Abstract
We recently developed a technique for rapidly and reversibly inhibiting protein function through light-inducible sequestration of proteins away from their normal sites of action. Here, we adapt this method for inducible inactivation of Bem1, a scaffold protein involved in budding yeast polarity. We find that acute inhibition of Bem1 produces profound defects in cell polarization and cell viability that are not observed in bem1Δ. By disrupting Bem1 activity at specific points in the cell cycle, we demonstrate that Bem1 is essential for the establishment of polarity and bud emergence but is dispensable for the growth of an emerged bud. By taking advantage of the reversibility of Bem1 inactivation, we show that pole size scales with cell size, and that this scaling is dependent on the actin cytoskeleton. Our experiments reveal how rapid reversible inactivation of protein function complements traditional genetic approaches. This strategy should be widely applicable to other biological contexts.
Keywords: cell polarity, budding yeast, Bem1, optogenetics, cell size
Polarization of cell signaling and growth is essential for a wide variety of cell types. Yeast cells must restrict their growth to a small region of the cell cortex in order to bud or mate. This process of polarization is fast, occuring over a period of about 30 minutes. In budding yeast, cell polarity is orchestrated by the small membrane-bound GTPase Cdc42. Active Cdc42 polarizes at the nascent bud site, where it directs assembly of actin cables that serve as tracks for polarized secretion of the machinery that builds the bud.1 Cdc42 activation is both necessary2 and sufficient2,3 to specify polarity. Cdc42 activation is mediated by its guanine nucleotide exchange factor (GEF), Cdc24, and other regulatory proteins such as Bem1.4
Because polarization in yeast is fast, a full understanding of the process requires techniques for observation and perturbation that operate on a similarly rapid timescale. Traditionally, polarity signaling has been studied using loss-of-function experiments with slow, irreversible techniques like genetic knockouts or mutations. While these techniques are powerful, they have several limitations. First, constitutive genetic knockouts cannot be used to study essential genes. Second, it is impossible to use these constitutive perturbations to determine when a protein acts during a dynamic process. Third, cells are able to compensate for loss of a gene of interest over long timescales, either by upregulating related genes or paralogs5 or through other processes like post-translational modifications or aneuploidy.6 It takes many cell generation times to make a knockout strain, giving ample time for these adaptations to occur.
We have developed an acute, rapid, and reversible optogenetic technique7 in order to overcome these limitations and study dynamic processes like yeast polarity. Our technique sequesters protein away from its normal site of action by trapping the soluble pool, building on previous work with light- or chemical-gated dimerizer techniques.8–12 Tools like temperature-sensitive (ts) mutations13 and degrons14–16 can be used to study dynamic processes, but both have drawbacks. Generating ts alleles is time consuming, and may not be possible for all genes. Degrons are significantly slower than our system, operating on a scale of minutes rather than seconds. Degron systems are reversible, but on a much slower timescale, limited by the kinetics of protein synthesis. Our system is beneficial in cases where a ts allele is difficult to generate or where extra speed is needed.
In this work, we investigate yeast polarity signaling using our previously developed optogenetic system.7 In this case, we inactivate the protein by inducibly sequestering it away from its normal site of action, a technique we call Depletion with Light (DeLight). Using this tool, we find that inactivation of Bem1 by sequestration prevents polarization and budding, resulting in cells that grow isotropically until they burst. Next, we use the DeLight system to sequester Bem1 at different times during the polarization process and find that Bem1 is required for polarization and bud emergence but not for bud growth. Finally, we use the enlarged cells generated by Bem1 sequestration to demonstrate that pole size scales with cell size.
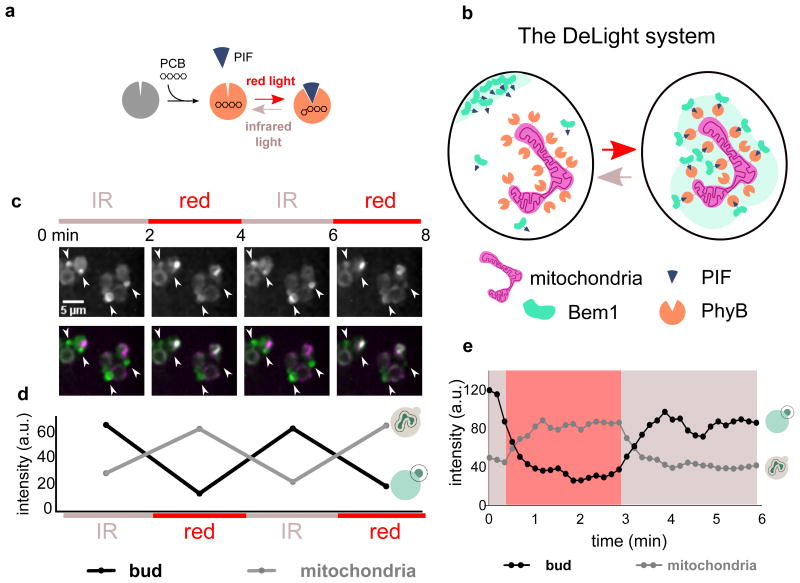
The DeLight system consists of two protein components, Phytochrome B (PhyB) and phytochrome interacting factor (PIF).17,18 The light-sensitive protein PhyB is localized to a subcellular compartment where it can act as a light-regulated “anchor” for proteins fused to its binding partner, PIF. PhyB is only light responsive when bound to the small molecule chromophore phycocyanobilin (PCB), which can be delivered exogenously to yeast cells19 (Fig. 1a). When cells containing both protein components and PCB are exposed to red light, PhyB changes conformation, and the PIF-tagged protein is recruited to the anchor and depleted from the cytosol and plasma membrane. This interaction can be reversed with infrared light (Fig. 1b).7
Figure 1. The DeLight system can be used to inducibly sequester Bem1.
(a) PhyB is a photoreceptor that interacts with a small molecule chromophore, PCB. PhyB interacts with Phytochrome Interacting Factor (PIF) in the presence of red light and dissociates in the presence of infrared light. (b) By anchoring PhyB at the mitochondria of budding yeast, a PIF-tagged protein can be rapidly and reversibly sequestered away from its normal site of action using red and infrared light. We have named this approach Depletion with Light, or DeLight. (c) Images of PhyB-mCherry-Tom7 (anchor) and Bem1-YFP-PIF (polarity regulator) strains treated with PCB and exposed to red and infrared light, alternating on a two-minute interval; corresponds to Movie 1. White arrowheads indicate small buds or poles. Fluorescence images are a maximum intensity projection of z-stacks. Color bars indicate periods of exposure to red light (Bem1 sequestered at mitochondria) and infrared light (Bem1 released from mitochondria). (d) Intensity of Bem1-YFP-PIF measured at the bud versus mitochondria during red and infrared light exposure. (e) Timecourse of Bem1 sequestration and release, measured every 10 seconds.
In budding yeast, several putative feedback loops modulate Cdc42 activation at the nascent bud site, including a positive feedback loop centering on the scaffold protein Bem1.20–22 Bem1 binds to both Cdc24 (the GEF for Cdc42) as well as Cla4 (a p21 activated kinase), forming a trimeric complex. Cla4 and Bem1 can both bind to active Cdc42,23 enriching the Bem1-GEF complex at sites of Cdc42 activity. This increased local GEF recruitment activates more Cdc42, which in turn recruits more Bem1 complex, forming a positive feedback loop that amplifies Cdc42 signal. While this feedback loop appears to be important in activating Cdc42, the bem1Δ strain is viable, with relatively mild polarity and growth defects, suggesting that other parallel pathways may be able to compensate for loss of Bem1. Much of the argument for the importance of Bem1 stems from the observation that bem1Δ is synthetic lethal with rsr1Δ, a landmark protein involved in directing the location of the bud site, indicating that the Bem1-dependent positive feedback loop may be more important when cells must break symmetry.23 However, recent evidence indicates that the bem1Δ rsr1Δ double mutant is viable in some genetic backgrounds.24 Here we sought to revisit the role of Bem1 in yeast polarity using our DeLight system. We hoped to clarify if and when Bem1 function is required for polarity in a manner that would not be confounded by the compensation that may accompany long-term inactivation of Bem1.
We set out to establish that the DeLight system can be used to inducibly inactivate Bem1. For complete depletion of Bem1, every Bem1 protein in the cell must have a PIF tag. Therefore, we tagged Bem1 with mCitrine and PIF at the endogenous locus and verified that localization was unaffected by the PIF tag. As expected, Bem1-PIF polarizes to the nascent bud site and the cortex of growing buds (Fig. S1). Next, we sought a localized PhyB that could efficiently sequester Bem1-PIF from the cytoplasm and the plasma membrane. We initially used a PhyB-mCherry-Htb2 construct from our previous work,7 reasoning that sequestering Bem1 inside the nucleus would be most likely to remove it from its relevant effectors. However, persistent recruitment of YFP-PIF alone to the PhyB-mCherry-Htb2 construct slowed cell growth and altered cell size and morphology, possibly due to misregulation of histone function (Fig. S2).
We sought a new PhyB-anchor that would provide efficient sequestration of PIF-tagged proteins without off-target effects. Sequestration of proteins at the mitochondria has previously been used to inactivate proteins.12,25 The mitochondrial outer membrane protein Tom7, a component of the translocase of outer membrane (TOM) complex, has its N terminus facing the cytosol, making it suitable for fusion with PhyB, since PhyB only tolerates C-terminal fusions.26 Our pADH-PhyB-mCherry-Tom7 construct expressed well, localized to mitochondria, and was well-tolerated by yeast cells (Fig. 1c). Unlike the PhyB-Htb2 anchor strains, the PhyB-Tom7 anchor strains were unaffected by constitutive recruitment of YFP-PIF (Fig. S2). This result demonstrates that mitochondrial sequestration with PhyB-Tom7 is a good anchor for use with the DeLight system.
To determine whether our mitochondrial anchor was functional in sequestering Bem1-PIF, we added the PhyB-Tom7 construct to Bem1-YFP-PIF cells. After incubating cells with PCB to render the PhyB light responsive, we exposed the cells to successive rounds of red and infrared light, acquiring a 3D stack of images of Bem1-PIF between each light exposure. We observed reversible depletion of Bem1 from poles and buds and reversible enrichment of Bem1-PIF at the mitochondria (Fig. 1c, d Movie 1). Both enrichment at the mitochondria and sequestration from the bud are rapid, with a t1/2 of roughly 30 seconds, and the interaction can be reversed on a similar timescale (Fig. 1e).
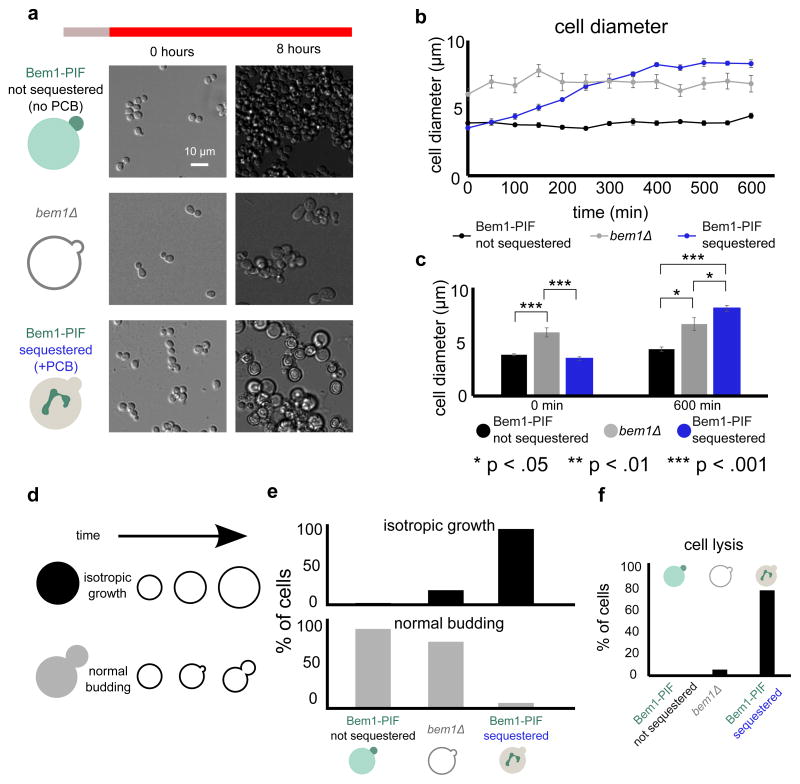
Deletion of BEM1 generates enlarged cells displaying morphological abnormalities (Fig. 2a) that grow more slowly than a wild-type strain.27 Acute sequestration of Bem1 at the mitochondria generates a morphological phenotype that is significantly more profound than bem1Δ. Bem1-PIF sequestered cells fail to bud and instead grow isotropically until they are about twice as wide as control cells that have not been incubated with PCB and therefore do not sequester Bem1. These non-sequestered cells are indistinguishable from wild type cells (Fig. S3). The Bem1 sequestered cells grow even larger than bem1Δ cells (Fig. 2b, c, Movie 2). After ten hours of red light exposure, controls without PCB have an average diameter of 4.4 ± 0.2 μm (mean ± SEM), whereas Bem1-PIF depleted cells have an average diameter of 8.3 ± 0.3 μm. In contrast, bem1Δ cells have an average diameter of 6.8 ± 0.6 μm (Fig. 2c). If we assume the cell is a sphere, this corresponds to a 6.6 fold increase in volume for Bem1-PIF sequestered cells relative to control cells, compared to a 3.6 fold increase in volume for bem1Δ relative to the control.
Figure 2. Bem1 sequestration with DeLight prevents cells from budding.
(a) bem1Δ and Bem1 DeLight strains (+PCB, Bem1-sequestered or −PCB, not sequestered) were pre-treated with infrared light and then switched to red light to activate sequestration at t=0 and imaged in DIC for a period of 10 hours; see Movie 2. (b) Cell diameter for each condition, measured every five minutes (error bars = standard error of mean, SEM; 10 cells measured for each condition at each timepoint). (c) Average cell diameters at the beginning and end of the experiment (error bars = SEM, 10 cells measured for each condition at each timepoint). (d) Schematic of phenotypes for cell scoring. (e) Quantification of movies from (b). Cells were scored for phenotypes based on the schematic in (d). (f) Quantification of cell lysis during the 10-hour red light exposure. For panels (e) and (f), >100 cells were scored for each condition. Acute inhibition of Bem1 with DeLight produces more profound defects in budding and cell viability than genetic nulls of Bem1.
95% of Bem1-PIF sequestered cells grow isotropically and do not form new buds after exposure to red light. In contrast, only 17% of bem1Δ cells grow isotropically (Fig. 2e). In the acutely-sequestered Bem1-PIF cells, this isotropic growth phenotype often causes the cells to lyse; 79% of cells lyse by ten hours of sequestration, as opposed to 6% of bem1Δ cells and 0% of unperturbed cells (Fig. 2f).
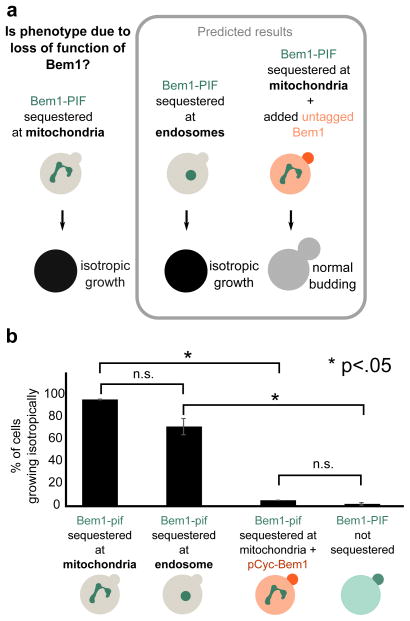
In the above experiments, it is possible that the phenotypes observed upon Bem1-PIF recruitment result from a gain-of-function phenotype at the mitochondria rather than loss of Bem1 activity at polarity sites. We performed two sets of experiments to discriminate between these possibilities (Fig. 3a). First, we investigated whether the phenotype depended on the intracellular site of Bem1 sequestration. If sequestration induced loss of function, recruitment to other organelles should generate a similar phenotype. As an additional test, we added an additional untagged (non-sequesterable) copy of Bem1. This should rescue the budding phenotype if it is due to loss of function. In contrast, if the budding phenotype reflects gain of function at the mitochondria, it would not be replicated by sequestering Bem1 to other anchors and would not be rescued by adding an additional wildtype copy of Bem1.
Figure 3. The Bem1 sequestration phenotype is due to loss of function of Bem1.
(a) The phenotype generated by sequestration of Bem1-PIF to the mitochondria could arise in two ways: gain of function of Bem1 at the mitochondria or loss of function due to sequestration away from its normal site of action. These two models can be distinguished by recruiting Bem1 to another organelle (endosome) and by adding a non-sequesterable copy of Bem1. Predicted results are shown for Bem1 loss of function (gain of function predicts opposite phenotypes). (b) Quantification of phenotypes from movies (Movie 3). >100 cells were scored for each condition. The phenotypes are consistent with mitochondrial sequestration inducing acute loss of Bem1 function.
To investigate the consequences of Bem1 sequestration to a different organelle, we recruited Bem1 to the endosome using PhyB-mCherry-Snf7. This anchor, developed in our previous work, efficiently sequesters PIF-tagged protein but is not fully reversible, making it less suitable for more complex time-varying experiments, but appropriate for comparison with PhyB-mCherry-Tom7 in this case7. Sequestration to the endosome via PhyB-Snf7 produced a budding phenotype similar to the one generated by sequestration of Bem1 to the mitochondria via PhyB-Tom7. As with mitochondrial sequestration of Bem1, the majority of cells (70%) did not form new buds after sequestering Bem1 at the endosome and instead grew isotropically until they burst (Fig. 3b, Movie 3). The phenotypes are qualitatively similar, but the magnitude of the defect is somewhat smaller for Snf7 sequestration of Bem1-PIF, possibly due to differing expression levels or accessibility to PIF between Snf7 and Tom7.
To further validate that Bem1 sequestration to the mitochondria causes a loss of protein function, we tested the ability of an additional untagged copy of Bem1 to rescue the phenotype. We performed Bem1-YFP-PIF sequestration to the mitochondria in the presence of an additional untagged copy of Bem1 driven by the pCyc promoter. The non-sequesterable copy of Bem1 rescued the phenotype, with only 4% of cells growing isotropically, compared to 95% for cells in which sequestration was active and in which all copies of Bem1 had the PIF tag (Fig. 3b, Movie 3). The rescued cells grew at a similar rate to unperturbed cells, with an average time between buds of 112 ± 6 min for the rescued cells and 108 ± 2 min for the unperturbed cells. In contrast, the bem1Δ cells had a time between buds of 133 ± 18 min.
While these results argue that the Bem1 sequestration phenotype is likely to be due to loss of function of Bem1, it could also be due to sequestration of other Bem1 binding partners. For example, Cdc24, the Cdc42 GEF, binds Bem1 and could be sequestered along with Bem1 at the mitochondria. In this case, we would expect an additional copy of Cdc24 to rescue the phenotype, similar to the rescue we observe for adding additional wildtype Bem1 (Fig. 3b). However, adding an additional copy of Cdc24 fails to rescue the growth and polarity phenotypes; these cells behave similarly to the Bem1 sequestered cells without added Cdc24 (Fig. S4).
Taken together, our observations that (1) the Bem1 sequestration phenotype does not depend on the site of sequestration and (2) the Bem1 sequestration phenotype can be rescued by non-sequesterable Bem1 argue that the observed phenotype is due to acute loss of function of Bem1 rather than gain of function at the anchor location. Because this acute phenotype is stronger than the bem1Δ phenotype, our data suggest that the bem1Δ strain may have undergone compensation, either by genetic changes or post-transcriptional regulation. Similar isotropic growth phenotypes have been observed in cases where cells cannot polarize secretion, specifically with temperature-sensitive alleles of myosin, tropomyosin, and formins.28,29
Acute sequestration provides a platform to examine the effects of inactivating Bem1 before compensation can occur and may help to clarify conflicting data regarding the role of Bem1. We find that Bem1 is essential for polarity, not just redundant with other components. Previous work indicated that Bem1 was only essential in a symmetry-breaking background, where the landmark protein Rsr1 was deleted23. Our results indicate that Bem1 is crucial even when cells still contain landmark proteins.
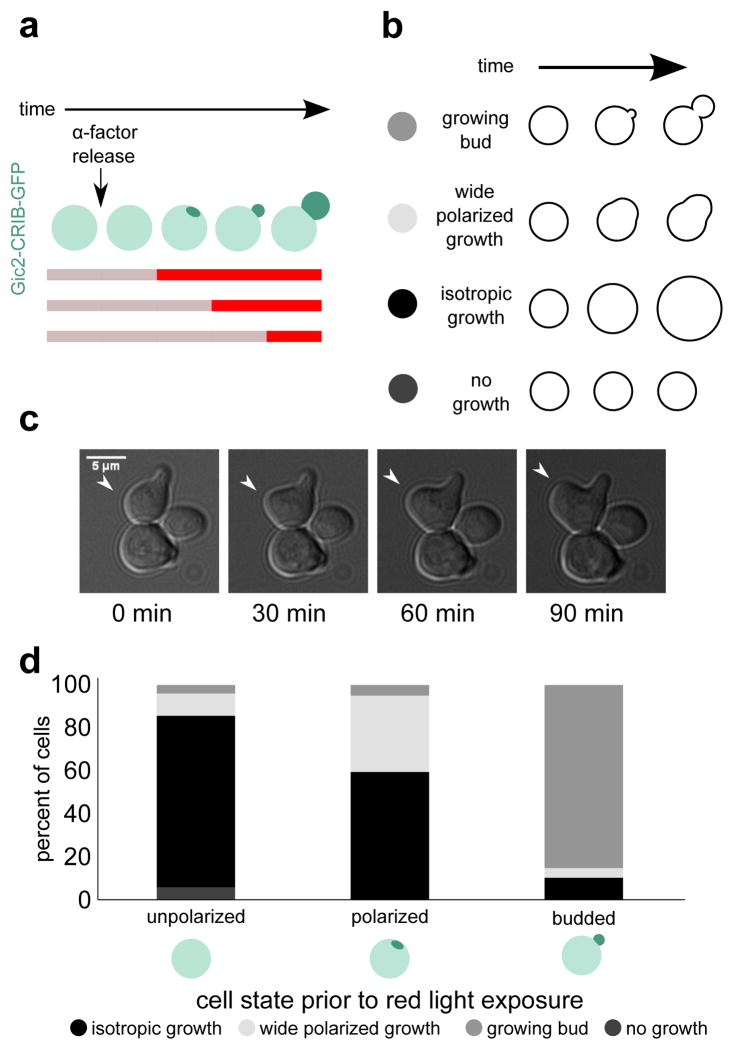
Next, we used the temporal control of DeLight to investigate when Bem1 is required for polarization of cell growth. Cells were arrested with α-factor to generate a synchronized population of unpolarized cells. Consistent with previous reports,30 most cells polarize the Cdc42 activity reporter Gic2-CRIB within an hour of release by α-factor washout. (Fig. S5). Cells were exposed to infrared light (no sequestration) until 45, 60, or 75 minutes after release, then switched to red light to sequester Bem1 to the mitochondria (Fig. 4a). Cells were scored for polarity by imaging the Gic2-CRIB activity reporter prior to switching to red light. Unpolarized cells were strongly affected by Bem1-PIF sequestration; 80% of cells that were unpolarized when switched to red light did not subsequently develop polarity and instead grew isotropically (Fig. 4d). Cells that were polarized but had not yet formed a bud were also affected, with 60% of cells growing isotropically following Bem1 sequestration. However, cells that had undergone bud emergence before Bem-PIF sequestration were largely unaffected, with only 11% of cells growing isotropically.
Figure 4. Bem1 localization is required for polarization and bud emergence but not bud growth.
Here we use temporal control of DeLight to determine when Bem1 function is required for budding. (a) Schematic of experimental setup. The colored bar indicates periods of red and infrared light exposure. (b) Phenotypes for cell scoring. (c) Example of wide polarized growth phenotype. (d) Percent of cells that exhibit each phenotype, binned by polarization state at the time of red light exposure. Polarization state is scored using the CRIB-GFP reporter. Bem1 is required in the early phase of the polarization process (pole formation and bud emergence) but is dispensable for bud growth following emergence.
In addition to the isotropic growth phenotype, we also observed cells that showed polarized growth but with abnormal morphology; the region of growth was much wider than a normal bud, and lacked the constriction seen in a bud neck (Fig. 4c). 11% of cells that were unpolarized at the time of Bem1-PIF sequestration had the wide polarized growth phenotype. In cells that were already polarized but had not yet budded, 36% percent of cells showed wide polarized growth. In cells that had already formed a bud, the occurrence of wide polarized growth was much lower, with only 4.5% of cells showing this phenotype (Fig. 4d).
From these data, we conclude that Bem1 is required for the establishment of polarity as well as initiation of bud growth. In contrast, Bem1 is largely dispensable for bud growth once the bud has emerged.
Because acute loss of Bem1 yields cells that grow isotropically and attain much larger size than control cells, we reasoned that we could use these large cells to investigate whether the size of the pole scales with the size of the cell. The pole is shaped by diffusion of activated Cdc42, active transport to the pole, and endocytosis.31 Many mutations generate enlarged yeast cells, including Bem1; bem1Δ cells are in the largest 5% of mutants in the yeast whole-genome deletion collection,32 and pole size could be measured in these backgrounds with persistently altered cell size. However, the reversibility of our system provides a major advantage: we use depletion of Bem1-PIF to generate large cells, but once they are moved to infrared light, they recover wild-type Bem1 activity and can polarize (Fig. S6). This allows us to measure pole size in isogenic cells that are large or small. In addition, cells incubated in red light are larger on average, but significant variation exists in the population, allowing comparison between large and small cells that have been treated identically.
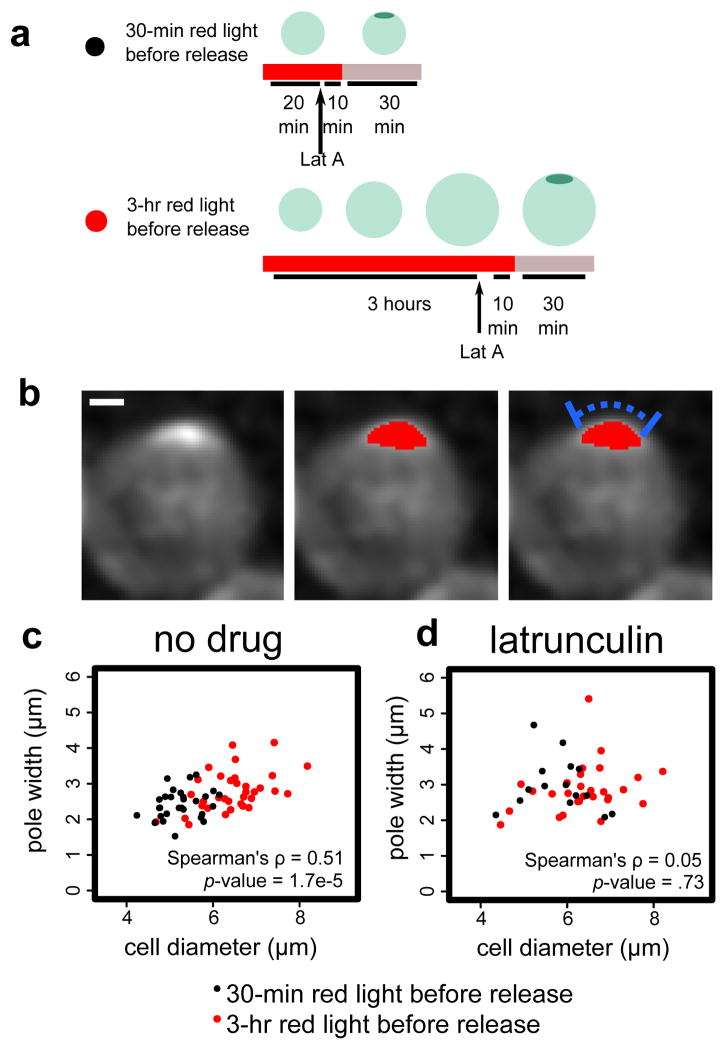
To generate enlarged cells, Bem1-PIF PhyB-Tom7 cells with the Gic2-CRIB-2xGFP reporter were exposed to red light for three hours, which is sufficiently long to significantly enlarge the cells but not cause cell lysis (Fig. 5a). After switching to infrared light, most cells polarized within five minutes, confirming reversibility (Fig. S6). As a control for the Bem1 sequestration and release, Bem1-PIF PhyB-Tom7 cells were exposed to red light for 30 minutes, not long enough to significantly alter cell size, and then switched to infrared light. In this context, cells were not enlarged, and most cells polarized within five minutes after switching to infrared light.
Figure 5. Pole width scales with cell size in an actin-dependent manner.
We use the reversibility of DeLight to investigate how pole size scales with cell size. (a) We treat Bem1 DeLight cells with red light for 3 hrs to generate enlarged cells, or 30 min to retain wild-type cell size, and then release Bem1 to allow cells to polarize. (b) Example of pole measurement. Image shown is a maximum intensity projection of a z-stack of CRIB-GFP, a Cdc42 activity reporter. Red overlay is the thresholded pole. Blue bar indicates pole width measurement. Scale bar is 1 μm. (c) and (d) Pole width vs. cell diameter with no drug (c) versus latrunculin, an actin depolymerizing drug (d). Correlation is indicated by the Spearman’s rho value. Black points are measurements from 30-min cells and and red points measurements from 3-hr cells (enlarged). n=63 and n=44 for no drug and latrunculin, respectively.
We measured pole width in each cell population by tracing the perimeter of the cell from end to end of the thresholded pole, based on the Gic2-CRIB-2xGFP Cdc42 activity reporter (Fig. 5b). In the pooled population of enlarged and normal-sized cells, the width of the pole is correlated with cell diameter (Spearman’s ρ =0.51, p-value <0.001). Importantly, for cells with similar diameters, there is no significant difference in pole size between the two red light durations (t-test, p-value = 0.97, cells between 5.5 and 6 μm diameter). Since pole width is thought to be governed by active transport and endocytosis at the pole31, we reasoned that pole width scaling with cell size would require actin polymer. Indeed, in cells treated with Latrunculin A to depolymerize actin, the correlation between cell diameter and pole size is lost (Spearman’s ρ =0.05, p-value = 0.73) (Fig. 5c). The enlarged cells generated by our light-gated perturbation were essential to observing pole width scaling; there is not enough endogenous variation in cell size to detect a correlation (Spearman’s ρ =0.31, p-value = 0.10). Particularly in Latrunculin A treated cells, and to a lesser degree in wildtype cells, the poles are quite dynamic, and it is not always clear which timepoint represents a stable pole to measure. Therefore, for each cell, we selected the timepoint with the brightest pole intensity for width measurement. We tested other timepoint selection criteria and found that each method generated similar results (Fig. S7).
In summary, we have refined the DeLight system, generating a new anchor for protein sequestration, and applied this system to inactivate Bem1 rapidly and reversibly. Acute sequestration of Bem1 prevents budding, and generates a phenotype significantly stronger than genetic deletion of Bem1. We used the DeLight system’s speed to probe the temporal regulation of polarity by Bem1 and found that Bem1 is necessary for polarization and bud emergence, but not for bud growth. Finally, we used the reversibility of the DeLight system to uncover pole size scaling with cell size.
While significant previous work has established the importance of the signaling surrounding Bem1 in yeast polarity, the viability of the bem1Δ strain indicated that Bem1 was not essential for polarization unless it was combined with mutations in other polarity components such as Rsr1. Here, we show that acute depletion of Bem1 prevents cells from polarizing, suggesting that Bem1 is essential for polarization under these conditions. Compensation over the relatively slow timecourse of generating the bem1Δ mutant likely enables survival of the knockout strain. Our acute perturbation permits analysis of the cells before compensation can occur.
We show that the requirement for cytoplasmic Bem1 is restricted to the polarization and bud emergence phases and that sequestering Bem1 once the bud has emerged does not prevent further bud growth. This result could extend our current understanding of cell cycle regulation of bud growth. Cdc42 is activated in late G1 and must polarize in order for the cell to form a bud. It continues to polarize through bud emergence until G2, when growth in the bud switches from polarized to isotropic. In G2, Bem1 and Cdc24 lose their localization to the bud tip, and active Cdc42 disperses to the entire bud cortex, suggesting that polarization of these signaling components is important for polarized growth but not for isotropic growth of the bud33. This transition is known as the apical-isotropic switch. Importantly, previous experiments have primarily relied on observation of protein localization, rather than perturbation of protein function during specific phases of this multi-stage process. Techniques for assaying necessity of Bem1 and other polarity proteins before and after the apical-isotropic switch have been lacking. Because we do not use any cell cycle reporters, our results only establish that Bem1 localization is necessary before but not after bud emergence, but lack the time resolution to further clarify whether Bem1 specifically acts at the apical-isotropic switch. In future work, our system could be used in conjunction with cell cycle reporters or mutants to determine whether the change in Bem1 necessity occurs concurrently with the switch to isotropic growth.
Our finding that pole size scales with cell size raises several intriguing questions. What is the mechanism by which cells scale their pole size? Does increased pole size lead to increased daughter cell size, and if so, for how many generations does this altered cell size persist? More generally, the inducibly isotropically growing cells generated here could be used in other analyses of scaling in budding yeast.
Broadly, we have demonstrated the application of the light-gated sequestration system to a dynamic and rapid cellular process, cell polarity. The DeLight system is generally applicable to a wide range of signaling cascades. Previously, we have applied the system to the mitotic cyclin Clb27, and we envision that it can be used to probe the function of many other proteins in dynamic processes ranging from cell cycle regulation to yeast polarity and beyond.
Materials and Methods
Yeast strain construction and growth conditions
Standard yeast strain construction methods were used throughout. All strains were constructed in congenic w303 (MATa his3-11,15 trp1-1 leu2-3 ura3-1 ade2-1). PhyB-mCherry-Snf7 and PhyB-mCherry-Htb2 were constructed in our previous work.7 To select a new anchor for light-induced protein sequestration, we used the yeast GFP localization collection (yeastgfp.yeastgenome.org)34 to identify proteins that localized to the mitochondria and were expressed at high levels. This candidate list of proteins was narrowed by excluding those with overexpression phenotypes, since the PhyB-anchor constructs are expressed on top of the endogenous protein. Finally, we reviewed sequence and topology information for each protein to identify proteins for which the N-terminus was facing the cytosol, because PhyB only tolerates fusions on its C-terminus. From this list, Tom7 was the most favorable candidate. Tom7 was cloned from the genome and inserted in the PhyB-mCherry backbone using Gibson enzymatic assembly.35 Endogenous Bem1 was tagged at the C-terminus with mCitrine and PIF using an optimized PCR-based tagging cassette36 modified with a PIF addition. The pADH-Gic2-CRIB-2xGFP (KAN) plasmid was a gift from Jessica Walter. Additional copies of pcyc-Bem1 and pcyc-Cdc24 were added using integrating plasmids at the TRP locus.
For all time-lapse microscopy experiments, cells growing exponentially in synthetic defined complete medium (Sunrise) were vortexed and plated on Concanavalin A coated coverslip-bottom 384-well plates (Brooks), which were then washed to remove floating cells. For optogenetic experiments, cells were incubated with a final concentration of 31.25 μM (0.25 μl of 12.5 mM stock in 100 ul culture) PCB (purified ourselves37,38 or purchased from Santa Cruz Biotechnology, Inc.) for 2–4 hours, then pelleted and resuspended in fresh media without PCB before plating. For α-factor synchronization and release experiments, cells were incubated with 100 uM α-factor (Zymo) for 90 minutes with or without PCB, washed, and then released into fresh synthetic medium.
Microscopy and optogenetic control
The experiments in Figures 1, 4, and 5 were performed on two spinning disk confocal microscopes, both based on a Nikon Eclipse Ti inverted microscope with a motorized stage (ASI). The first microscope used a Diskovery spinning disk confocal equipped with a 100 μm pinhole disk (Spectral Applied Research), 405, 440, 488, 514 and 561 nm laser wavelengths (LMM5, Spectral Applied Research), a 60x 1.49NA TIRF Apo objective (Nikon), and a Zyla sCMOS camera (Andor). The second microscope used a Yokogawa CSU-X1 spinning disk confocal, an MLC400B monolithic laser combiner with laser lines at 405, 488, 561, and 640nm (Agilent), a 60x 1.4NA Plan Apo VC objective (Nikon), and a Clara interline CCD (Andor).
The experiments in Figures 2 and 3 were performed on a Nikon Eclipse Ti inverted microscope equipped with a Lambda XL Broad Spectrum Light Source (Sutter), a 20x 0.75NA Plan Apo objective (Nikon) or a 60x 1.4NA Plan Apo VC objective (Nikon), FITC and TRITC filter cubes (Chroma), and a Clara interline CCD (Andor). A motorized stage (ASI) allowed automated sampling of multiple XY positions.
For all experiments, microscope settings, dichroic positions, filters, shutters, and cameras were controlled by NIS Elements (Nikon).
For optogenetic experiments, red and infrared light were provided by placing filters on top of the condenser in the brightfield lightpath18 (650 nm 20 nm-bandpass filter, Edmund Optics, near-IR RG9 glass filter, Newport). To provide constant illumination of the sample, the transmitted light shutter was left continuously open during imaging. In order to illuminate multiple wells of a 384-well black glass-bottom imaging plate (Brooks), the condenser was lowered as close as possible to the plate, and the field and aperture diaphragms were opened fully. In this configuration, nine wells received uniform simultaneous illumination, allowing imaging of multiple strains and controls in the same experiment.
Fluorescence images were computationally denoised in collaboration with John Sedat (UCSF), using an algorithm built into the Priism image analysis package.39
All image analysis was performed in Fiji.40 Intensity measurements in Figure 1 were performed on background-subtracted images using manually drawn ROIs. The Cell Counter (Kurt De Vos, University of Sheffield) plugin was used for manual scoring and counting of cell types. Pole widths were measured in Fiji. First, the image background was subtracted using Fiji’s built-in rolling-ball background subtraction algorithm. Next, cytoplasmic background was measured in a hand-drawn ROI and then subtracted from the whole image. The maximum pixel intensity was measured and the image was thresholded using 25% of the maximum intensity value. Then, the arc around the cell periphery from end to end of the pole was measured using the curved line tool. The timepoint for with the highest maximum intensity value was used for each cell. This process was semi-automated with a custom-written macro.
Data analysis, statistical analysis, and graphing were performed in Microsoft Excel and R (www.R-project.org/).41
Supplementary Material
Figure S1. PIF-tagged Bem1 localizes normally to poles and buds. White dotted lines indicate cell periphery (traced from DIC images). Bem1-mCitrine-PIF fluorescence shown.
Figure S2. Prolonged recruitment of YFP-PIF to PhyB-mCherry-Htb2 anchor generates a cell growth artifact, while recruitment to of YFP-PIF to PhyB-mCherry-Tom7 and PhyB-mCherry-Snf7 does not. Cells were incubated with continuous red light. All strains contain pADH-PIF-YFP and pADH-PhyB-mCherry fused to the indicated anchors. Time between buds was measured by counting the number of frames between the first and second visible bud emergence events. Error bars represent SEM. 10–15 cells were measured for each condition.
Figure S3. Bem1-PIF DeLight cells without sequestration behave identically to wild-type cells. (a) Cell size over time measured from DIC images. Cells were not treated with PCB, but were exposed to red light as in DeLight experiments. (b) Cell size at the beginning and end of the movie is not significantly different between non-sequestered Bem1 DeLight cells and wild-type cells (t-test, p-value >0.05). In addition, cells were scored for isotropic growth, with less than 1% of cells growing isotropically in each condition, and for cell lysis, with no cells lysing in either condition.
Figure S4. Additional Cdc24 does not rescue the Bem1 DeLight phenotype. (a) Cell size over time measured from DIC images. Bem1-PIF sequestered + Cdc24 strain contains a pCyc-Cdc24 construct integrated at the TRP locus, but is otherwise identical to the Bem1-PIF sequestered strain. Integration of the construct was verified by colony PCR across both junctions. (b) Cell scoring for isotropic growth vs. normal growth/budding.
Figure S5. Cells polarize active Cdc42 after release from alpha factor arrest. Cells were washed after incubating with alpha factor for 90 minutes. Polarity was scored using the CRIB-2xGFP Cdc42 activity reporter.
Figure S6. Sequestering Bem1-PIF to the mitochondria reversibly prevents polarization of active Cdc42. Cells were exposed to red light for 3 hours and then switched to infrared light to release Bem1-PIF. Cells polarize and grow normally after release.
Figure S7. Pole size scales with cell size, and scaling is lost with latrunculin treatment, regardless of method used to select measurement timepoint. Pole size for each cell was measured over 3–10 timepoints. A single timepoint per cell was plotted. This timepoint was selected by one of the following criteria: signal to noise (highest ratio of pole intensity to cytoplasmic intensity), brightest pole, widest pole, narrowest pole, or median pole width.
Acknowledgments
We would like to thank Xiaojing Yang for the PhyB anchor strains and fruitful collaboration, and John Sedat for image denoising collaboration. The bem1Δ strain was a gift from Michelle Lu in David Drubin’s lab. The C-terminal tagging plasmids are from Kurt Thorn, and the Gic2 CRIB-2xGFP reporter is from Jessica Walter. We thank Oliver Hoeller, Brian Graziano, Justin Farlow, and the Weiner lab for critical reading and feedback, and Adam Jost for help with R and statistical analysis. This work was supported by an American Heart Association Predoctoral Fellowship (APJ), an Achievement Rewards for College Scientists scholarship (APJ), and NIH grants R01GM084040, R01GM096164, and R21EB017399 (ODW).
Footnotes
Supporting Information Available: Figures S1–S7 and Movies 1–3 are available as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etienne-Manneville S. Cdc42--the centre of polarity. J Cell Sci. 2004;117:1291–300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 3.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–84. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–72. [PubMed] [Google Scholar]
- 5.DeLuna A, Springer M, Kirschner MW, Kishony R. Need-based up-regulation of protein levels in response to deletion of their duplicate genes. PLoS Biol. 2010;8:e1000347. doi: 10.1371/journal.pbio.1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–93. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Jost APT, Weiner OD, Tang C. A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol Biol Cell. 2013;24:2419–30. doi: 10.1091/mbc.E13-03-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Park H, Kyung T, Kim NY, Kim S, Kim J, Do Heo W. Reversible protein inactivation by optogenetic trapping in cells. Nat Methods. 2014;11:633–636. doi: 10.1038/nmeth.2940. [DOI] [PubMed] [Google Scholar]
- 9.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–52. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 2010;7:206–8. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–32. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Robinson MS, Sahlender DA, Foster SD. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell. 2010;18:324–31. doi: 10.1016/j.devcel.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- 14.Dohmen R, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science (80-) 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–22. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 16.Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013;30:341–51. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–4. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 18.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–4. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 20.Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276:7176–86. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- 21.Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wedlich-Soldner R, Wai SC, Schmidt T, Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol. 2008;18:1719–26. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slaughter BD, Unruh JR, Das A, Smith SE, Rubinstein B, Li R. Nonuniform membrane diffusion enables steady-state cell polarization via vesicular trafficking. Nat Commun. 2013;4:1380. doi: 10.1038/ncomms2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bear JE, Loureiro JJ, Libova I, Fässler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–28. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 26.Schleiff E, Becker T. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol. 2011;12:48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- 27.Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139:731–43. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–51. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143:1931–45. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- 30.Smith SE, Rubinstein B, Mendes Pinto I, Slaughter BD, Unruh JR, Li R. Independence of symmetry breaking on Bem1-mediated autocatalytic activation of Cdc42. J Cell Biol. 2013;202:1091–106. doi: 10.1083/jcb.201304180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–22. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 33.Howell AS, Lew DJ. Morphogenesis and the cell cycle. Genetics. 2012;190:51–77. doi: 10.1534/genetics.111.128314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 35.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 36.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–70. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 37.Toettcher JE, Gong D, Lim WA, Weiner OD. Light control of plasma membrane recruitment using the Phy-PIF system. Methods Enzymol. 2011;497:409–23. doi: 10.1016/B978-0-12-385075-1.00017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–34. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlton PM, Boulanger J, Kervrann C, Sibarita JB, Salamero J, Gordon-Messer S, Bressan D, Haber JE, Haase S, Shao L, Winoto L, Matsuda A, Kner P, Uzawa S, Gustafsson M, Kam Z, Agard DA, Sedat JW. Fast live simultaneous multiwavelength four-dimensional optical microscopy. Proc Natl Acad Sci U S A. 2010;107:16016–22. doi: 10.1073/pnas.1004037107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Core Team. R: A language and environment for statistical computing. Found Stat Comput 2014 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PIF-tagged Bem1 localizes normally to poles and buds. White dotted lines indicate cell periphery (traced from DIC images). Bem1-mCitrine-PIF fluorescence shown.
Figure S2. Prolonged recruitment of YFP-PIF to PhyB-mCherry-Htb2 anchor generates a cell growth artifact, while recruitment to of YFP-PIF to PhyB-mCherry-Tom7 and PhyB-mCherry-Snf7 does not. Cells were incubated with continuous red light. All strains contain pADH-PIF-YFP and pADH-PhyB-mCherry fused to the indicated anchors. Time between buds was measured by counting the number of frames between the first and second visible bud emergence events. Error bars represent SEM. 10–15 cells were measured for each condition.
Figure S3. Bem1-PIF DeLight cells without sequestration behave identically to wild-type cells. (a) Cell size over time measured from DIC images. Cells were not treated with PCB, but were exposed to red light as in DeLight experiments. (b) Cell size at the beginning and end of the movie is not significantly different between non-sequestered Bem1 DeLight cells and wild-type cells (t-test, p-value >0.05). In addition, cells were scored for isotropic growth, with less than 1% of cells growing isotropically in each condition, and for cell lysis, with no cells lysing in either condition.
Figure S4. Additional Cdc24 does not rescue the Bem1 DeLight phenotype. (a) Cell size over time measured from DIC images. Bem1-PIF sequestered + Cdc24 strain contains a pCyc-Cdc24 construct integrated at the TRP locus, but is otherwise identical to the Bem1-PIF sequestered strain. Integration of the construct was verified by colony PCR across both junctions. (b) Cell scoring for isotropic growth vs. normal growth/budding.
Figure S5. Cells polarize active Cdc42 after release from alpha factor arrest. Cells were washed after incubating with alpha factor for 90 minutes. Polarity was scored using the CRIB-2xGFP Cdc42 activity reporter.
Figure S6. Sequestering Bem1-PIF to the mitochondria reversibly prevents polarization of active Cdc42. Cells were exposed to red light for 3 hours and then switched to infrared light to release Bem1-PIF. Cells polarize and grow normally after release.
Figure S7. Pole size scales with cell size, and scaling is lost with latrunculin treatment, regardless of method used to select measurement timepoint. Pole size for each cell was measured over 3–10 timepoints. A single timepoint per cell was plotted. This timepoint was selected by one of the following criteria: signal to noise (highest ratio of pole intensity to cytoplasmic intensity), brightest pole, widest pole, narrowest pole, or median pole width.