Abstract
Purpose
Quantitative T1ρ imaging is beneficial for early detection for osteoarthritis, but has seen limited clinical use due to long scan times. This work evaluates the feasibility of accelerated T1ρ mapping for knee cartilage quantification using a combination of compressed sensing (CS) and data-driven parallel imaging (ARC).
Methods
A sequential combination of ARC and CS, both during data acquisition and reconstruction, was used to accelerate the acquisition of T1ρ maps. Phantom, ex vivo (porcine knee) and in vivo (human knee) imaging was carried out on a GE 3T MR750 scanner. T1ρ quantification post CS-accelerated acquisition was compared with non CS-accelerated acquisition for various cartilage compartments.
Results
Accelerating image acquisition using CS did not introduce major deviations in quantification. The coefficient of variation for the root mean squared error (CV(RMSE)) increased with increasing acceleration, but for in vivo measurements, it stayed under 5% for net acceleration factor up to 2, where the acquisition was 25% faster than the reference (only ARC).
Conclusion
To the best of our knowledge this is the first implementation of CS for in vivo T1ρ quantification. These early results show that this technique holds great promise in making quantitative imaging techniques more accessible for clinical applications.
Keywords: compressed sensing, T1ρ, cartilage, ARC, MRI
INTRODUCTION
Osteoarthritis (OA) is a major public heath problem in the United States, affecting nearly 27 million people(1). Cartilage degeneration and the ensuing loss of joint function, as a result of OA, is a leading cause of work disability and reduced quality of life. As such, non-invasive early detection of cartilage degeneration is of increasing clinical importance. Recent studies have shown the advantages of quantitative MRI (2–4) over conventional qualitative MRI, particularly for early detection, since the biochemical changes measured by these techniques occur much sooner than the morphological changes seen with conventional MRI. This work focuses on quantification of T1ρ relaxation times.
Cartilage degeneration is triggered by damage to the collagen-proteoglycan (PG) matrix. The T1ρ parameter reflects the changes in this matrix, which acts as a motion-restrictive environment for the surrounding water molecules. T1ρ relaxation rate (1/T1ρ) has been shown to decrease linearly with decreasing PG content(5,6). Additionally, in vivo studies have demonstrated a connection between T1ρ measurements and OA, with higher cartilage T1ρ values for patients with OA(7,8).
Quantitative T1ρ imaging requires the acquisition of multiple images with different spin-lock times (TSLs) to generate the T1ρ maps. This translates to long scan times, thus limiting the widespread clinical use of this technique. Reducing the number of TSLs acquired is one way of reducing the acquisition times, but this compromises the T1ρ quantification accuracy and precision(9). Another commonly used technique for accelerating the acquisition is parallel imaging(10–12), but in this case the acceleration factor is limited by the number of coils used.
Compressed sensing (CS) is still a relatively new technique for accelerating image acquisition(13,14). It relies on the inherent sparsity and compressibility of MR data to overcome under-sampling induced artifacts in the acquired data(15). CS MRI has predominantly been shown to be beneficial in applications that rely on anatomical image quality(16), and only recently have there been some publications exploring its use in quantitative MR parameter mapping(17–19). However, with regard to application of CS to accelerate T1ρ mapping, the literature is still lacking. Recently, Zhu et al(20) showed very promising results in simulated data for brain and spine imaging. The task is even more challenging for knee cartilage imaging due to the small size of the structures. Thus the goal of this work is to evaluate the feasibility of accelerated T1ρ mapping for knee cartilage quantification, and our chosen approach is a combination of CS and data-driven parallel imaging (ARC).
METHODS
Sequential Combination of Compressed Sensing and Data-Driven Parallel Imaging
CS and ARC can be combined in either an integrated(21,22) or a sequential(14,23) manner, with each having distinct advantages. In the sequential implementation, acceleration is clearly split between CS and ARC, allowing better conditioning for both methods than in the integrated approach with full acceleration, though the integrated approach could be more efficient(22). Additionally, with the sequential combination, CS can be used with any exiting ARC method with minimum modifications. This work employs a sequential combination of ARC and CS, both during k-space data acquisition and image reconstruction(23). The two sequential steps are illustrated in Fig 1. In the first step, the k-space is under-sampled following the strategy used for ARC. This intermediate sampling pattern is composed of a uniformly under-sampled grid in the outer k-space regions (higher frequency) and a fully-sampled central region (low frequency) called auto-calibration signal (ACS).
Figure 1.
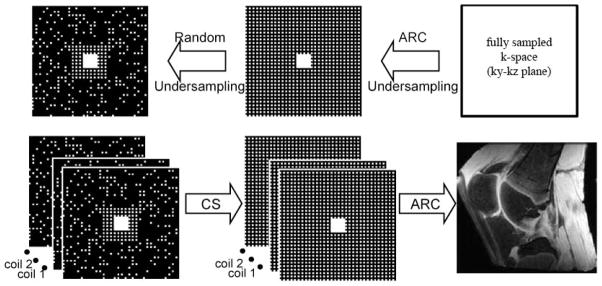
The sampling pattern was defined in two consecutive steps (top row): regular under sampling with ARC followed by random undersampling for CS. The center of the k-space remained fully sampled. The image reconstruction was also performed in two consecutive steps (bottom row): first CS followed by ARC reconstruction.
In the second step, the remaining k-space points on the uniformly under-sampled grid are further under-sampled in a random fashion. The sampling pattern used is Gaussian pseudo-random distribution with standard deviation in each direction equal to the width of the random matrix in that direction. Since CS works better with full sampling near the center of k-space, no under-sampling is used over an area slightly larger than the ACS region. This resulting k-space sampling pattern is used for the acquisition of all the different repetitions for the T1ρ-weighted gradient echo sequence.
The acquired data sets are then reconstructed with the reversed sequential combination of CS and ARC. The k-space data are masked with a grid of uniformly undersampled points and used in the first step of the reconstruction. Since the aliased images from each coils are still sparse(14), the CS algorithm is used to fill in the missing randomly undersampled points on the uniformly undersampled grid. The objective function for the CS reconstruction is
| [1] |
where the derivatives ∇y and ∇z are implemented as nearest-neighbor finite differences of the complex image m, || ||1 is the L1-norm implemented as a sum over all image pixels,
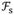 denotes the Fourier transform and y is the measured k-space data. A non-linear conjugate gradient (CG) solver with 15 iterations is used to minimize the penalty (L1-norm in Eq. [1]) for each coil (13). The inequality constraint in Eq. [1] is imposed by substituting the measured k-space data back into the estimated k-space data at the measured locations following each CG iteration.
denotes the Fourier transform and y is the measured k-space data. A non-linear conjugate gradient (CG) solver with 15 iterations is used to minimize the penalty (L1-norm in Eq. [1]) for each coil (13). The inequality constraint in Eq. [1] is imposed by substituting the measured k-space data back into the estimated k-space data at the measured locations following each CG iteration.
The first step yields a uniformly undersampled k-space. In the second step, the ACS points are combined with the now uniformly undersampled k-space and ARC reconstruction is applied to fill in the remaining k-space points. The reconstruction used is similar to GRAPPA (24) but fills k-space in (x,ky,kz) space for slightly greater efficiency (25). Fully sampled k-space is thus obtained for each coil and the respective images are combined using sum of squares across coils.
Data Acquisition
The scanning was carried out on a GE 3T MR750 scanner (GE Healthcare, Milwaukee, WI) with an 8-channel phased array knee coil (Invivo, Gainsville, FL).
All T1ρ MR images were acquired with the 3D MAPSS pulse sequence(26). The sequence is composed of two parts: magnetization preparation using a series of spin-lock pulse clusters for generating the T1ρ contrast, followed by segmented 3D SPGR for data acquisition. The pulse sequence parameters were set to match the protocol employed in a number of research studies; FOV = 14 cm, matrix resolutions 256x128, 20 to 24 slices, slice thickness 4 mm, Views Per Segment (VPS) = 64, time of recovery = 1.2 s, time of spin-lock (TSL) = 0/2/4/8/12/20/40/80 ms and spin-lock frequency = 500 Hz was used.
As previously described, the sampling pattern was obtained by combining two under-sampling strategies: 2-fold ARC under-sampling as well as different random under-sampling accelerator factors. Table 1 shows the different combination of accelerations used in this work. Net acceleration is calculated based on the actual imaging time for the accelerated acquisition with respect to the time required for a fully-sampled acquisition.
Table 1.
Combination of ARC and CS used during acquisition and the corresponding net acceleration obtained. ‘X’ indicates the parameters used for the various acquisitions.
| ARC | CS | Net acceleration | Phantom | Ex vivo | In vivo | |
|---|---|---|---|---|---|---|
| Fully sampled | 1 | 1 | 1 | X | ||
| ARC only | 2 | 1 | 1.5 | X | X | X |
| ARC & CS | 2 | 2 | 1.93 | X | X | X |
| 2 | 3 | 2.2 | X | X | X | |
| 2 | 4 | 2.31 | X | |||
| 2 | 5 | 2.35 | X |
The ACS region covered the central 32 k-space lines in phase encoding direction. The size of the region where no random under-sampling was applied was 64x10 (ky-kz) at the center of the k-space.
T1ρ acquisitions were first performed on a phantom set made of 6 tubes (25mm diameter) filled with different concentrations (2% to 4%) of agarose gel. Ex vivo acquisition was performed on porcine knee (n = 1) and in vivo acquisitions on healthy volunteers (n = 2).
The study was approved by the Committee for Human Research at our institution, and informed consent was obtained from all volunteers.
Simulation
The raw k-space data of a fully sampled acquisition was used to simulate a larger number of acceleration factors. The full k-space was under-sampled using the same strategies as the ones used on the MRI scanner: 2 fold ARC under-sampling combined with random under-sampling. Acceleration factors between 1.65(only ARC) and 2.77 (ARC+CS) were used in this simulation. The images were reconstructed using the same reconstruction algorithm employed on the MRI scanner.
Reconstruction and data analysis
The acquired data were directly reconstructed on the MRI scanner and the DICOM images were exported for analyses. The raw k-space data sets for fully sampled acquisitions were saved and used in simulations. All algorithms used for analysis were implemented in Matlab (Mathworks, Natick, MA).
For the phantom images, circular ROI (20mm-diameter) were placed in the center of each tube on the 5 central slices of the acquisition.
For the knee images, each acquisition was registered to the first acquired data set (only accelerated with ARC). Additionally, in case of in vivo acquisitions, individual T1ρ-weighted images were first registered to the images with TSL=0. This was carried out in order to correct for motion artifacts during T1ρ quantification. The rigid registration was performed using the VTK CISG Registration Toolkit (27). The knee cartilage was then segmented in different compartments on the first echo image acquired with TSL=0 ms. The 6 different compartments are: lateral/medial femoral condyles (LFC/MFC), the lateral/medial tibia (LT/MT), patella (PAT) and trochlea (TRO). The segmentation was performed semi-automatically using an in-house developed software (28). In addition to the cartilage compartments, a small region in the homogeneous part of the gastrocnemius muscle was also segmented. Only one segmentation per subject was performed since inter scan motion was corrected by the registration process.
T1ρ maps were generated using voxel-by-voxel 3 parameter mono-exponential fit
| [2] |
The mean value and standard deviation within each defined ROIs was calculated. The fidelity of the CS-accelerated acquisition to the reference acquisition (only ARC) was estimated using the coefficient of variation of the root-mean-square error (CV(RMSE)) calculated across the ROIs. The choice for the reference acquisition was based on existing protocols for human knee cartilage quantification, that use parallel imaging (only ARC) to reduce scan time and have been shown to have image quality comparable to fully-sampled acquisitions (11).
RESULTS
T1ρ values between 29 ms and 60 ms were measured in phantoms with different agarose concentrations. When comparing the CS-accelerated acquisitions, with net accelerations of 1.9 and 2.2, to the reference acquisition (only ARC), the difference in T1ρ values was below 2.6% and 3.3% respectively. The CV(RMSE) calculated across the 6 phantoms was equal to 2% and 2.4% respectively.
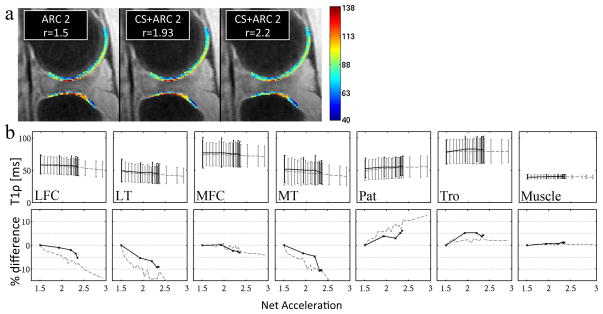
Figure 2(a) shows the results for the ex vivo pig knee acquisition, where T1ρ color maps are overlaid on gray-scale images (TSL = 0) for 3 different acceleration factors. The T1ρ quantification in the different compartments performed in these acquisitions (solid line) and in simulations (dashed line) is presented in Fig. 2(b). The T1ρ and percentage difference between the accelerated acquisitions and reference acquisition (only accelerated with ARC) are displayed for the different compartments. The T1ρ values had a greater than 5% difference for acceleration factors larger than 1.93 in the LT and TRO compartments, 2.32 in MT and PAT compartments, and 2.35 in LFC compartment. In the MFC and muscle compartment the difference stayed below 5% and was equal to 3.06% and 1.21% respectively for acceleration factor of 2.35. The CV(RMSE) for the four accelerated acquisitions (Net acceleration of 1.93, 2.2, 2.31, and 2.35) was 3.68%, 4.21%, 5.86%, 6.22% respectively across the 6 cartilage compartments.
Figure 2.
(a) T1ρ color maps of the lateral tibia and femoral condyle measured in the ex vivo experiment. The color maps overlay on the image corresponding to TSL=0 (gray scale). (b) Top row: T1ρ measured in the simulation (dashed line) and in the ex vivo acquisition (solid line). Bottom row: Percent difference to the acquisition accelerated with ARC only.
The acquisition times of the in vivo measurement were between 7.25 min and 6.33 min. Following each acquisition, a first set of images was reconstructed within 10s, to allow a quick image quality check. The CS calculation was skipped for this first reconstruction and the empty k-space lines were zero-filled. The second set of images correctly reconstructed with CS was obtained within 10 mins. Figure 3(a) shows images and T1ρ map obtained from one volunteer. The T1ρ color map of the LFC and LT compartments are overlaid on the first echo image. In vivo T1ρ quantification in three acquisitions performed with different acceleration factors on two volunteers is presented in Fig. 3(b). The CV(RMSE) across the 6 cartilage compartments and both volunteers was 4.7 and 9.6% for acquisitions with net acceleration of 1.9 and 2.2 respectively.
Figure 3.
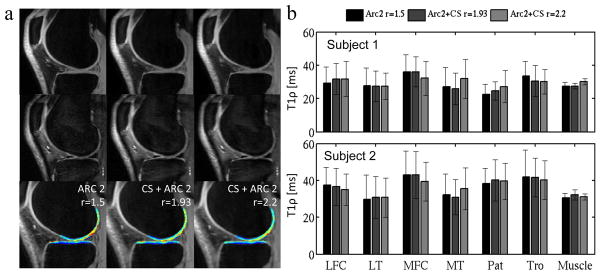
(a) In vivo images acquired with the T1ρ MAPSS sequence. TSL=0ms (top row) TSL=40 (middle row) and T1ρ color map (bottom row) calculated from the 8 set of data acquired with 8 different TSL. (b) In vivo T1ρ quantification in the 7 compartments for the two different volunteers.
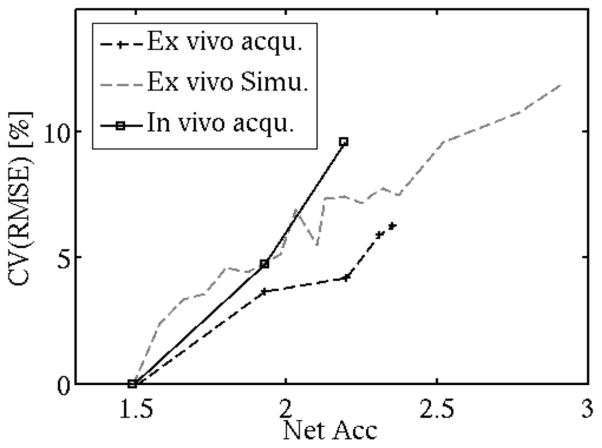
The CV(RMSE) calculated for the simulations, the ex vivo, and the in vivo acquisitions is presented in Fig. 4.
Figure 4.
CV(RMSE) calculated across all cartilage compartments for the ex vivo acquisitions, the simulations and in vivo acquisitions.
DISCUSSION
The data acquisition and reconstruction for CS and data-driven parallel imaging (ARC) has been successfully implemented on a clinical MRI scanner.
Two sets of images were reconstructed for these acquisitions. The first set was not used for any of the analysis but was useful to decide whether the scan had been successfully acquired or needed reacquiring. The images contained artifacts due to random under-sampling but their quality was sufficient for detecting errors during prescription or major motion artifacts during acquisition. The second set of images, obtained 10 minutes later, were used for the analysis.
Accelerating the phantom acquisition with CS did not introduce major deviations in quantification. Maximum difference of 3.3% was measured for acquisitions with acceleration factor of 2.2 when compared with the non CS-accelerated acquisition. These values were obtained in 20 mm circular ROIs in 25 mm wide cylindrical phantoms. The signal was homogenous within the ROIs and in their surrounding, thus explaining the good results obtained in the phantom.
In the ex vivo experiment, the T1ρ values in the muscle show similar results as in the phantom experiment. When the ROI is surrounded by similar tissue, as in the case of muscle, the T1ρ value does not change much with acceleration. The error stayed under 1.21% in the muscle compartment for all the acquisitions. The difference in T1ρ values between non CS-accelerated and CS-accelerated acquisitions is higher when measured in the cartilage. This difference exceeded 5% in two compartments at an acceleration factor of 1.93. However the overall CV(RMSE) across all compartment stayed below 5% for acceleration lower than 2.31.
For increasing accelerations, the simulation results follow a similar global trend as that from the acquired data but there are deviations for each compartment. One of the potential sources of difference between the simulations and the acquisitions could be the noise in the original data. In the simulation, the different acceleration images were reconstructed from the same acquired data set and had the same noise, whereas in the latter case, they were separate acquisitions and hence the noise was different.
The consistent results in the phantom and in the muscle on one hand, and the larger deviation in the cartilage compartments on the other, suggests that though the filtering introduced by CS reduces the noise in the image, it unfortunately also introduces signal leakage between neighbor pixels which results in variation of the T1ρ quantification.
The in vivo acquisitions have higher quantification error compared to the ex vivo acquisitions due to mild motion artifacts and potential mis-registration. Nevertheless no obvious problem was visually detected. Due to long acquisition times only two CS accelerated datasets were acquired for each subject. The limited number of acquisitions makes it difficult to predict the relation between evolution of T1ρ values in vivo and acceleration factor, but nevertheless the CV(RMSE) across all cartilage compartment follow similar trend as for the ex vivo simulation and acquisition (Fig. 4). In all these measurements the CV(RMSE) increases with acceleration. It crosses the 5% limit for acceleration factor of 2. With this acceleration factor the acquisition is 25% faster than in the existing protocols with ARC-only acquisition.
To the best of our knowledge this is the first work demonstrating in vivo implementation of CS for T1ρ quantification. We have shown the feasibility of this approach, though there is a room for much improvement. With this current algorithm the gain in acquisition time is relatively limited. There are a number of different algorithms and sparsity domains that can be implemented and used within the CS framework for further improvements - both in scan time reduction as well as reconstruction accuracy.
In this implementation each echo was reconstructed independently. The redundancy of signal from anatomical structures between echoes was not used to accelerate the acquisition. This dimension has great potential for accelerating the acquisition but due to the non-linear nature of exponential decays, its implementation requires more work.
This work also showcases the challenges of using CS for quantitative imaging of the cartilage. The cartilage is a thin structure surrounded by tissues having different signal properties. Thus any filtering of the image induced by the reconstruction has a major impact on the quantification, and needs to be taken into account while choosing the algorithm. Furthermore, the iterative reconstruction produces a solution by considering the entire image. This solution may not be optimal for the cartilage, which represents only a small proportion of entire volume. There are CS methods that target specific regions of the image that might be extended to be beneficial here(29).
When using parallel imaging techniques for proton MRI, the penalty for acquiring fewer signals is a loss of SNR in the final image by a factor of the square root of the acceleration factor. CS reconstruction is inherently a denoising procedure and thus there is little or no SNR decrease depending on acceleration(13,23) but the real penalty comes with the filtering of the images and can manifest as loss of low-contrast features. The artifacts can be subtle and vary with different implementation methods.
In conclusion, CS for quantitative T1ρ MRI is still very much an open field, but we believe that advanced acceleration techniques such as ours, where we combine CS and PI for scan time reduction hold great promise in making quantitative imaging techniques more accessible for clinical applications.
Acknowledgments
Grant Support:
NIH P50 AR060752, GE Healthcare
References
- 1.Helmick CG, Felson DT, Lawrence RC, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Arthrit Rheum. 2008;58:15–25. doi: 10.1002/art.23177.. [DOI] [PubMed] [Google Scholar]
- 2.Binks DA, Hodgson RJ, Ries ME, Foster RJ, Smye SW, McGonagle D, Radjenovic A. Quantitative parametric MRI of articular cartilage: a review of progress and open challenges. Brit J Radiol. 2013;86:20120163–20120163. doi: 10.1259/bjr.20120163.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roemer FW, Crema MD, Trattnig S, Guermazi A. Advances in imaging of osteoarthritis and cartilage. Radiology. 2011;260:332–354. doi: 10.1148/radiol.11101359.. [DOI] [PubMed] [Google Scholar]
- 4.Gold GE, Chen CA, Koo S, Hargreaves BA, Bangerter NK. Recent advances in MRI of articular cartilage. AJR Am J Roentgenol. 2009;193:628–638. doi: 10.2214/AJR.09.3042.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duvvuri U, Reddy R, Patel SD, Kaufman JH, Kneeland JB, Leigh JS. T1rho-relaxation in articular cartilage: effects of enzymatic degradation. Magn Reson Med. 1997;38:863–867. doi: 10.1002/mrm.1910380602. [DOI] [PubMed] [Google Scholar]
- 6.Regatte RR, Akella SVS, Borthakur A, Kneeland JB, Reddy R. Proteoglycan Depletion–Induced Changes in Transverse Relaxation Maps of Cartilage. Acad Radiol. 2002;9:1388–1394. doi: 10.1016/S1076-6332(03)80666-9.. [DOI] [PubMed] [Google Scholar]
- 7.Duvvuri U, Charagundla SR, Kudchodkar SB, Kaufman JH, Kneeland JB, Rizi R, Leigh JS, Reddy R. Human knee: in vivo T1(rho)-weighted MR imaging at 1. 5 T--preliminary experience. Radiology. 2001;220:822–826. doi: 10.1148/radiol.2203001662.. [DOI] [PubMed] [Google Scholar]
- 8.Regatte RR, Akella SVS, Wheaton AJ, Lech G, Borthakur A, Kneeland JB, Reddy R. 3D-T1rho-relaxation mapping of articular cartilage: in vivo assessment of early degenerative changes in symptomatic osteoarthritic subjects. Acad Radiol. 2004;11:741–749. doi: 10.1016/j.acra.2004.03.051.. [DOI] [PubMed] [Google Scholar]
- 9.Zhao F, Deng M, Yuan J, Teng G-J, Ahuja AT, Wang Y-XJ. Experimental evaluation of accelerated T1rho relaxation quantification in human liver using limited spin-lock times. Korean J Radiol. 2012;13:736–742. doi: 10.3348/kjr.2012.13.6.736.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn Reson Med. 2008;60:895–907. doi: 10.1002/mrm.21728.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo J, Li X, Banerjee S, Han E, Majumdar S. Parallel imaging of knee cartilage at 3 Tesla. J Magn Reson Imaging. 2007;26:1001–1009. doi: 10.1002/jmri.21122.. [DOI] [PubMed] [Google Scholar]
- 12.Bolbos RI, Zuo J, Banerjee S, Link TM, Benjamin Ma C, Li X, Majumdar S. Relationship between trabecular bone structure and articular cartilage morphology and relaxation times in early OA of the knee joint using parallel MRI at 3T. Osteoarthr Cartilage. 2008;16:1150–1159. doi: 10.1016/j.joca.2008.02.018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391.. [DOI] [PubMed] [Google Scholar]
- 14.Liang D, Liu B, Wang J, Ying L. Accelerating SENSE using compressed sensing. Magn Reson Med. 2009;62:1574–1584. doi: 10.1002/mrm.22161. [DOI] [PubMed] [Google Scholar]
- 15.Lustig M, Donoho DL, Santos JM, Pauly JM. Compressed Sensing MRI. Signal Processing Magazine, IEEE. 2008;25:72–82. doi: 10.1109/MSP.2007.914728.. [DOI] [Google Scholar]
- 16.Vasanawala SS, Murphy MJ, Alley MT, Lai P, Keutzer K, Pauly JM, Lustig M. Practical parallel imaging compressed sensing MRI: Summary of two years of experience in accelerating body MRI of pediatric patients. IEEE; 2011. pp. 1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velikina JV, Alexander AL, Samsonov A. Accelerating MR Parameter Mapping Using Sparsity-Promoting Regularization in Parametric Dimension. Magn Reson Med. 2013;70:1263–1273. doi: 10.1002/mrm.24577.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C, Graff CG, Clarkson EW, Bilgin A, Altbach MI. T2 mapping from highly undersampled data by reconstruction of principal component coefficient maps using compressed sensing. Magn Reson Med. 2012;67:1355–1366. doi: 10.1002/mrm.23128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doneva M, Börnert P, Eggers H, Stehning C, Sénégas J, Mertins A. Compressed sensing reconstruction for magnetic resonance parameter mapping. Magn Reson Med. 2010;64:1114–1120. doi: 10.1002/mrm.22483.. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y, Zhang Q, Liu Q, Wang Y-XJ, Liu X, Zheng H, Liang D, Yuan J. PANDA-T1ρ: Integrating principal component analysis and dictionary learning for fast T1ρ mapping. Magn Reson Med. 2014 doi: 10.1002/mrm.25130.. [DOI] [PubMed] [Google Scholar]
- 21.Lustig M, Alley M, Vasanawala S. L1 SPIR-iT: autocalibrating parallel imaging compressed sensing. Proceedings of the International Society for Magnetic Resonance in Medicine. 2009:379. [Google Scholar]
- 22.Otazo R, Kim D, Axel L, Sodickson DK. Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion MRI. Magn Reson Med. 2010;64:767–776. doi: 10.1002/mrm.22463.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King K, Xu D, Brau AC, Lai P, Beatty PJ, Marinelli L. A new combination of compressed sensing and data driven parallel imaging. Proceedings of the International Society for Magnetic Resonance in Medicine. 2010:4881. [Google Scholar]
- 24.Beatty PJ, Brau AC, Joshi SM, Michelich CR, Bayram E, Nelson TE, Herfkens RJ, Brittain JH. A method for autocalibrating 2D-accelerated volumetric parallel imaging with clinically practical reconstruction times. Proceedings of the International Society for Magnetic Resonance in Medicine. 2007:1749. [Google Scholar]
- 25.Brau ACS, Beatty PJ, Skare S, Bammer R. Comparison of reconstruction accuracy and efficiency among autocalibrating data-driven parallel imaging methods. Mag Reson Med. 2008;59:382–395. doi: 10.1002/mrm.21481.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Han ET, Busse RF, Majumdar S. In vivo T(1rho) mapping in cartilage using 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (3D MAPSS) Magn Reson Med. 2008;59:298–307. doi: 10.1002/mrm.21414.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284.. [DOI] [PubMed] [Google Scholar]
- 28.Carballido-Gamio J, Bauer JS, Lee Keh-Yang, Krause S, Majumdar S. Combined Image Processing Techniques for Characterization of MRI Cartilage of the Knee. IEEE; 2006. pp. 3043–3046. [DOI] [PubMed] [Google Scholar]
- 29.Sharma SD, Nayak K. Region of Interest Compressed Sensing. Proceedings of the International Society for Magnetic Resonance in Medicine. 2009:2816. [Google Scholar]