Abstract
Resistance substantially limits the depth and duration of clinical responses to targeted anticancer therapies. Through the use of complementary experimental approaches, investigators have revealed that cancer cells can achieve resistance through adaptation or selection driven by specific genetic, epigenetic, or microenvironmental alterations. Ultimately, these diverse alterations often lead to the activation of signaling pathways that, when co-opted, enable cancer cells to survive drug treatments. Recently developed methods enable the direct and scalable identification of the signaling pathways capable of driving resistance in specific contexts. Using these methods, novel pathways of resistance to clinically approved drugs have been identified and validated. By combining systematic resistance pathway mapping methods with studies revealing biomarkers of specific resistance pathways and pharmacological approaches to block these pathways, it may be possible to rationally construct drug combinations that yield more penetrant and lasting responses in patients.
Characterizing resistance to targeted therapies
Intrinsic and acquired resistance place substantial limits on the clinical effectiveness of targeted therapies (1). Intrinsic resistance can be visualized through “waterfall” plots, which show the maximum percentage change in the size of target lesions in a cohort of individual patients following initial drug treatment. In a typical example, a fraction of patients will fail to respond by standard criteria (e.g., Response Evaluation Criteria in Solid Tumors (RECIST)), while another fraction of patients will respond but in an incomplete fashion. For the case of targeted therapies considered to yield high rates of initial response, like the RAF and MEK inhibitors used in patients with BRAF mutant melanoma, the percentage of patients that fail to meet RECIST criteria is often less than 30%, although the majority of those who meet these criteria nevertheless fail to achieve complete responses (2). Further, even in patients who respond initially to targeted therapies, the development of acquired resistance is nearly universal. Collectively these resistance phenomena yield a scenario in which patients treated with most currently approved targeted therapies derive progression-free and overall survival benefits on the scale of weeks to months, but rarely longer (1, 2).
Determining the key alterations driving resistance to targeted therapies is likely to be the first step toward improving their potencies and durabilities. A suite a complementary experimental approaches have been developed to address this problem. These methods, summarized below, have revealed that a wide range of events can drive resistance by modifying the target of a drug or the signaling network or cell state on which a drug acts (1).
Target-centric approaches, most commonly using sequencing or mutagenesis, have identified mutations, amplifications, and alternative splicing events in the genes encoding the targets of kinase inhibitors. Included in this group are alternative splicing events in BRAF, which drive acquired resistance in a substantial fraction of patients treated with RAF inhibitors (3), as well as the “gatekeeper” mutations in BCR-ABL (e.g., T315I) and EGFR (e.g., T790M), both of which are the targets of third-generation inhibitors with increased potency in these contexts (4–6).
Hypothesis-driven approaches, including those that leverage our understanding of the key architectural features of oncogenic signaling networks in cancer cells, have also been a valuable source of insight into the mechanisms of drug resistance. These approaches typically make use of cell lines in which resistance has been evolved through step-wise selection in the presence of drug treatment, although drug-sensitive cell lines, cell lines with varying levels of intrinsic resistance, and primary cells derived from human or mouse tumors have also been used in these studies. Hypothesis-driven studies have revealed important resistance alterations based, for example, on drug target bypass, signaling pathway feedback, compensatory survival signaling, and reversible changes in cellular differentiation state (7–12).
More recently, large, unbiased efforts have sought to reveal genomic or epigenomic alterations associated with resistance. For example, drug screens across large panels of genomically annotated cell lines have identified cell lines harboring “sensitizing” mutations (for example, BRAF in melanoma) that nevertheless exhibit variable levels of sensitivity to the cognate kinase inhibitor (13). Analyses of secondary mutations, expression patterns, and other features of these cell line panels suggest potential mechanisms underlying variations in drug sensitivity (i.e. intrinsic resistance) (14, 15). Separately, next-generation sequencing of matched pre-treatment and post-relapse patient tumors has identified, in a fully unbiased fashion, alterations associated with resistance that can be credentialed in subsequent validation assays (2, 16).
Finally, large, unbiased functional screening efforts have led to the identification of novel resistance alterations. For example, genome-scale gain-of-function screens using libraries of lentivirally delivered open reading frames (ORFs) or short guide RNAs (to direct CRISPR/Cas9-based transcriptional activation), and analogous loss-of-function screens using genome-scale short hairpin RNA (shRNA) or sgRNAs (to direct CRISPR/Cas9-based gene knockouts), have identified genes whose overexpression or inhibition can drive resistance (17–20). Creative screening approaches utilizing growth factors or combinatorial cell culture techniques have also identified potential microenvironmental mediators of drug resistance which have subsequently been validated in cells and patient-derived samples (21, 22). Finally, large-scale drug screens have uncovered novel mechanisms of resistance using pharmacological probes that reverse resistance (23, 24). In one particularly exciting example, biopsy samples were obtained from patients with EGFR or ALK mutation positive lung adenocarcinomas who developed resistance to EGFR or ALK inhibitors, respectively. These samples were converted to cell lines, then subjected to drug screens to identify drugs that resensitize patient-derived tumor cells in vitro and in vivo. By matching sensitizing drug combinations with sequencing, specific resistance alterations could be functionally credentialed in individual tumors (24). Given recent advances in conditional reprogramming (25) and organoid culture (26) methods, which enable the stable propagation of primary cells from tumor biopsies, it is anticipated that methods like this will become increasingly common in the study of drug resistance.
Systematic identification of the signaling pathways controlling resistance
Although the results above clearly indicate that we possess the tools necessary to identify the prominent adaptive, genetic, epigenetic, and microenvironmental alterations that drive resistance to targeted therapies, they also raise concerns. Cataloguing the constellation of alterations that can drive resistance to even a single drug in a single disease context is an enormous undertaking, as evidenced by the dozens of published papers reporting such events. Perhaps more importantly, if resistance can be independently driven by so many different alterations, how can it be controlled?
A potential solution to these challenges comes from the observation that diverse alterations appear to drive resistance by co-opting a smaller, concerted set of oncogenic pathways (Table 1). For example, in BRAF mutant melanomas treated with RAF inhibitors, mutations in NRAS, MEK, and ERK, amplification and alternative splicing of BRAF, and alternative regulation of MAP-3-kinases like COT and C-RAF each have the potential to independently drive resistance by reactivating the Ras-mitogen-activated protein kinase (MAPK) pathway. Similarly, mutations or altered expression of IGF-1R, PIK3CA, PTEN, and AKT may also be individually sufficient to drive resistance to these drugs, but in all cases they appear to do so through their activation of the PI3K (phosphatidylinositol-3-kinase) pathway (2). Consistent with this idea, a broad survey of the literature suggests that resistance to targeted therapies is typically associated with the activation of one or more of the canonical pathways controlling growth, survival, differentiation, and apoptosis in cancer cells.
Table 1.
Examples of common resistance alterations and pathways for selected targeted therapies.
| Drug | Target (cancer type) | Common resistance alterations | Key resistance pathways |
|---|---|---|---|
| vemurafenib | BRAF (melanoma) |
NRAS, MEK, ERK mut. BRAF amp. or splicing IGF1R exp., PIK3CA mut., PTEN loss NOTCH1 exp./activation |
Ras-MAPK pathway Ras-MAPK pathway PI3K pathway Notch1 pathway |
| gefitinib/erlotinib | EGFR (lung) |
EGFR secondary mut. EGFR, MET amp. PIK3CA mut. |
Ras-MAPK, PI3K pathways Ras-MAPK, PI3K pathways PI3K pathway Other pathways (EMT, lineage commitment) |
| cetuximab/panitumumab | EGFR (colorectal) |
KRAS, NRAS, BRAF mut. EGFR mut. or amp. ERBB2, MET amp. |
Ras-MAPK pathway Ras-MAPK pathway Ras-MAPK pathway |
| crizotinib | ALK (lung) |
EML4-ALK amp. or secondary mut. EGFR, KIT, IGF1R, ERBB2, ERBB3 amp. or exp. |
Ras-MAPK, PI3K pathways Ras-MAPK, PI3K pathways |
| lapatinib/trastuzumab | HER2 (breast) | EGFR, ERBB3, ERBB4, AXL, IGF1R, MET or EPHA2 exp. TGFBRI, ERα exp. HER2 mut. or splicing PIK3CA mut., PTEN loss RELA exp. |
ERBB-PI3K, Ras-MAPK pathways TGFβ, ERα pathways ERBB-PI3K pathway PI3K pathway NF-κB pathway |
| quizartinib | FLT-3 (acute myeloid leukemia) | FLT-3 secondary mut. | Pathways downstream of FLT-3 (e.g., Ras-MAPK, PI3K, STAT) |
| ruxolitinib | JAK2 (myeloproliferative neoplasms) | JAK2/JAK1 or JAK2/TYK2 heterodimerization JAK2 secondary mut. KRAS, NRAS mut. |
JAK-STAT pathway JAK-STAT pathway PI3K, Ras-MAPK pathways |
| imatinib | BCR-ABL (chronic myeloid leukemia) |
BCR-ABL secondary mut. BCR-ABL amp. |
Pathways downstream of BCR-ABL (e.g., Ras-MAPK, PI3K, STAT) |
mut. = mutation; amp. = amplification; exp. = expression
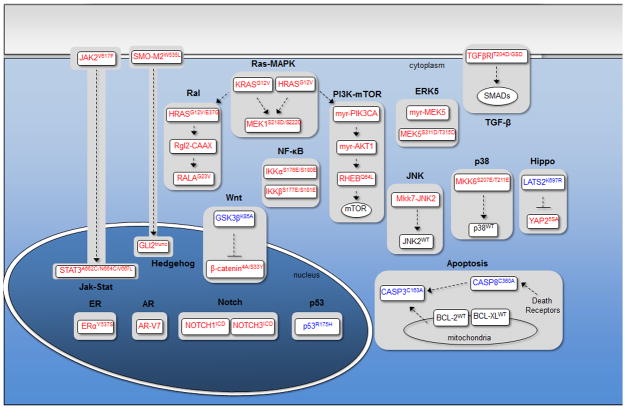
On the basis of these observations, we reasoned that a systematic effort to identify the pathways having potential to confer functional resistance to targeted therapies may accelerate progress in the field. We constructed an initial list of 17 signaling pathways that have been frequently implicated in the oncogenic processes above, and for each pathway, a set of 1–3 mutant complementary DNAs (cDNAs) were identified representing core nodes in each pathway whose overexpression should render the pathway constitutively active (or, in a minority of cases, inactive). Pathway-modulating cDNAs were assembled, barcoded, and cloned into a PGK promoter-driven lentiviral expression vector, then validated through sequencing and functional assays to ensure proper engagement of targeted pathways (Figure 1a) (27).
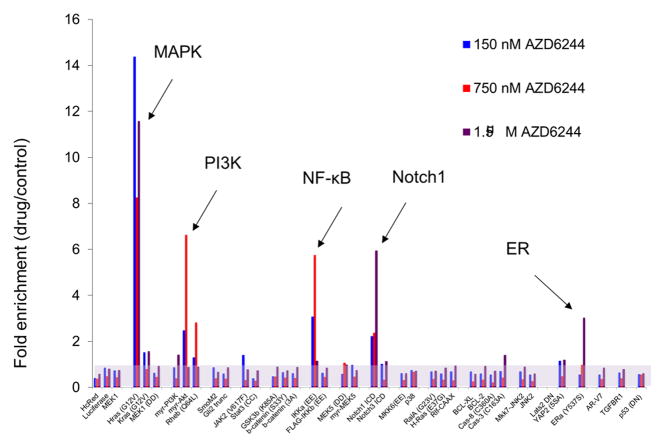
Figure 1.
Systematic identification of signaling pathways conferring functional resistance to drugs. (a) Library of pathway modulating cDNAs. Activating constructs are listed in red boxes, dominant negative constructs are listed in blue boxes, and wild-type constructs are listed in black boxes. (b) Results of a proof-of-concept screen in UACC-62 cells (BRAF mutant melanoma) for pathways whose activation can confer functional resistance to a MEK1/2 inhibitor (AZD6244).
To identify pathways whose modulation is sufficient to drive therapeutic resistance in vitro, we used a positive selection pooled screening protocol with sequencing-based deconvolution. In BRAFV600E melanoma cells, this method identified five major pathways capable of driving resistance to a MEK1/2 inhibitor (AZD6244), including the well-documented Ras-MAPK, PI3K, and NF-κB pathways along with the previously unreported Notch1 and the estrogen receptor (ERα) pathways (Figure 1b). On the strength of these findings, a large number of screening assays have been performed to identify potential pathways of resistance to a range of targeted therapies. Globally, these results suggest that certain pathways, including Ras-MAPK, PI3K, and Notch1, are frequently capable of driving resistance in many contexts, while other pathways appear to confer resistance in a context-specific fashion. Further, for a given drug, the number of screened pathways capable of conferring resistance by this assay is modest: typically 5 or fewer (27).
Using these methods, a series of novel pathways of resistance have been nominated and then subsequently confirmed in resistant cell lines, tumor models, and/or patient samples. For example, Ras-driven AKT activation may contribute to the ubiquitous intrinsic resistance observed in patients with JAK2-mutant myelofibrosis treated with JAK inhibitors, and combination therapies involving JAK inhibitors paired with either AKT or downstream BCL-XL inhibitors reverse resistance in multiple models (28). In a second example, intrinsic resistance to RAF/MEK inhibitors in BRAF-mutant melanoma is controlled by combinations of the PI3K, Notch1, and ERα pathways, and co-inhibition of these pathways uniformly sensitizes a panel intrinsically resistant cell lines to these drugs (27). As a third example, Notch1 activation can drive acquired resistance to RAF/MEK inhibitors in numerous evolved cellular models of BRAF-mutant melanoma, and Notch1 activation is observed in the subset of human patients with acquired resistance whose tumors lack evidence of resistance driven by the established Ras-MAPK and PI3K pathways (27). As a final example, Notch1 activation also identifies a subset of patients with ER+ breast cancer who fail to derive benefit from treatment with the selective ER modulator tamoxifen, and acquired resistance to tamoxifen can be reversed in vivo using a γ-sectretase inhibitor that blocks Notch1 signaling (27).
Outlook
Pathway-centric gain-of-function screening adds to the repertoire of methods by which mechanisms of resistance can be identified and may help to streamline our ability to connect diverse resistance-conferring alterations to the underlying pathways through which they act. Importantly, as with other methods, putative resistance pathways identified using pathway-centric screening must be confirmed through the demonstration that pathway inhibition can reverse resistance in independent cell lines or tumors before they can be confirmed as bona fide resistance pathways (27, 28). Pathway-centric screening may have particular advantages owing to its use of validated reagents which specifically activate or inhibit their cognate pathways, thus potentially minimizing false positives and false negatives in screening assays, and its use of small libraries of lentiviral cDNAs, which enable a single researcher to perform many screening assays across cell lines and drug treatments in parallel (27).
A potential limitation of this approach is its ability to sample only a restricted set of signaling pathways. To address this limitation, it will be important to expand the library of cDNA constructs to include those encoding common oncogenic mutations and activators or inhibitors of additional pathways (for example, metabolic, transcriptional, and epigenetic) as well as additional nodes within each pathway. Toward this goal, we have initiated a two-year project with the National Cancer Institute’s Ras Program that will expand the cDNA library by several-fold, and all sequence-validated reagents from this effort will be made publicly available at low cost. These reagents may also be complemented by the use of libraries of mutant alleles developed by other groups (29, 30). Finally, although our applications of pathway-centric screening have thus far been limited primarily to drug resistance, it is straightforward to envision additional applications. For example, two groups recently reported elegant chemogenomic mapping efforts involving mutant cDNA libraries (29, 30), and assays used to study drug dependencies, in vivo tumorigenesis, and various cancer hallmark properties can potentially be married with pathway-centric screening.
In the coming years, the key pathways of resistance to a range of targeted therapies used in distinct contexts are likely to be defined. At that point, however, major scientific challenges will remain. For example, it will likely be necessary to monitor minimal disease states with high resolution, identify the specific resistance pathways activated in patients’ tumors using credentialed biomarkers, define safe and effective strategies to block resistance pathways in vivo, and then construct methods to anticipate and pharmacologically adapt to tumor evolution over time.
Acknowledgments
Research in the Wood laboratory is supported by the NIH Building Interdisciplinary Research Careers in Women’s Health Program, the Harry J. Lloyd Trust, Golfers Against Cancer, the Stewart Trust, the V Foundation for Cancer Research, the Ovarian Cancer Research Fund, the Duke University School of Medicine, and the Duke Cancer Institute. I apologize to those colleagues whose valuable contributions could not be cited here owing to space limitations.
Footnotes
Conflicts of interest
The author discloses no potential conflicts of interest.
References
- 1.Glickman MS, Sawyers CL. Converting cancer therapies into cures: lessons from infectious diseases. Cell. 2012;148(6):1089–98. doi: 10.1016/j.cell.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solit DB, Rosen N. Towards a unified model of RAF inhibitor resistance. Cancer Discov. 2014;4(1):27–30. doi: 10.1158/2159-8290.CD-13-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. New Engl J Med. 2015;372(18):1689–99. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 5.Sequist LV, Soria JC, Goldman JW, Wakelee HA, Gadgeel SM, Varga A, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. New Engl J Med. 2015;372(18):1700–9. doi: 10.1056/NEJMoa1413654. [DOI] [PubMed] [Google Scholar]
- 6.Cortes JE, Kim DW, Pinilla-Ibarz J, le Coutre P, Paquette R, Chuah C, et al. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. New Engl J Med. 2013;369(19):1783–96. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19(1):58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 9.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4(120):120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstein JN. Drug discovery: Cell lines battle cancer. Nature. 2012;483(7391):544–5. doi: 10.1038/483544a. [DOI] [PubMed] [Google Scholar]
- 14.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4(7):816–27. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood KC, Konieczkowski DJ, Johannessen CM, Boehm JS, Tamayo P, Botvinnik OB, et al. MicroSCALE screening reveals genetic modifiers of therapeutic response in melanoma. Sci Signal. 2012;5:rs4. doi: 10.1126/scisignal.2002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4(1):94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504(7478):138–42. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3(3):350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487(7408):505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda A, et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013;3(12):1416–29. doi: 10.1158/2159-8290.CD-13-0350. [DOI] [PubMed] [Google Scholar]
- 24.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480–6. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180(2):599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A, et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci Signal. 2014;7:ra121. doi: 10.1126/scisignal.aaa1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winter PS, Sarosiek KA, Lin KH, Meggendorfer M, Schnittger S, Letai A, et al. Ras effector pathways drive resistance to JAK inhibitors by suppressing BAD-mediated apoptosis. Sci Signal. 2014;7:ra122. doi: 10.1126/scisignal.2005301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martins MM, Zhou AY, Corella A, Horiuchi D, Yau C, Rakshandehroo T, et al. Linking tumor mutations to drug responses via a quantitative chemical-genetic interaction map. Cancer Discov. 2015;5(2):154–67. doi: 10.1158/2159-8290.CD-14-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muellner MK, Uras IZ, Gapp BV, Kerzendorfer C, Smida M, Lechtermann H, et al. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol. 2011;7(11):787–93. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]