a functional rim to bind to Hfq and be regulated by Class II sRNAs. Additionally, in our previous assays of the effects of various Hfq mutants on function [table 1 of Zhang et al (2013)], it was striking that rim mutants were very defective for Class II sRNA-dependent regulation (ChiX regulation of chiP; McaS regulation of flhD), while the rim mutants were not as defective for regulation by Class I sRNA (particularly for RyhB).
We thus predicted Class I mRNA targets require distal face binding and might have decreased Hfq interactions in distal site mutants like K31A, while Class II mRNA targets require rim binding and would be defective in Hfq interactions in rim mutants like R16A. This prediction was examined using the previous genome-wide co-IP experiments with wild-type, R16A, and K31A Hfq (table S4 of Zhang et al (2013)). mRNAs known to be regulated by Class I or Class II sRNAs or both were selected and examined for the ratio of the mRNA captured by co-IP with R16A Hfq to that captured by K31A Hfq. In agreement with our predictions, the summary of the co-IP data suggests that Class II target mRNAs are somewhat enriched for those with a low ratio of R16AIP/K31AIP (Table EV2; ratio < 0.5), the opposite of what was observed for the Class II sRNAs (Table EV1).
The possibility of alternative mRNA-binding sites for Class II targets was tested using our chimeric ChiX-RyhB sRNAs. If a Class II target such as chiP has sequences other than ARN that recognize Hfq, it seemed possible that the chimeric ChiX-RyhB sRNA, while inactive on wild-type sodB, would be able to regulate an mRNA that includes these alternative Hfq-binding sites. We thus created a chimeric target translational fusion in which the region for chiP pairing to ChiX was replaced with the region for sodB pairing to RyhB (Fig6C). The chiP+sodBbp-lacZ fusion was then assayed for repression by ChiX, RyhB, and the chimeras described above (Fig6D). As expected, ChiX no longer regulated the fusion as it no longer has the ability to base-pair with the target mRNA. In contrast, wild-type RyhB as well as all of the chimeras were able to regulate the chiP+sodBbp-lacZ fusion. The regain of function by the chimeras indicates that possible alterations in secondary structure are not the reason for the reduced regulation of the sodB-lacZ fusion. The regulation of the chiP+sodBbp-lacZ fusion was still fully Hfq dependent (Appendix Fig S5C). Strikingly, the presence of the ARN motifs no longer interfered with regulation. These observations are consistent with this Class II-like target having sites other than ARN motifs for Hfq interaction and regulation.
Deletion of the internal UA-rich potential Hfq rim-binding site in the sRNAs only slightly lessened regulation by RyhB and ChiX-RyhB-ChiX (Fig6D). The results are consistent with the ability of ChiX deleted for ARN motifs to still regulate chiP (Fig4D). Overall, the physiological role of the ARN motif in Class II sRNAs may be to stabilize the sRNA to make it a more efficient regulator, and one that is not turned over with each use. At the same time, the presence of ARN motifs in Class II sRNAs means that mRNA targets of these sRNAs use other motifs to bind Hfq.
UA-rich sequences and the Hfq rim are needed for regulation of Class II target mRNAs
The importance of ARN sequences for regulation by Class I sRNAs has been demonstrated for a number of mRNAs (Salim & Feig, 2010; Soper et al, 2010; Beisel et al, 2012; Salim et al, 2012). The experiment in Fig6 demonstrated that a site within chiP, a target of the Class II sRNA ChiX, is needed for regulation by sRNA derivatives carrying an ARN motif. In addition, our previous results showed that the R16A mutant, with an inactivated rim-binding site, was defective for regulation by Class II sRNAs (Zhang et al, 2013). We thus hypothesized that mRNAs need UA-rich rim-binding sites in order to be targets of Class II sRNAs. To test this hypothesis, we examined the ability of RyhB to regulate the sodB-lacZ (carrying an ARN distal site binding motif) and chiP+sodBbp-lacZ (carrying a potential rim-binding site) fusions in WT, R16A, and Y25D mutants. The results of these assays show a complete change in the requirement for distal versus rim residues dependent on the nature of the target (compare Fig7A and B). RyhB regulated the sodB-lacZ fusion well in the R16A rim mutant, but less well in the Y25D distal site mutant (Fig7A), as previously reported (Zhang et al, 2013). However, regulation of the chiP+sodBbp-lacZ fusion was lost in the R16A mutant but was even better in the Y25D mutant than in hfq+ cells (Fig7B). This increased activity in the Y25D mutant may reflect the effects of blocking the binding of other RyhB targets, which should all carry ARN motifs, to Hfq; RyhB levels increase (Fig1), and no other target can compete for pairing with RyhB.
Figure 7.
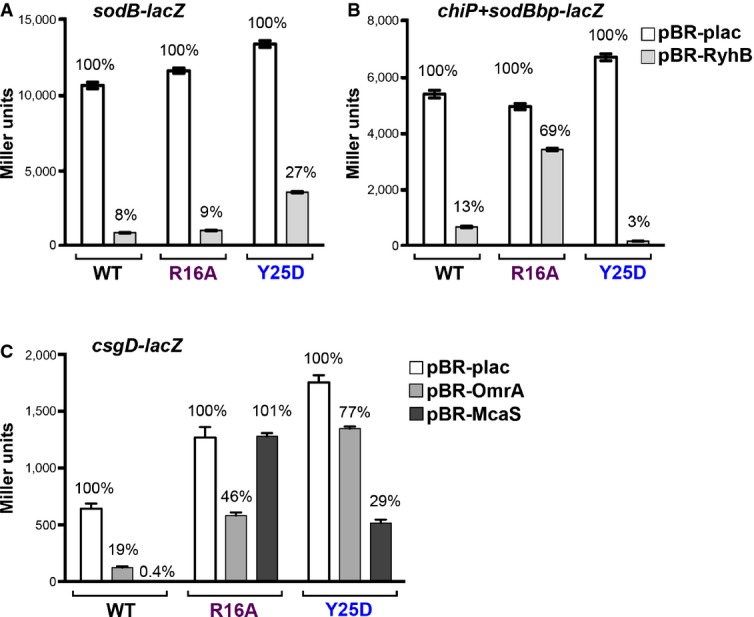
- Derivatives of NRD537, each with PBAD-sodB-lacZ [WT hfq (DJS2683), hfqR16A (DJS2686), and hfqY25D (DJS2687)] and carrying a plasmid expressing wild-type RyhB (pBR-RyhB) or a vector control (pBR-plac), were grown and assayed as in Fig4D.
- Derivatives of DJS2985, each with PBAD-chiP+sodBbp-lacZ [WT hfq (DJS3009), hfqR16A (DJS3010), and hfqY25D (DJS3011)] and carrying a plasmid expressing wild-type RyhB (pBR-RyhB) or a vector control (pBR-plac). Samples were treated as in Fig4D.
- Derivatives of MPK0379, each with PBAD-csgD-lacZ ΔmcaS::kan ΔpgaA::cat [WT hfq (DJS3017), hfqR16A (DJS3018), and hfqY25D (DJS3019)] and carrying a plasmid expressing wild-type McaS (pBR-McaS), wild-type OmrA (pBR-OmrA), or a vector control (pBR-plac), were grown and assayed as in Fig4D except that the LB contained 0.02% arabinose.
Data information: Data are average of three assays, and error bars denote the standard deviation of the mean. Percent activity compared to the vector control strain is indicated. Source data are available online for this figure.
Some mRNAs are regulated by both Class I and Class II sRNAs; possibly these mRNAs contain both ARN and UA-rich binding sites. One such target mRNA is csgD, regulated by multiple sRNAs, including the Class I sRNA OmrA and the Class II sRNA McaS (Holmqvist et al, 2010; Jørgensen et al, 2012; Thomason et al, 2012). UA-rich sequences and an ARN motif are seen in the 5′ UTR of csgD (Appendix Fig S6A). The dependence of OmrA and McaS on rim and distal Hfq sites was tested for csgD expression (Fig7C). In agreement with the results for regulation of sodB and chiP+sodBbp, regulation by the Class I sRNA OmrA was most defective in the distal site mutant, while regulation by Class II RNA McaS was defective in the rim mutant.
Examination of chiP (target of ChiX) and eptB (target of MgrR) identified possible UA-rich sites within these targets. Mutations in these sites were constructed for chiP, the chiP+sodBbp chimera, and for eptB (Appendix Fig S6B) and tested for regulation by the corresponding sRNAs (Fig8). In each case, regulation was reduced by deletion of the UA-rich region in the mRNA, consistent with the proposal that Hfq binding to the target via the rim is important for regulation by Class II sRNAs and that these UA sequences mediate that binding. In all cases, the ΔUA mutant had a somewhat lower level of expression in the absence of the sRNA (white bars in each set), particularly for chiP+sodBbp-lacZ (Fig8B), indicating the UA sequence (or Hfq binding) may also contribute to mRNA stability or translation.
Figure 8.
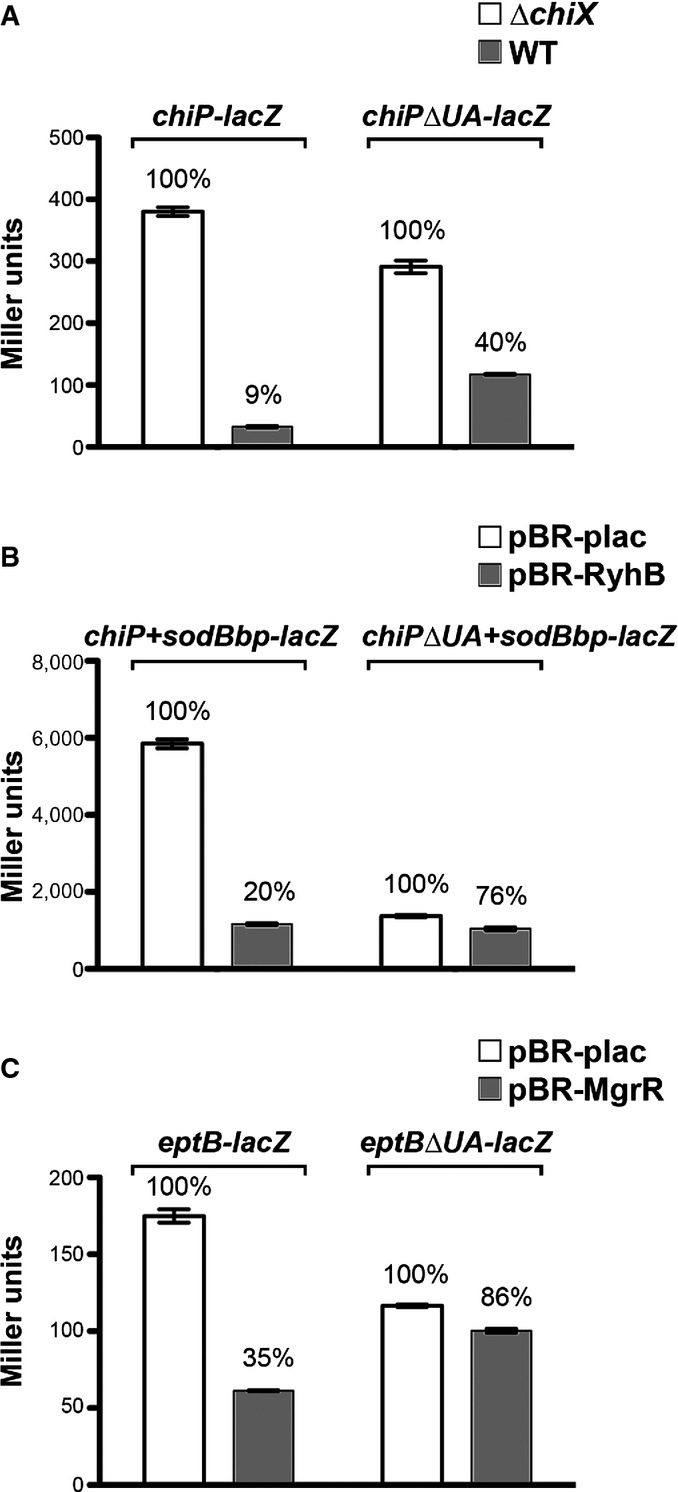
- β-Galactosidase activity measured in DJS2979 (PBAD-chiP-lacZ), DJS2982 (PBAD-chiPΔUA-lacZ), and the ΔchiX derivatives (DJS2991 and DJS2994, respectively). Samples were treated as in Fig4D.
- β-Galactosidase activity measured in DJS2985 (PBAD-chiPsodBbp-lacZ) or DJS2998 (PBAD-chiPΔUA+sodBbp-lacZ) carrying a plasmid expressing wild-type RyhB (pBR-RyhB) or a vector control (pBR-plac). Samples were treated as in Fig4D.
- β-Galactosidase activity measured in DJS3003 (PBAD-eptB-lacZ ΔmgrR::kan) or DJS3004 (PBAD-eptBΔUA –lacZ ΔmgrR::kan) carrying a plasmid expressing wild-type MgrR (pBR-MgrR) or a vector control (pBR-plac). Samples were treated as in Fig4D, except that the LB contained 0.2% arabinose.
Finally, we asked whether addition of an ARN-binding site to an mRNA deleted for the rim-binding site (and therefore no longer well-regulated by an sRNA) could restore regulation, presumably by restoring the interaction with Hfq. While the location and extent of the ARN motifs will undoubtedly affect activity and translation, the well-characterized ARN motifs from rpoS (Soper & Woodson, 2008) were inserted at the site of the mutated UA-rich rim-binding site in chiP+sodBbp (Fig9A), and the ability of both RyhB and the ChiX-RyhB sRNAs to regulate this chimeric target was tested. The results in Fig9B were consistent with our expectations; introduction of the rpoS ARN into the chiPΔUA+sodBbp mRNA improved regulation by RyhB (from 1.4-fold to 2.2-fold), but not regulation by ChiX-RyhB (1.4-fold to 1.2-fold).
Figure 9.
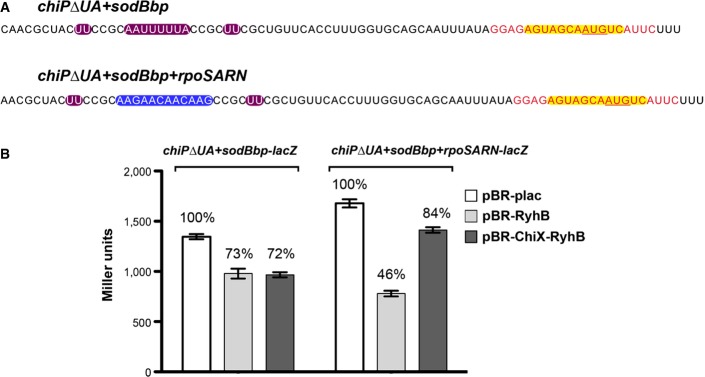
- Sequences from the 5′ UTR of PBAD-chiPΔUA+sodBbp-lacZ and PBAD-chiPΔUA+sodBbp+rpoSARN-lacZ fusions. Color codes show the sequence for the base-pairing region to the mRNA target sodB to the sRNA RyhB highlighted in yellow, remaining sequence of the mutated UA element from chiP in purple, and ARN motifs from the 5′ UTR of rpoS in blue. Black text represents bases from chiP, and red text represents bases from sodB.
- β-Galactosidase activity measured in DJS2998 (PBAD-chiPΔUA+sodBbp-lacZ) or DJS3015 (PBAD-chiPΔUA+sodBbp+rpoSARN-lacZ) carrying a plasmid expressing wild-type RyhB (pBR-RyhB), ChiX-RyhB (pDJS2219), or a vector control (pBR-plac). Samples were treated as in Fig4D, except that the LB contained 100 μM IPTG.
Source data are available online for this figure.
Discussion
The work reported here has demonstrated an unexpected flexibility in the ways in which Hfq interacts with sRNAs and mRNAs to bring about pairing. These alternative modes of Hfq action lead to different outcomes, in terms of the fate of the sRNA, the requirements of the mRNA targets of these sRNAs, and possibly the physiological roles for the sRNAs. While we emphasize two very different behaviors of sRNAs (designated as Class I and Class II sRNAs) and the effects of these different binding modes on target requirements, it is clear that there is a continuum of properties spanning from purely “Class I-like” to purely “Class II-like” sRNAs.
Class I sRNAs use proximal and rim sites of Hfq and base pair with mRNAs with distal face-binding sites
The majority of E. coli Hfq-binding sRNAs (16/24 examined) belong to Class I. Based on the work here, Class I sRNAs are dependent on the proximal face and rim sites of Hfq for their intrinsic stability, presumably reflecting a role for each of these faces in binding the sRNA (Fig3). Binding to these faces is consistent with previous in vivo and in vitro work (Schumacher et al, 2002; Mikulecky et al, 2004; Link et al, 2009). However, the rim-binding site in Class I sRNAs was not essential for activity, and mutations in the rim of Hfq (R16A) were not defective for regulation by RyhB (Figs6 and 7, and Zhang et al, 2013). In these assays, RyhB overexpression may overcome the decreased stability of the sRNA. Further studies under conditions of limiting sRNA expression are needed to understand the physiological effect of sRNA binding to the rim.
Previous work suggested that A-rich sequences (ARN motifs) found in mRNAs would bind to the distal face of Hfq and that this binding was necessary for proper regulation (Soper & Woodson, 2008; Salim & Feig, 2010; Beisel et al, 2012; Salim et al, 2012; Peng et al, 2014). Here, we show that ARN motifs introduced into the Class I RyhB sRNA blocked the ability of the chimeric RNA to regulate the target mRNA (Figs5D and 6B). These results are all consistent with a model in which, for Class I sRNAs, binding of mRNAs to the distal face is essential for pairing and regulation (Fig 3). Blocking access of the mRNA to the distal face either by mutation (Y25D) or by including ARN motifs in the sRNA itself both blocks sRNA function and stabilizes the sRNA. This also implies that, at least under the conditions assayed, an mRNA with ARN motifs cannot successfully compete with an sRNA for productive binding to the same Hfq hexamer, when both contain ARN motifs.
Class I sRNAs, while stable in hfq+ cells in the absence of transcription (measured in the presence of rifampicin), are unstable when transcription is ongoing (Fig2 and Massé et al, 2003). The stabilization of these sRNAs by a distal site mutant defective in target binding (Figs2 and EV2) supports the model that turnover is coupled to pairing. Such a coupled turnover process provides an OFF switch for regulation by these sRNAs, ensuring rapid disappearance of the sRNA in the absence of the signals that allow transcription. In fact, tight transcriptional regulation in response to environmental and nutritional signals has been identified for most Class I sRNAs (reviewed in Hoe et al, 2013). These sRNAs may thus be considered emergency response regulators that are generally only needed for a limited time.
Class II sRNAs use proximal and distal sites of Hfq and base pair with mRNAs with rim-binding sites
A subset of Hfq-binding sRNAs showed a very different pattern of accumulation in hfq mutants; these were defined as Class II sRNAs. The four sRNAs assigned to this group are dependent upon the proximal and distal sites for full accumulation. For this class of sRNAs, rim mutations led to stabilization and distal mutants led to instability, the opposite of what was observed for Class I sRNAs. These results suggest a model in which Class II sRNAs bind to the proximal and distal sites (Fig3). Consistent with these sRNAs binding to the distal site of Hfq, ARN motifs were found in regions predicted to be single stranded in each of the Class II sRNAs; deleting or improving ARN sequences affected sRNA stability (Fig4, Appendix Fig S2). Our results are consistent with the results of others showing distal site interactions for RprA and OxyS (Updegrove et al, 2008; Olejniczak, 2011), both in the intermediate class of sRNAs in our tests, and distal face binding to A-rich sequences in ChiX (Ellis et al, 2015; Małecka et al, 2015). We observed that deletion of the ARN motifs only partially affected function of the two Class II sRNAs tested under overexpression conditions (Fig4D and H). Thus, as for the rim-binding site with the Class I sRNAs, this second site of sRNA interaction with Hfq affected Class II sRNA stability more than activity. When a Class II sRNA deleted for its ARN motifs is expressed in single copy, however, this loss of stability is correlated with a decrease in regulation (Appendix Fig S4).
Given that the Class II sRNAs have distal site-binding motifs, we imagined that their target mRNAs might not be free to bind to the distal site, and in fact, there were no clear ARN motifs in some Class II target mRNAs. Our results indicate a critical role for the Hfq rim for regulation of these mRNA targets. Cells expressing the HfqR16A mutant inhibited Class II regulation, as well as the ability of a Class I sRNA to regulate a chimeric target that depends on a rim-binding site (the chiP+sodBbp fusion in Fig7). In addition, rim mutants stabilized MgrR and CyaR in washout experiments (Fig2), suggesting, in parallel with Class I behavior, that barring target mRNA binding may inhibit sRNA decay by inhibiting pairing.
Rim binding has been observed in Hfq crystals with the RydC Class I sRNA (Dimastrogiovanni et al, 2014) and appears to select UA-rich sequences (Sauer et al, 2012). UA-rich regions were identified in chiP, a target of ChiX, and eptB, a target of MgrR; mutation of these sites significantly interfered with regulation by these Class II sRNAs and blocked the ability of a chimeric ChiX-RyhB RNA to regulate chiP+sodBbp (Fig8). While our data indicate a requirement for Hfq binding to targets is conserved for Class I and Class II sRNAs, it is less clear whether the geometry of binding (relative arrangement of pairing region and binding regions) is different for the targets of the two sRNA classes. However, inserting ARN sites in place of rim-binding sites in chiP+sodBbp modestly restored regulation by RyhB (no ARN sites), but not regulation by ChiX-RyhB (carrying ARN motifs) (Fig9), suggesting flexibility in location of Hfq-binding motifs. Similar flexibility has been seen in experiments in which Hfq-binding regions in the rpoS leader were moved to other locations in the mRNA (Peng et al, 2014).
The distal face binding of Class II sRNAs is likely to have implications for their functions. It is noteworthy that Class II, particularly ChiX and McaS, show greater overall stability in washout experiments (Fig2). Unlike the tested Class I sRNAs, ChiX is not degraded upon binding most of its mRNAs (Figueroa-Bossi et al, 2009; Overgaard et al, 2009a). This suggests that tethering of the ChiX and other Class II sRNAs to the proximal and distal faces is not easily displaced upon pairing with the mRNA target. The loss of coupled degradation means that there is no intrinsic mechanism for shutting off regulation by these sRNAs. ChiX sRNA is turned off by an RNA decoy, made under specific growth conditions, that interacts with the sRNA, destabilizing the sRNA terminator and leading to degradation of the sRNA. In growing cells, ChiX levels are high enough to fully repress its target(s); repression is only relieved in response to signals that induce the decoy, making ChiX more of a silencer than an emergency response sRNA. The same may be true for other Class II sRNAs. Although McaS is induced by specific stationary phase growth conditions (Thomason et al, 2012), its stability suggests it will only disappear slowly after growth conditions change. Possibly, a decoy RNA or adapter protein that increases the degradation of this sRNA remains to be found. Consistent with a silencing role for McaS, cells lacking the sRNA have altered biofilm formation, a process of a long duration. The two other Class II sRNAs, CyaR and MgrR, were somewhat unstable in washout experiments, suggesting that interactions with Hfq proximal and/or distal sites may not be as strong. However, MgrR, whose expression is regulated by the PhoQP two-component system, is unusually insensitive to repression by magnesium and thus silences the eptB target mRNA under many growth conditions (Moon & Gottesman, 2009). In fact, the EptB protein is only synthesized when the promoter for eptB is stimulated (by σE), and the newly transcribed mRNA outstrips the repression by MgrR (Moon et al, 2013). Thus, overcoming Class II sRNA regulation could also occur by specifically inducing target synthesis; as soon as inducing signals disappear, sRNA regulation would resume. These features are consistent with Class II sRNAs acting more as silencers and requiring mechanisms other than target-mediated sRNA turnover for relief of the silencing.
sRNAs and mRNAs have a continuum of rim and distal face-binding sites, but some conserved features are required for effective regulation
For much of this study, we have classified the sRNAs as one class or the other, but as noted for ArcZ, RprA, OxyS, and SgrS, there is likely to be a continuum for sRNAs and for mRNAs as well, with stronger or weaker binding to the Hfq rim or distal face depending on the location and length of ARN and UA-rich sequences. In addition, some mRNAs are regulated by both Class I and Class II sRNAs. For instance, csgD is subject to negative regulation by a number of Class I sRNAs (OmrA, OmrB, RydC, RprA, GcvB) as well as the Class II sRNA McaS (reviewed in Mika & Hengge, 2014), and we find that regulation by a Class I sRNA is disrupted more by a distal site mutant then a rim mutant, while the reverse is true for the Class II sRNA (Fig7).
Our experiments suggest that a Class I sRNA, while it usually uses targets with ARN motifs, can also regulate targets with rim sites. For example, we observed RyhB repression of the chiP-sodBbp target (Fig6D). This suggests that target rim sites may be able to bind in concert with or displace sRNA rim binding. However, we suggest that Class II targets do not easily regulate mRNAs that solely have ARN motifs. Presumably both classes of sRNAs will be able to act if both ARN and rim motifs are present. However, in some cases, an sRNA from one class may interfere with recognition of a critical mRNA site for the other class.
Despite the continuum of binding sites, a number of characteristics are required for all effective regulation. sRNAs are anchored at two types of sites within Hfq, with the proximal site likely the primary one. The secondary interactions (rim for Class I and distal for Class II) contribute to stability but are not essential for in vivo function, at least when the sRNAs are abundant. However, these secondary interactions will limit Hfq accessibility to mRNAs. As a consequence, Class II sRNAs can exclude Class I targets from Hfq, as seen in Fig6. Coupled with the observation that some Class II sRNAs are less subject to displacement after pairing, these findings may explain why ChiX was most effective in competing with other sRNAs for regulation (Moon & Gottesman, 2011). It is less clear whether the rim interactions of Class I sRNAs can exclude Class II mRNAs. Our results also demonstrate that targets of both classes of sRNA require interactions with Hfq. Rapid decay of the sRNA is only observed when the Hfq-binding sites for the mRNA are present, although not every sRNA will decay with each use. We assume that rapid decay reflects displacement from Hfq after pairing, but exactly how this occurs remains to be explored. Finally, the requirement that the mRNA target requires Hfq-binding sites distinct from those the cognate sRNA binds provides strong support for the regulation occurring on a single hexameric Hfq ring.
We have focused on the tertiary complex formed between Hfq, an sRNA, and its mRNA targets. This complex also interacts with other proteins such as ribonuclease E and polynucleotide phosphorylase (De Lay & Gottesman, 2011), and it is likely that these interactions are impacted by the different modes of sRNA binding. Although we have not explored it here, different RNA-binding configurations additionally impose topological constraints that may or may not facilitate base pairing. Overall, the flexibility in Hfq binding to both sRNAs and their mRNA targets allows for increased permutations for sRNA-mediated regulation as well as a hierarchy of use. It will be interesting to see whether this flexibility is conserved for other organisms as well as for the eukaryotic Sm and Lsm protein counterparts.
Materials and Methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study and their construction are listed in Appendix Tables S1 and S2. All strains used in this study are derivatives of the E. coli K12 strain MG1655. Mutations were introduced into strains by λ-Red-mediated recombineering and P1 transduction as described (Zhang et al, 2013). The sequences of the oligonucleotides obtained from Integrated DNA Technologies (IDT) used for strain and plasmid construction are given in Appendix Table S3.
Strains carrying pBAD-translational lacZ fusions were constructed in PM1205 (Mandin & Gottesman, 2009). This strain carries a counter-selectable sacB gene as well as a catR gene inserted between pBAD and lacZ at the chromosomal lacZ site, it also carries a temperature-inducible mini-lambda carrying λ-Red recombination genes at the λ att site. For the fusions constructed for this study, gBlocks gene fragments (IDT) with 40 bp of homology to the PBAD promoter followed by the specific gene sequence starting at the transcription start site through codon 9, followed by 40 bp of homology to lacZ, were introduced into the PM1205 chromosome using λ-Red recombination (Yu et al, 2000). Successful recombinants were obtained by selection for growth in the presence of sucrose, and the resulting fusions were verified by PCR and sequencing.
Plasmids expressing sRNAs and chimeric sRNAs of interest were constructed by annealing complementary primers with AatII and EcoR1 sites for ligation into the digested pBR-plac vector (Guillier & Gottesman, 2006). The ligation products were transformed into strain DJ624, and plasmids were selected on Luria–Bertani (LB) plates containing ampicillin. Inserts were confirmed by sequencing.
Bacterial growth conditions
Bacterial strains were grown at 37°C in LB rich. Ampicillin (50–100 μg/ml) and kanamycin (30–50 μg/ml) were added where appropriate. IPTG was added at a final concentration of 100 μM or 10 μM, and L-arabinose was added at a final concentration of 0.002% or 0.2%.
To examine RNA stability, cells were grown in LB with ampicillin (100 μg/ml) at 37°C to an OD600 of ∼ 0.4, and 100 μM IPTG was added for 15 min. Aliquots (15 ml) of culture were pipetted into a clean flask (+ rifampicin samples) or across a filter membrane (wash out samples). Rifampicin (250 μg/ml final concentration) was added to the culture in the clean flask. The culture on the filter membrane was washed with two volumes of LB and then resuspended in 15 ml LB with ampicillin. Cultures were returned to 37°C, and samples were removed at the intervals indicated.
RNA isolation
Total RNA was extracted by the hot phenol method described previously (Massé et al, 2005) or using the TRIzol Reagent (Invitrogen) followed by isopropanol precipitation.
Northern blot analysis
Northern blot analysis was performed by fractionating indicated amounts of total RNA for each sample on 8% polyacrylamide urea gels containing 8 M urea in 1× TBE buffer at 300 V for 90 min. The RNA was transferred to a Zeta-Probe GT membrane (Bio-Rad) at 20 V for 16 h in 0.5× TBE. The RNA was cross-linked to the membrane by UV irradiation, and the membrane was probed with the 5′-32P-end-labeled primer specific to the indicated genes as listed in Appendix Table S3 in ULTRAhyb-Oligo Hybridization Buffer (Ambion) at 45°C overnight and washed as in Zhang et al (2003). Phosphoimages of the Northern membranes were analyzed by using GE Typhoon imaging scanner and ImageJ software. Graphs were plotted with Prism software. All quantitation is shown in the Source Data for each figure.
Primer extension analysis
Primer extension analysis was performed using oligonucleotides listed in Appendix Table S3 specific to the indicated genes as previously described (Zhang et al, 1998). RNA samples (5 μg of total RNA) were incubated with 2 pmol of 5′-32P-end-labeled primer at 80°C and then slow-cooled to 42°C. After the addition of dNTPs (1 mM each) and AMV reverse transcriptase (10 U, Life Sciences Advanced Techologies Inc.), the reactions were incubated in a 10-μl reaction volume at 42°C for 1 h. The reactions were terminated by adding 10 μl of Stop Loading Buffer, and cDNA products then were fractionated on 8% polyacrylamide urea gels containing 8 M urea in 1× TBE buffer at 70 W for 70 min.
β-Galactosidase assays
Unless stated otherwise, samples were grown in LB medium containing 100 μg/ml ampicillin, 10 μM IPTG, and 0.002% arabinose at 37°C to early stationary phase (OD600 ∼ 1.0–1.2), and assayed for β-galactosidase activity using o-nitrophenyl-β-D-galactopyranoside as a substrate. All assays were carried out in triplicate with data shown in Source Data for each figure.
Acknowledgments
We thank N. Majdalani as well as S. Woodson and members of her laboratory for helpful discussions. We thank N. De Lay, T. Updegrove, J. Chen, and other members of our laboratories for their comments on the manuscript. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research and in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Author contributions
Studies were planned by DJS, AZ, SG and GS and carried out by DJS and AZ. DJS, AZ, SG and GS carried out data analysis and manuscript preparation.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Source Data for Expanded View and Appendix
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Source Data for Figure 9
References
- Balbontín R, Fiorini F, Figueroa-Bossi N, Casadesús J, Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol Microbiol. 2010;78:380–394. doi: 10.1111/j.1365-2958.2010.07342.x. [DOI] [PubMed] [Google Scholar]
- Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011;17:1172–1189. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimastrogiovanni D, Fröhlich KS, Bandyra KJ, Bruce HA, Hohensee S, Vogel J, Luisi BF. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. eLife. 2014;3:e05375. doi: 10.7554/eLife.05375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Trussler RS, Haniford DB. Hfq binds directly to the ribosome binding site of IS10 transposase mRNA to inhibit translation. Mol Microbiol. 2015;96:633–650. doi: 10.1111/mmi.12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- de Haseth PL, Uhlenbeck OC. Interaction of Escherichia coli host factor protein with oligoriboadenylates. Biochemistry. 1980;19:6138–6146. doi: 10.1021/bi00567a029. [DOI] [PubMed] [Google Scholar]
- Hoe CH, Raabe CA, Rozhdestvensky TS, Tang TH. Bacterial sRNAs: regulation in stress. Int J Med Microbiol. 2013;303:217–229. doi: 10.1016/j.ijmm.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Holmqvist E, Reimegård J, Sterk M, Grantcharova N, Römling U, Wagner EGH. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen MG, Nielsen JS, Boysen A, Franch T, Møller-Jensen J, Valentin-Hansen P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol Microbiol. 2012;84:36–50. doi: 10.1111/j.1365-2958.2012.07976.x. [DOI] [PubMed] [Google Scholar]
- Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Małecka EM, Stróżecka J, Sobańska D, Olejniczak M. Structure of bacterial regulatory RNAs determines their performance in competition for the chaperone protein Hfq. Biochemistry. 2015;54:1157–1170. doi: 10.1021/bi500741d. [DOI] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol. 2009;72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 2014;11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Moon K, Gottesman S. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol. 2009;74:1314–1330. doi: 10.1111/j.1365-2958.2009.06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Six DA, Lee HJ, Raetz CR, Gottesman S. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol Microbiol. 2013;89:52–64. doi: 10.1111/mmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011;50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA. 2011;108:13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol Microbiol. 2009a;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- Overgaard M, Kallipolitis B, Valentin-Hansen P. Modulating the bacterial surface with small RNAs: a new twist on PhoP/Q-mediated lipopolysaccharide modification. Mol Microbiol. 2009b;74:1289–1294. doi: 10.1111/j.1365-2958.2009.06943.x. [DOI] [PubMed] [Google Scholar]
- Panja S, Schu DJ, Woodson SA. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013;41:7536–7546. doi: 10.1093/nar/gkt521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Soper TJ, Woodson SA. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J Mol Biol. 2014;426:275–285. doi: 10.1016/j.jmb.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AA, Johansen J, Nielsen JS, Overgaard M, Kallipolitis B, Valentin-Hansen P. A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol Microbiol. 2009;72:566–577. doi: 10.1111/j.1365-2958.2009.06688.x. [DOI] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KE, Orans J, Kovach AR, Link TM, Brennan RG. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014;42:2736–2749. doi: 10.1093/nar/gkt1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS ONE. 2010;5:e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim NN, Faner MA, Philip JA, Feig AL. Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 2012;40:8021–8032. doi: 10.1093/nar/gks392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci USA. 2012;109:9396–9401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 2013;10:610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrero P, Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol. 2012;38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason MK, Fontaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove TB, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry. 2008;47:11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- Updegrove TB, Wartell RM. The influence of Escherichia coli Hfq mutations on RNA binding and sRNA•mRNA duplex formation in rpoS riboregulation. Biochim Biophys Acta. 2011;1809:532–540. doi: 10.1016/j.bbagrm.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang L, Wu J, Gong Q, Shi Y. Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA. Nucleic Acids Res. 2013;41:5938–5948. doi: 10.1093/nar/gkt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. Lsm proteins and Hfq: life at the 3′ end. RNA Biol. 2013;10:592–601. doi: 10.4161/rna.23695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol. 2013;425:3678–3697. doi: 10.1016/j.jmb.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Source Data for Expanded View and Appendix
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Source Data for Figure 9