Abstract
For many years now, researchers have known of a sensory appendage on the surface of most differentiated cell types, called primary cilium. Primary cilia are both chemo-and mechano-sensory in function and have an obvious role in cell cycle control. Because of this, it has been thought that primary cilia are not found on rapidly proliferating cells, as e.g. cancer cells. Here we report using immunofluorescent staining for the ciliary protein Arl13b that primary cilia are frequently found on HeLa (human epithelial adenocarcinoma) and other cancer cell lines such as MG63 (human osteosarcoma) commonly used for cell culture studies, and that the ciliated population is significantly higher (ave. 28.6% and 46.5%, respectively in starved and 15.7 to 18.6% in un-starved cells) than previously anticipated. Our finding impacts the current perception of primary cilia formed in highly proliferative cells.
Keywords: Arl13b, Cilia, Ciliopathies, Cancer cells, HeLa cells, Primary cilia
1. Introduction
Primary cilia are single non-motile cilia found on the surface of non-dividing or quiescent cells (Pan and Snell, 2007). First discovered by Zimmerman in 1898 (Zimmermann, 1898), the existence of primary cilia was known for some time, however without knowledge of their function. Most early descriptions of this cellular projection were accomplished through electron microscopy, but recent advances in immunofluorescent microscopic techniques including antibodies directed against proteins found primarily in motile and primary cilia have allowed for further analysis of the primary ciliary role.
Primary cilia are thought to be both mechano- and chemosensory and to function in coordinating several signaling pathways e.g. sonic hedgehog, Wnt, and RTK (Barral et al., 2012; Christensen et al., 2012; Goto et al., 2013; Mukhopadhyay and Rohatgi, 2014; Oh and Katsanis, 2013; Satir et al., 2010). Ciliopathies, the dysfunction or lack of cilia, have been implicated in obesity, diabetes, situs inversus, polydactyly, Joubert, orofaciodigital and Bardet-Biedl syndromes, and other developmental complications (Pan et al., 2005; Satir et al., 2010). Like other cilia, the axoneme is structurally formed by stable microtubules composed primarily of acetylated- and glutamylated- tubulin, but unlike other cilia, which have a 9+2 configuration, primary cilia have a 9+0 arrangement (Satir and Christensen, 2007; Satir et al., 2010). The cilium projects from the basal body, which originates from the active mother centriole. Because of these structural roots, the primary cilium is expected to be involved in the regulation of progression into the cell cycle (Goto et al., 2013; Jackson, 2011; Pan and Snell, 2007; Plotnikova et al., 2008; Plotnikova et al., 2009; Pugacheva et al., 2007; Tucker et al., 1979). Additionally, evidence for the primary cilium’s role in regulating the cell cycle has been published describing the localization of several critical cell cycle proteins to the cilium, including Aurora A which functions in deciliation and prevention of cilium regeneration (Goto et al., 2013; Inoko et al., 2012; Pugacheva et al., 2007).
In general, cancer cells are thought to have lost their ability to form primary cilia, since by definition cancer is the loss of the cells’ ability to control growth and results in cells entering the cell cycle aberrantly (Hassounah et al., 2012; Seeley et al., 2009; Yuan et al., 2010). However, implementing antibodies directed against a protein called Arl13b (ADP-ribosylation factor-like 13b), a small GTPase found in the axoneme of cilia, here we describe the frequent presence of primary cilia on HeLa and MG63 (human epithelial adenocarcinoma and osteosarcoma, respectively) cancer cells. Arl13b is a small GTPase whose cellular localization is restricted to the axoneme of cilia and to some extent to some actin rich structures of migrating cells (Casalou et al., 2014; Caspary et al., 2007; Duldulao et al., 2009; Sun et al., 2004). Arl13b is known to function in the maintenance of ciliary structure, however in a not yet completely understood fashion. Mutations in the Arl13b gene, which lead to Joubert syndrome, a disease that manifests itself in brain malformations, ocularmotor apraxia, kidney cysts, and polydactyly, are also poorly understood and can sometimes lead to the loss of cilia (Delling et al., 2013; Higginbotham et al., 2013; Juric-Sekhar et al., 2012; Miertzschke et al., 2014)
2. Materials and Methods
2.1. Cell culture
HeLa, human epithelial adenocarcinoma cells (CCL-2, two different lots, 59681574 and 60143948 purchased on 10/12/2012 and 3/24/2014, respectively), mouse embryo NIH3T3 fibroblasts (CRL-1658), MG63 human osteosarcoma cells (CRL-1427), and MC3T3-E1 subclone 4 mouse pre-osteoblasts (CRL-2593) were purchased from American Type Culture Collection (ATCC, Manassas,VA). All cell types were maintained at 37°C in a 5% CO2 atmosphere and 100% humidity in Dulbecco’s modified Eagles medium (Sigma, St. Louis, MO) (HeLa and NIH3T3), E-modified Eagles medium (MG63), or alpha-modified Eagles medium (Gibco/Invitrogen, Grand Island, NY) (MC3T3-E1) completed with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA), 1% L-glutamine (HyClone, Logan, UT) and 1% penicillin/streptomycin (Corning, Corning, NY) according to vendor’s instructions.
2.2. Immunofluorescence staining
Cells were seeded onto poly-L-lysine (Sigma, St. Louis, MO) coated cover slips placed into 3.5cm diameter tissue culture plastic dishes and cultured for 2 to 3 days in complete medium to approximately 60 to 70% confluence or in a series of dilutions of suspended cells resulting in a range in confluency of approximately 20% - 80% and then fixed or were serum starved for 24 hours (Li et al., 2011) before fixation. Cells were fixed in either 3.7% formaldehyde (in 1×PBS) for 15 min at room temperature (RT) followed by permeabilization in ice-cold methanol (5 minutes), or were fixed and permeabilized directly in ice-cold methanol (5 min). Then, cells were blocked in 5% FBS/1×PBS at RT for 30 min. Primary antibodies directed against anti-Arl13b (mouse monoclonal-NeuroMab, Davis, CA Cat. # 73-287; at 1:200), anti-acetylated-tubulin (rabbit polyclonal, Sigma, St. Louis, MO cat # 6-11B-1; at 1:250.), anti-PCNA (rabbit polyclonal, Santa Cruz, Dallas, TX Cat. #Sc-7907; at 1:200.) and anti-γ-tubulin (rabbit polyclonal, Sigma, St. Louis, MO Cat. # T3559; at 1:500) were diluted in blocking solution and incubated with cells for 1h at RT. Cells were then incubated in secondary antibodies Alexa488-conjugated goat-anti-mouse and Alexa568-conjugated goat-anti-rabbit (Molecular Probes/Invitrogen, Grand Island, NY, Cat. # A11029 and A11036, respectively) diluted at 1:200 for 1h at RT. Nuclei were stained using DAPI (Molecular Probes, Eugene, OR, Cat. # D1306). Slides were imaged using a Nikon Eclipse TE2000-E inverted fluorescence microscope equipped with 40× and 60× Plan Apo, NA 1.4 oil immersion objectives and a forced-air-cooled Photometrics CoolSnap HQ CCD camera (Roper Scientific, Duluth, GA). Images were captured using MetaVue (Molecular Devices, Sunnyvale, CA) software version 6.1r5.
2.3. Quantitative Image analyses
Images of 7 randomly chosen areas on each coverslip (n=5) were acquired by scanning the coverslips in the UV excitation channel (DAPI, showing blue emitting nuclei). Next, the filter cubes were rotated to the blue excitation channel (showing green Arl13b emission), focused and imaged. Both authors evaluated and analyzed the images independently. HeLa cells were counted from two different lots from ATCC (lot # 59681574 and 60143948) totaling 1845 counted cells and 551 counted cilia. Variation in counts between evaluators was below 15%. MetaVue software was used to count cell nuclei to determine the total number of cells present in the field of view. Similarly, the cilia in the same field of view were counted to determine the number of ciliated cells in the population. The same analysis protocol was used for NIH3T3 cells (n=3), MG63 cells (n=2), and MC3T3-E1 cells (n=3); 175, 56, and 185 cells were counted respectively.
3. Results and Discussion
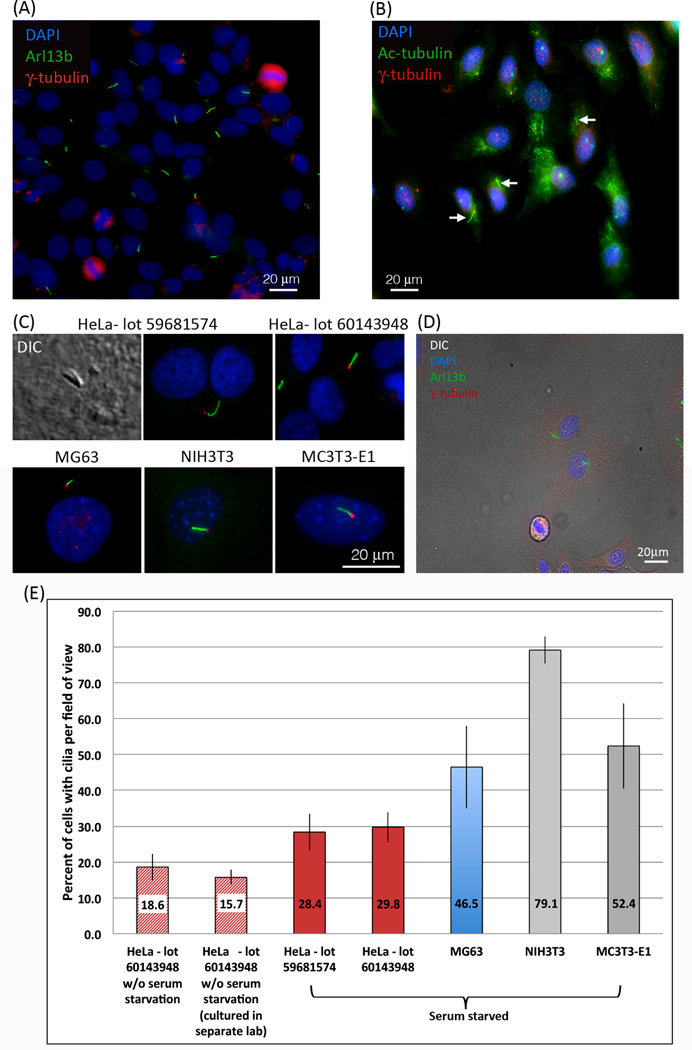
Since primary cilia contribute to cell cycle control and are usually present during the G0 and G1 phase of the cell cycle (Goto et al., 2013; Inoko et al., 2012; Li et al., 2011; Pan and Snell, 2007), we arrested HeLa cells growing on cover glasses at the G1/S phase by serum starvation to enrich for cells containing cilia as described by Li et al. (Li et al., 2011). Unexpectedly and contrary to earlier reports (Alieva et al., 1999; Alieva and Vorobjev, 2004), we frequently detected primary cilia on Hela cancer cells, as shown by DIC imaging and indicated by staining positive for Arl13b, a specific protein marker for ciliary axonemes (Figure 1A, C and D). Additionally, the base of each primary cilium stained positive for γ-tubulin (Figure 1A, C and D), indicating that the cellular projection was rooted at a centriole, another characteristic of primary cilia (Wheatley et al., 1996). Although unlikely, to exclude the possibility that our HeLa cell cultures (lot 59681574) were unintentionally contaminated with another cell type (Lorsch et al., 2015; Yu et al., 2015), we purchased a second vial of a later batch of HeLa cells from ATCC, the main repository of the U.S. (lot 60143948), and investigated cilia formation after the cells had been sub-cultured only once. No significant differences in the overall number of ciliated cells from the two different lots of HeLa cells were detected (Figure 1C, E). To quantify the number of HeLa cells that were ciliated under our standard starvation cell culture conditions, we counted cell nuclei and cilia, and then compared the number of ciliated cells to the total number of cells. Of the 1845 HeLa cells counted, 28.6% +/− 5% (Figure 1E) stained positive for both Arl13b and γ-tubulin indicating the presence of a primary cilium (Figure 1A,C and D).
Figure 1. Primary cilia are frequent on HeLa cancer cells.
(A, C) Two different lots of HeLa cells purchased from ATCC stained positive for Arl13b (green), a protein localized with high specificity to the axonemes of cilia. At the base of the cilia, centrioles visualized using an antibody targeting γ-tubulin (red) are clearly detectable as well. Cell nuclei are stained with DAPI (blue). (B) A more traditional stain for primary cilia, acetylated-tubulin (green) and γ-tubulin (red), also stains primary cilia (arrows) but not as distinctly as with antibodies directed against Arl13b. (C) Primary cilia were also detected using DIC. NIH3T3 cells, reported as ‘the gold standard’ for primary ciliated cells, and MC3T3-E1 cells both exhibited a similar staining pattern to HeLa cells. (D) Fluorescent Arl13b (green), γ–tubulin (red), and DAPI (blue) stains merged with a DIC image demonstrate that even at very low density HeLa cells form primary cilia. (E) Quantitative analysis of primary cilia detected in different lots of starved and un-starved HeLa cells, as well as in starved MG63 osteosarcoma, NIH3T3 fibroblasts, and MC3T3-E1 pre-osteoblast cells. The number of HeLa cells that stain positive for primary cilia that we detected is significantly higher (28.6% after starvation, 18.6% in un-starved cells) than had been reported previously (<2%). In NIH3T3 and MC3T3-E1 comparable values to previously published reports were detected (Alieva et al., 1999; Malone et al., 2007; Schneider et al., 2005).
To investigate whether other cancer cells also form primary cilia, we cultured, and stained human MG63 osteosarcoma cells. Also on these cells we frequently detected primary cilia with 46.5% +/− 11.5% of cells containing primary cilia (Figure C, E). Mouse NIH3T3 fibroblasts and mouse MC3T3-E1 pre-osteoblasts reported previously to exhibit primary cilia in a large number of cells were grown and stained in parallel as a positive control (Figure 1C). We determined that 79.5% +/− 3.7% and 52.4% +/− 11.9% of NIH3T3 fibroblasts and MC3T3-E1 pre-osteoblasts, respectively were ciliated (Figure 1E), similar to published numbers (Alieva et al., 1999; Malone et al., 2007; Schneider et al., 2005). As another control, we used a more traditional stain for primary cilia, acetylated-tubulin and γ-tubulin (Figure 1B). This stain, while commonly used by others, in our hands was less cilium specific since, in addition to staining ciliary acetylated-tubulin it also stained acetylated-tubulin in the cytoplasm. Thus, the additional stain of acetylated cytoskeletal microtubules may have obscured cilia present on some cells and this may have contributed to our enhanced efficiency in detecting primary cilia in HeLa and MG63 cancer cells using Arl13b staining.
To exclude the possibility that we were inducing a quiescent G0–like state causing the HeLa cells to aberrantly form cilia, we grew HeLa cells without serum starvation. These HeLa cells formed less cilia than serum starved cells, 18.6% +/− 3% and 15.7% +/− 2.1% compared to 28.6% +/− 5% (Figure 1E), however they formed significantly more primary cilia than has been reported previously (none or <2%) (Alieva et al., 1999; Alieva and Vorobjev, 2004; Inoko et al., 2012). This result is in agreement with our current understanding of primary cilia formation in that serum starvation increases the number of ciliated cells. Additionally, as culture conditions including culture facilities, growth media, passage numbers, etc. may affect cell culture behavior, we analyzed HeLa cells for Arl13b expression that were independently purchased by another laboratory from ATCC (lot # 60143948) and were cultured without serum starvation in their facility using their own consumables (medium, culture dishes, coverglasses, etc.). Also in these cells a significant amount of primary cilia were detected (15.7% +/− 2.1%), similar to what we detected previously in the HeLa cell batches that we had cultured (Figure 1E).
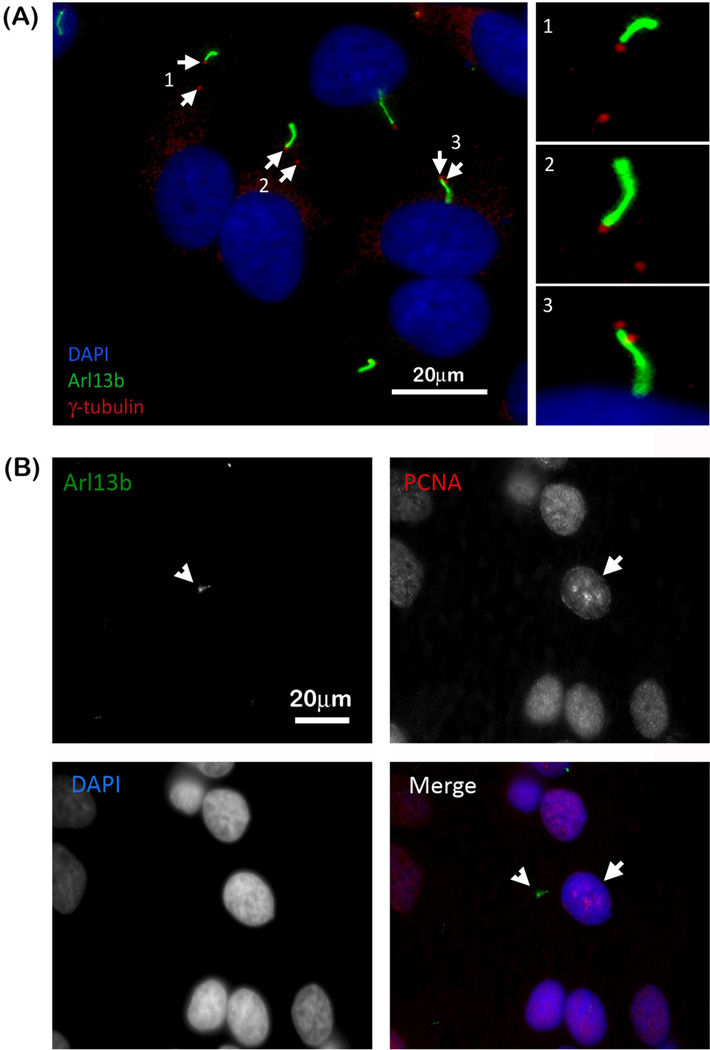
As described above we provide evidence that HeLa as well as other cancer cell lines form primary cilia under starved and un-starved culture conditions. To exclude the possibility that we induced primary cilia formation by growing HeLa cells at a too high confluency unintentionally inducing an arrested stage (although an unlikely assumption as cancer cells such as HeLa are not contact inhibited and continue to grow in three dimensions when reaching total confluency), we performed a series of serial dilutions ranging from approximately 20% to 80% confluency. We did not observe a difference in the number of cells containing primary cilia as depicted by the representative merged DIC and fluorescence image of the lowest dilution shown in Figure 1D. Since the presence of duplicated centrosomes indicates that the cells are in late S- or G2 phase of the cell cycle, we counted the number of cells containing primary cilia that had two centrosomes. Approximately 13% of serum starved HeLa cells containing a primary cilium also had 2 centrosomes (a representative image is shown in Figure 2A, centrosomes depicted with arrows), suggesting that actively dividing HeLa cells were also ciliated. To further support this conclusion, we stained HeLa cells for Arl13b as well as PCNA (proliferating cell nuclear antigen). PCNA is a DNA clamp that promotes polymerase δ interaction with DNA during the S-phase of the cell cycle to increase processivity (Herce et al., 2014). While PCNA is found in the nucleus at all stages of the cell cycle, it becomes concentrated and localized in foci of active DNA synthesis during S-phase forming a specific, punctate staining pattern within the nucleus (Herce et al 2014). As shown in Figure 2B, HeLa cells that exhibited the distinctive PCNA S-phase staining pattern (Figure 2B, depicted with arrow) in addition stained positive for Arl13b (Figure 2B, depicted with arrowhead), further suggesting the presence of primary cilia in cells that are in the S-Phase of the cell cycle. Taken together, these results suggest that in addition to quiescent cells (G0), actively proliferating HeLa cells exhibit primary cilia.
Figure 2. Actively proliferating HeLa cells also have primary cilia.
(A) Arl13b (green) and γ-tubulin (red) staining reveals several HeLa cells with two centrioles (depicted with arrows) indicating that these cells are in either the S- or G2- phase of the cell cycle. Higher magnifications of the regions containing the cilium and centrioles are to the right. (B) Arl13b staining (depicted with arrowhead) together with PCNA staining further indicates that proliferating cells have primary cilia as indicated by the distinctive punctate PCNA S-phase staining pattern in the nuclei of cells (depicted with arrow) containing primary cilia. Cell nuclei are stained with DAPI (merged panel: Arl13b, green; PCNA, red; nuclei, blue).
Recently, Emoto et al. reported that both primary pancreatic cells isolated from tumor biopsies as well as PANC-1 and CFPAC-1 cells, pancreatic cancer cell lines, stained positive for primary cilia. The authors reported that 25 of 100 tissue biopsies (25%) not only stained positive for primary cilia but that these samples also correlated with increased lymph node metastasis and decreased patient survival (Emoto et al., 2014). Additionally, Yuan et al. described that cells of healthy breast epithelial tissue typically had primary cilia while cells of cancerous epithelial tissue lacked primary cilia, except in the case of a very invasive and metastatic cancer type called basal B subtype (Yuan et al., 2010). These two examples suggest that cancer cells with primary cilia may be more aggressive in nature than un-ciliated cancer cells. Knowing the history of HeLa cancer cells and their highly aggressive behavior, detecting primary cilia on these cells may be less of a surprise.
3.1. Conclusions
Because of the particular cellular localization of Arl13b to the cilium shaft and the high specificity of the commercially available mouse monoclonal antibodies, detection of primary cilia, as shown here in HeLa and MG63 cancer cells, as well as in other cell lines known to exhibit high numbers of primary cilia (NIH3T3, MC3T3-E1), became more feasible. Taken together, our results indicate that proliferating malignant cancer cells, such as human epithelial HeLa adenocarcinoma cells or MG63 osteosarcoma cells robustly exhibit primary cilia, similar to other non-cancer cell lines, and that these cells do not need to reach a state of quiescence (G0), but also exhibit primary cilia during G1-, S- and G2-phases of the cell cycle. Thus, our finding sheds new light on the expression of primary cilia in highly proliferative cells, such as cancer (reported here) and embryonic stem (ES) cells (Bangs et al., 2015; Kiprilov et al., 2008).
Acknowledgements
The Authors would like to thank Dr. Sarah Goetz for her guidance with Arl13b staining, Drs. Robert Skibbens, Susan Perry and Lynne Cassimeris for generously providing antibodies and cells, and BioS368 Cell Bio lab students for optimizing Arl13b immunofluorescence staining.
Funding
Research in the Falk lab is funded by National Institutes of Health (NIH-NIGMS) [R01 GM55725] and Lehigh University.
Abbreviations
- Arl13b
ADP-ribosylation factor-like 13b
- ATCC
American Type Culture Collection
- DIC
Differential Interference Contrast
- RTK
Receptor Tyrosine Kinase
- Wnt
Wingless-related integration site
References
- Alieva IB, Gorgidze LA, Komarova YA, Chernobelskaya OA, Vorobjev IA. Experimental model for studying the primary cilia in tissue culture cells. Membr Cell Biol. 1999;12:895–905. [PubMed] [Google Scholar]
- Alieva IB, Vorobjev IA. Vertebrate primary cilia: a sensory part of centrosomal complex in tissue cells, but a "sleeping beauty" in cultured cells? Cell Biology International. 2004;28:139–150. doi: 10.1016/j.cellbi.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Bangs FK, Schrode N, Hadjantonakis AK, Anderson KV. Lineage specificity of primary cilia in the mouse embryo. Nature Cell Biology. 2015;17:113–122. doi: 10.1038/ncb3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral DC, Garg S, Casalou C, Watts GF, Sandoval JL, Ramalho JS, Hsu VW, Brenner MB. Arl13b regulates endocytic recycling traffic. Proc Natl Acad Sci U S A. 2012;109:21354–21359. doi: 10.1073/pnas.1218272110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalou C, Seixas C, Portelinha A, Pintado P, Barros M, Ramalho JS, Lopes SS, Barral DC. Arl13b and the non-muscle myosin heavy chain IIA are required for circular dorsal ruffle formation and cell migration. Journal Cell Science. 2014;127:2709–2722. doi: 10.1242/jcs.143446. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Christensen ST, Clement CA, Satir P, Pedersen LB. Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. J Pathol. 2012;226:172–184. doi: 10.1002/path.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504:311–314. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duldulao NA, Lee S, Sun Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 2009;136:4033–4042. doi: 10.1242/dev.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, Masugi Y, Yamazaki K, Effendi K, Tsujikawa H, Tanabe M, Kitagawa Y, Sakamoto M. Presence of primary cilia in cancer cells correlates with prognosis of pancreatic ductal adenocarcinoma. Human Pathology. 2014;45:817–825. doi: 10.1016/j.humpath.2013.11.017. [DOI] [PubMed] [Google Scholar]
- Goto H, Inoko A, Inagaki M. Cell cycle progression by the repression of primary cilia formation in proliferating cells. Cell Mol Life Sci. 2013;70:3893–3905. doi: 10.1007/s00018-013-1302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassounah NB, Bunch TA, McDermott KM. Molecular pathways: the role of primary cilia in cancer progression and therapeutics with a focus on Hedgehog signaling. Clinical Cancer Research. 2012;18:2429–2435. doi: 10.1158/1078-0432.CCR-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herce HD, Rajan M, Lättig-Tünnemann G, Fillies M, Cardoso MC. A novel cell permeable DNA replication and repair marker. Nucleus. 2014;5:590–600. doi: 10.4161/nucl.36290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Guo J, Yokota Y, Umberger NL, Su CY, Li J, Verma N, Hirt J, Ghukasyan V, Caspary T. Arl13b-regulated cilia activities are essential for polarized radial glial scaffold formation. Nat Neurosci. 2013;16:1000–1007. doi: 10.1038/nn.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoko A, Matsuyama M, Goto H, Ohmuro-Matsuyama Y, Hayashi Y, Enomoto M, Ibi M, Urano T, Yonemura S, Kiyono T. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. Journal Cell Biology. 2012;197:391–405. doi: 10.1083/jcb.201106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK. Do cilia put brakes on the cell cycle? Nat Cell Biol. 2011;13:340–342. doi: 10.1038/ncb0411-340. [DOI] [PubMed] [Google Scholar]
- Juric-Sekhar G, Adkins J, Doherty D, Hevner RF. Joubert syndrome: brain and spinal cord malformations in genotyped cases and implications for neurodevelopmental functions of primary cilia. Acta Neuropathol. 2012;123:695–709. doi: 10.1007/s00401-012-0951-2. [DOI] [PubMed] [Google Scholar]
- Kiprilov EN, Awan A, Desprat R, Velho M, Clement CA, Byskov AG, Andersen CY, Satir P, Bouhassira EE, Christensen SrT. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J Cell Biol. 2008;180:897–904. doi: 10.1083/jcb.200706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Saito M, Chuang JZ, Tseng YY, Dedesma C, Tomizawa K, Kaitsuka T, Sung CH. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat Cell Biol. 2011;13:402–411. doi: 10.1038/ncb2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Collins FS, Lippincott-Schwartz J. Cell Biology. Fixing problems with cell lines. Science. 2015;346:1452–1453. doi: 10.1126/science.1259110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone AM, Anderson CT, Tummala P, Kwon RY, Johnston TR, Stearns T, Jacobs CR. Primary cilia mediate mechanosensing in bone cells by a calcium-independent mechanism. Proc Natl Acad Sci U S A. 2007;104:13325–13330. doi: 10.1073/pnas.0700636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miertzschke M, Koerner C, Spoerner M, Wittinghofer A. Structural insights into the small G-protein Arl13B and implications for Joubert syndrome. Biochem J. 2014;457:301–311. doi: 10.1042/BJ20131097. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Rohatgi R. G-protein-coupled receptors, Hedgehog signaling and primary cilia. Semin Cell Dev Biol. 2014;33:63–72. doi: 10.1016/j.semcdb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh EC, Katsanis N. Context-dependent regulation of Wnt signaling through the primary cilium. J Am Soc Nephrol. 2013;24:10–18. doi: 10.1681/ASN.2012050526. [DOI] [PubMed] [Google Scholar]
- Pan J, Snell W. The primary cilium: keeper of the key to cell division. Cell. 2007;129:1255–1257. doi: 10.1016/j.cell.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Pan J, Wang Q, Snell WJ. Cilium-generated signaling and cilia-related disorders. Lab Invest. 2005;85:452–463. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 2008;68:2058–2061. doi: 10.1158/0008-5472.CAN-07-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova OV, Pugacheva EN, Golemis EA. Primary cilia and the cell cycle. Methods Cell Biology. 2009;94:137–160. doi: 10.1016/S0091-679X(08)94007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. Journal Cell Science. 2010;123:499–503. doi: 10.1242/jcs.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Current Biology. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Seeley ES, Carriere C, Goetze T, Longnecker DS, Korc M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Research. 2009;69:422–430. doi: 10.1158/0008-5472.CAN-08-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- Tucker RW, Scher CD, Stiles CD. Centriole deciliation associated with the early response of 3T3 cells to growth factors but not to SV40. Cell. 1979;18:1065–1072. doi: 10.1016/0092-8674(79)90219-8. [DOI] [PubMed] [Google Scholar]
- Wheatley DN, Wang AM, Strugnell GE. Expression of primary cilia in mammalian cells. Cell Biology International. 1996;20:73–81. doi: 10.1006/cbir.1996.0011. [DOI] [PubMed] [Google Scholar]
- Yu M, Selvaraj SK, Liang-Chu MM, Aghajani S, Busse M, Yuan J, Lee G, Peale F, Klijn C, Bourgon R. A resource for cell line authentication, annotation and quality control. Nature. 2015;520:307–311. doi: 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
- Yuan K, Frolova N, Xie Y, Wang D, Cook L, Kwon Y-J, Steg AD, Serra R, Frost AR. Primary cilia are decreased in breast cancer: analysis of a collection of human breast cancer cell lines and tissues. J Histochem Cytochem. 2010;58:857–870. doi: 10.1369/jhc.2010.955856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann KW. Beiträge zür Kenntniss einiger Drüsen und Epithelien. Archiv für Mikroskopische Anatomie. 1898;52:552–706. [Google Scholar]