SUMMARY
Phase-separated states of proteins underlie ribonucleoprotein (RNP) granules and nuclear RNA-binding protein assemblies that may nucleate protein inclusions associated with neurodegenerative diseases. We report that the N-terminal low complexity domain of the RNA-binding protein Fused in Sarcoma (FUS LC) is structurally disordered and forms a liquid-like phase-separated state resembling RNP granules. This state directly binds the C-terminal domain of RNA polymerase II. Phase-separated FUS lacks static structures as probed by fluorescence microscopy, indicating they are distinct from both protein inclusions and hydrogels. We use solution nuclear magnetic resonance spectroscopy to directly probe the dynamic architecture within FUS liquid phase-separated assemblies. Importantly, we find that FUS LC retains disordered secondary structure even in the liquid phase-separated state. Therefore, we propose that disordered protein granules, even those made of aggregation-prone prion-like domains, are dynamic and disordered molecular assemblies with transiently formed protein-protein contacts.
INTRODUCTION
The phenomenon of liquid-liquid phase separation has garnered much attention due to recent observations of liquid-like behavior of several cellular punctate and droplet structures including ribonucleoprotein granules and the nucleolus (Hyman et al., 2014). Breakthrough studies have demonstrated that these non-membrane bound structures or “assemblages” (Toretsky and Wright, 2014) behave as liquid phases, forming spherical droplets which flow, fuse upon contact, and take on spherical shapes after fusion (Brangwynne et al., 2011). Ribonucleoprotein (RNP) granules are biological liquid phase-separated structures of particular interest due to dynamic formation of puncta in cell development and granules in cellular stress (Bentmann et al., 2012; Ryu et al., 2014). Curiously, many of the proteins known to segregate into RNP granules contain repetitive putatively disordered domains (Kato et al., 2012). A subset of these proteins, including twenty-nine found in humans, contain a disordered domain rich in polar and aromatic residues and nearly devoid of aliphatic and charged amino acids, resembling the aggregation-prone glutamine/asparagine-rich domains of yeast prion proteins such as Sup35 (King et al., 2012). Hence these domains are also referred to as “prion-like domains”. Although RNA-binding protein prion-like domains have no homology nor sequence similarity to the human prion protein that forms infectious protein aggregates in new variant Creutzfeldt–Jakob disease and bovine spongiform encephalopathy (“mad cow disease”), many of these proteins have been identified as the major components of cytoplasmic inclusions associated with subtypes of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Drepper et al., 2011; Kim et al., 2013; Neumann et al., 2006; Zou et al., 2012). Furthermore, hereditary forms of ALS are linked to missense and deletion mutations within these low complexity domains, suggesting a mechanistic link between uncontrolled self-association mediated by RNA-binding protein low complexity domains and neurodegenerative disease (Ling et al., 2013).
Fused in Sarcoma (FUS) is an RNA-binding protein associated with protein aggregation in ALS and FTD as well as chromosomal translocation in certain sarcomas and leukemias. FUS plays a role in RNA processing and localizes both to cytoplasmic RNP granules and transcriptionally active nuclear puncta (Ryu et al., 2014; Schwartz et al., 2014; Yamaguchi and Kitajo, 2012; Yang et al., 2014). The low complexity N-terminal domain of FUS (defined here as residues 1-163, FUS LC) is a highly conserved prion-like domain composed primarily of serine, tyrosine, glycine, and glutamine (QGSY-rich) and contains only two charged residues. The twenty-four tyrosine residues are arranged in unusual repeats with a consensus sequence of [S/G]Y[S/G] often followed by one to three glutamine or proline residues. The LC domain mediates protein interactions in both nuclear assemblies and cytoplasmic RNP granules associated with processes spanning transcriptional regulation, pre-mRNA splicing, and mRNA transport and stability (Lagier-Tourenne et al., 2010; Yang et al., 2014). Although functionally essential, FUS LC drives the aggregation of FUS into protein inclusions in vitro and in models of ALS and FTD (Couthouis et al., 2011; Sun et al., 2011). Importantly, five missense or short deletion mutations located within the regions coding for FUS LC are linked to ALS (Belzil et al., 2009; Cruts et al., 2012; Ling et al., 2013; Ticozzi et al., 2009), including G156E which increases FUS aggregation propensity in vitro and in cell culture (Nomura et al., 2014). Additionally, more than a dozen related sarcomas and leukemias are caused by chromosomal translocations fusing the low complexity domain of FUS or that of two other human paralogs, RNA-binding protein EWS and TATA-binding protein-associated factor 2N (product of the TAF15 gene), to one of several DNA-binding domains, forming strong transcriptional activators (Riggi et al., 2007). Transcriptional activation by FUS LC may be due to the ability of phase-separated forms of FUS to bind the C-terminal tail of RNA polymerase II (Kwon et al., 2013; Schwartz et al., 2012).
Although the macroscopic behavior of protein-rich RNP granules and disease-associated inclusions has begun to be elucidated by microscopy both in vitro and in vivo (Brangwynne et al., 2009), the structural and mechanistic details of the protein domains as free monomers, in granule assemblies, and in disease aggregates remain uncharacterized. Therefore, in this work we seek to address several key questions: What conformations of FUS LC are sampled as a dispersed monomer in solution and in granule-associated forms? Are the contacts formed in the granule structured, adopting the highly ordered amyloid fibrillar form presumed to be present in hydrogel models of RNP granules, or do they remain highly disordered? Must FUS LC be in a fibrillar form in order to interact with the C-terminal domain of RNA polymerase II?
Here we use both microscopy and nuclear magnetic resonance (NMR) spectroscopy to characterize an in vitro granule model of the disordered domain of Fused in Sarcoma (FUS) and its interactions with the C-terminal domain of RNA polymerase II (CTD). We use differential interference contrast microscopy and fluorescence recovery after photobleaching techniques to observe the formation, diffusion and turnover of the phase-separated droplets structures. To understand the molecular details of FUS, we apply solution NMR spectroscopy to observe both the dispersed and phase-separated forms of FUS LC. By collapsing the spontaneously formed liquid droplets into a macroscopic phase using centrifugation, we directly characterize the structure and molecular motions of FUS LC within our in vitro models of FUS granules.
RESULTS
FUS N-terminal low complexity (LC) domain is structurally disordered
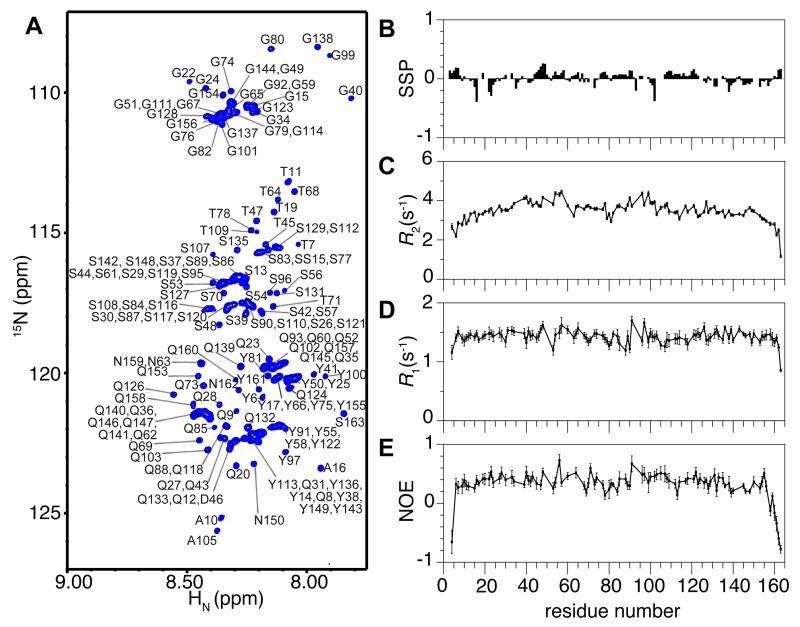
The N-terminal domain of FUS has been assumed to be intrinsically disordered based on its low complexity sequence. We analyzed soluble FUS LC (50 μM) using solution NMR spectroscopy in order to measure secondary structure population on a residue-by-residue level. Once the resonances of the FUS LC domain are assigned (i.e. matched to the specific atoms giving rise to the NMR signals) (Fig 1A), the 13C NMR chemical shifts available at each Cα and Cβ position are sensitive probes of secondary structure. With a well understood dependence on the backbone dihedral angles that change with secondary structure (Zhang et al., 2003), the difference in Cα and Cβ chemical shifts provides a sensitive estimate of the secondary structure propensity at each position. The lack of significant deviation between the observed shifts and those of a random coil reference demonstrate that the monomeric FUS LC domain is highly disordered, lacking significant population of α-helices or β-sheets (Figure S1A). These data can be combined with chemical shift values for backbone amide and carbonyl positions to generate residue-by-residue predictions of local secondary structure population using secondary structure propensity (SSP) (Marsh et al., 2006) and δ2D algorithms (Camilloni et al., 2012). The SSP scores are near 0 across the entire domain, indicating a lack of stable α-helical or β-sheet structure (Figure 1B). δ2D predicts predominant random coil structure with minor propensity for polyproline II helix across the domain and β-sheet structure at selected sites (Figure S1B). These data support our conclusion that the domain is predominantly disordered. Overlays of NMR spectra of FUS LC with full-length FUS indicate that chemical shift differences are small for the majority of residues in the LC domain (Figure S1C). Because NMR chemical shifts are sensitive reporters of structure, we conclude that the LC domain retains the same disordered structure in the monomeric full-length protein as in the isolated FUS LC domain. Although a previous study proposed that the monomeric LC domain populates significant (~49%) β-sheet structure based on decomposition of the circular dichroism (CD) spectrum using β-sheet, α-helix, and random coil reference spectra (Schwartz et al., 2013), these results were based on limited CD data (>200 nm) where spectral decomposition accuracy is poor. Indeed, the FUS LC domain CD spectra at our conditions replicate the previous CD data (Figure S1D) and share spectral features with other disordered, proline-containing domains (Hotta et al., 2014). Therefore, the previously predicted β-sheet content may simply be the result of the limited wavelength range leading to distorted secondary structure weighting.
Figure 1.
The FUS low complexity (LC, residues 1-163) domain is disordered as a monomer. (A) NMR spectrum (1H-15N heteronuclear single quantum coherence, HSQC) of the backbone amide region of FUS LC has a narrow chemical shift dispersion indicative of a disordered protein. Residue numbers are labeled (black). (B) Residue-specific secondary structure propensity (SSP) score of FUS LC indicate lack of local secondary structure formation. R2, R1, and (1H)-15N nuclear Overhauser effect (NOE) values (C, D, E, respectively) for the dispersed protein are consistent with disorder across the entire domain. Data are represented as mean +/− st dev. See also Figure S1.
Further evidence that the domain is uniformly disordered is provided by measurements of the protein backbone motions using NMR relaxation experiments (15N R2, 15N R1, heteronuclear NOE). Sensitive to motions on the picosecond-nanosecond timescale, the values of these dynamic observables as a function of residue position are highly uniform across FUS LC (Figure 1C-E). The lack of large variations in these dynamic observables due to slower rotational diffusion in structured areas indicates that no stable structured subregions of FUS LC are formed.
FUS LC stabilizes a liquid-liquid phase-separated state
While measuring the structural properties of the monomeric FUS LC, we noticed that our solutions of FUS turned opalescent or cloudy when placed on ice. This cloudiness rapidly dispersed at room temperature. Therefore, we investigated if the apparent opalescence is due to liquid-liquid phase separation of FUS proposed to be the underlying architecture of FUS-containing RNP granules and observed in vitro for other protein systems (Banjade and Rosen, 2014; Li et al., 2012; Nott et al., 2015). We used differential interference contrast microscopy to visualize the formation of liquid phase-separated FUS LC (Figure 2A), which resemble FUS-containing RNP granules previously observed in cells (Bentmann et al., 2012). These in vitro droplets flow, fuse and spontaneously return to a spherical shape (Movies S1 and S2) as observed for cytoplasmic RNP granules (Brangwynne et al., 2009).
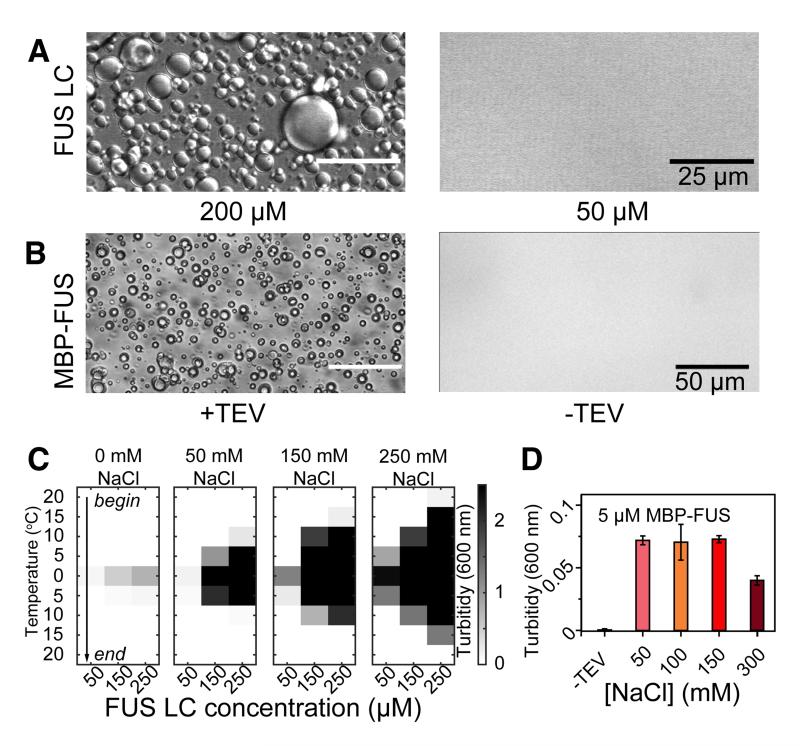
Figure 2.
The low complexity domain of FUS stabilizes phase separation that recruits the RNA polymerase II C-terminal domain. (A) Differential interference contrast (DIC) microscopy shows that the isolated FUS LC phase-separates at room temperature at high concentration (left) but not at low concentration (right). (B) 10 μM FUS full-length protein phase-separates only when liberated from an N-terminal maltose binding protein (MBP) fusion by addition of TEV protease (left) but not when incubated with buffer (right). (C) Phase separation of FUS LC as measured by optical density at 600 nm as a function of temperature and protein concentration, starting at 20 °C down to 0 °C. By increasing the temperature of the same samples back up to 20 °C, the phase-separated state dissipates, indicating the assembly is predominantly reversible. FUS LC phase separation is enhanced at increasing salt concentrations (left to right). (D) Phase separation of 5 μM full-length FUS after cleavage from a solubilizing MBP fusion (MBP-FUS) is unaffected by increasing NaCl concentration from 50 to 150 mM but reduced in 300 mM NaCl. Data are represented as triplicate mean +/− st dev. See also Figure S2.
Next we confirmed that the full-length FUS protein forms a liquid phase-separated state as observed for FUS LC. Full-length FUS (residues 1-526) is even more aggregation prone than the isolated LC domain (Sun et al., 2011) but is kept highly soluble by incorporation of an N-terminal fusion of maltose binding protein tag (MBP). Upon release of MBP by TEV cleavage leaving the native protein, samples as low as 2 μM full-length FUS assemble into an opalescent, phase-separated liquid (Figure 2B) resembling that formed by FUS LC. Phase separation proceeds rapidly after addition of TEV and the extent of phase-separation is greater at higher concentrations of full-length FUS (Figure S2A).
We then characterized the interactions that drive FUS liquid phase separation. FUS LC assembles more readily at higher protein concentrations (250 μM compared to 50 μM) (Figure 2C). Phase separation is enhanced at lower temperature but is rapidly reversible upon incubation at higher temperature (Figure 2C). We also tested the ability of increasing ionic strength to inhibit phase separation. Membraneless organelles of the highly charged low complexity domain of the nuage protein Ddx4 are stabilized by electrostatic (charge-charge or cation-pi) interactions between RG, FG, and E/D motifs and are dispersed at high ionic strength due to charge screening (Nott et al., 2015). In contrast to Ddx4, phase separation of FUS LC is in fact enhanced in the presence of sodium chloride (Figure 2C). Decreased solubility of FUS LC as a function of increasing salt concentration is likely due to salting out of the hydrophobic regions of the domains, notably the tyrosine residues. Interestingly, NMR spectra at 0 mM and 150 mM NaCl are nearly indistinguishable (Fig S2B), suggesting that FUS LC monomer structure is not altered substantially by salt.
We next interrogated the dependence of full-length FUS assembly on salt concentration. We find that the extent of phase separation is not significantly affected by an increase in NaCl from 50 mM to 150 mM but is reduced at the highest NaCl concentration tested (300 mM) (Figure 2D). The weak salt dependence of full-length FUS assembly suggests that, like for FUS LC phase separation, the interactions stabilizing full-length FUS liquid-liquid phase separation are not primarily electrostatic. However, full-length FUS assembly is not enhanced by increasing salt as observed for FUS LC and has a lower critical concentration for phase separation than FUS LC. Therefore, these data suggest that interactions outside of FUS LC contribute to phase separation of full-length FUS.
RNA enhances phase separation of FUS
Self-assembled forms of FUS are believed to be nucleated by RNA binding and subsequently recruit RNA polymerase II via direct LC domain interaction with tyrosine-containing CTD heptad repeats (Kwon et al., 2013; Schwartz et al., 2012). However, full-length FUS is a promiscuous RNA binder with little RNA sequence or structure preference (Wang et al., 2015) and the contribution of FUS LC to RNA binding is unknown. Therefore, we tested the ability of nonspecific eukaryotic RNA preparations (desalted solutions of torula yeast RNA) to bind to FUS LC and nucleate its assembly. Titration of up to 5 mg/ml of RNA (5:1 ratio of RNA:FUS LC by weight) did not result in phase separation of 50 μM FUS LC, conditions which resulted in phase separation of the prion-like domain of the related protein TDP-43 (data not shown). Additionally, no significant chemical shift or intensity differences in the NMR spectrum of FUS LC could be observed along this yeast RNA titration (Figure S3). Therefore, we find no evidence for direct interactions of RNA with monomers of FUS LC.
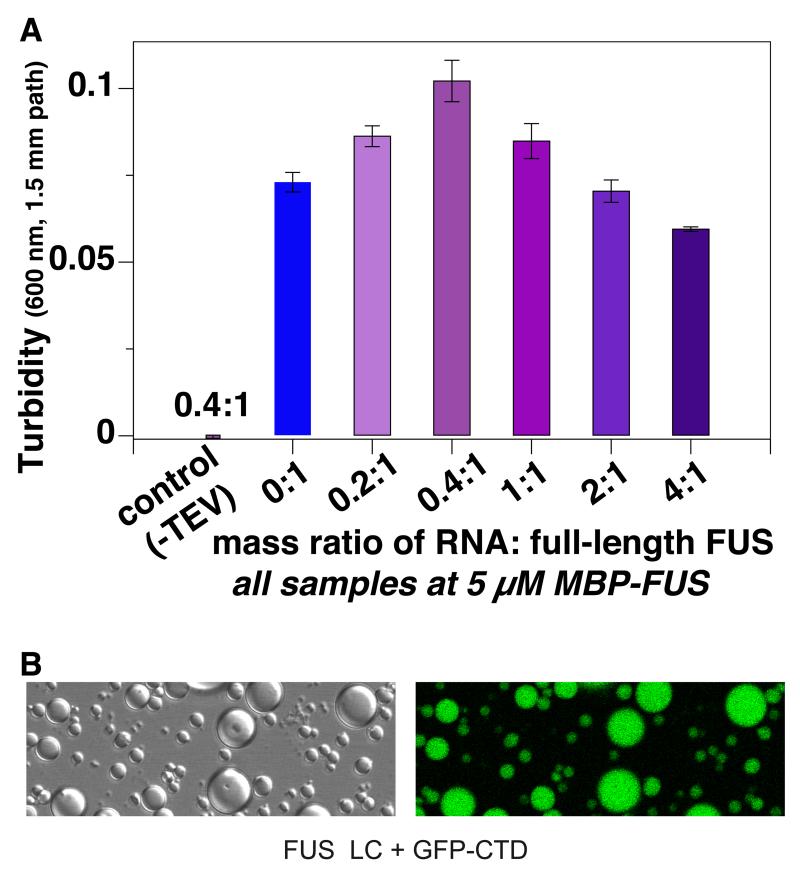
Because RNA enhances the formation of fibrous assemblies of full-length FUS presumably via binding to the RRM and zinc finger domains and RGG motifs (Schwartz et al., 2013), we tested if RNA can also enhance phase separation of full-length FUS. We observed the greatest extent of phase separation at an RNA:FUS ratio of 0.4:1 by mass (Fig 3A). The highest concentrations of RNA led to decreased phase separation, below that of FUS without RNA. These results mirror those of Cech and coworkers who observed that multiple FUS monomers can simultaneously bind substoichiometric amounts of RNA to induce formation of fibrous FUS assemblies, but that higher RNA amounts solubilize FUS (Schwartz et al., 2013). Taken together, these results support the view that the RNA-dependence of FUS phase-separation can be enhanced by FUS-RNA contacts but that the RNA contacts are not primarily mediated by the LC domain (Han et al., 2012; Kato et al., 2012; Kwon et al., 2013).
Figure 3.
Intermolecular interactions in phase separated FUS. (A) Phase separation of 5 μM FUS (after cleaving from MBP fusion by TEV protease, see Figure 2B) is enhanced by addition of torula yeast RNA up to a weight ratio of 0.4:1. (B) Fusions of GFP to the 26 degenerate heptad of the C-terminal domain of RNA polymerase II (GFP-CTD) localize to phase separated states of FUS LC. Data are represented as mean +/− st dev. See also Figure S3.
The C-terminal domain of RNA polymerase II nucleates FUS LC assembly and partitions into phase-separated state of FUS LC
Next, we tested if the 26 degenerate repeats of the C-terminal domain of the human RNA polymerase II (CTD) interact not only with fibrillar forms of FUS LC assembled into hydrogels as demonstrated by McKnight and coworkers but also with liquid phase-separated FUS LC. CTD expressed in E. coli is highly soluble and does not aggregate or phase separate at concentrations we tested (up to 500 μM). Surprisingly, addition of 50 μM CTD to samples of 350 μM FUS LC at 25 °C in salt-free buffer induces extensive phase separation as well as rapid aggregation and precipitation at conditions and concentrations where FUS LC and CTD alone are both monomeric and soluble. Lowering the amount of FUS LC to 50 μM equimolar concentrations resulted in stable liquid phase separation (droplet formation) at room temperature. We confirmed this interaction using fluorescence microscopy of CTD incorporating an N-terminal GFP fusion and observe that GFP-CTD localizes to FUS LC phase-separated states (Figure 3B). Taken together these data demonstrate that the C-terminal domain of RNA polymerase II can directly interact with FUS low complexity domain phase-separated states and can nucleate their assembly.
To further interrogate the interaction between FUS and CTD, we measured the effect of FUS LC phase separation on the NMR spectrum of CTD (Figure S3B) by comparing samples of 50 μM 15N CTD with and without 50 μM FUS LC in salt-free 20 mM MES pH 5.5 at 25 °C which spontaneously phase separate into a suspension of micron sized droplets (see above). Except for small chemical shift differences in 3 residues (Fig S3C-D), addition of FUS LC and the subsequent phase separation caused no changes in the NMR spectrum of 15N CTD. However, peak intensities in two-dimensional NMR spectra show that the CTD signal is nearly uniformly attenuated to ~75% of the control sample (Figure S3E). This loss of signal suggests that monomers of the CTD are incorporated into, or interact with, the FUS LC phase separated state. In other words, FUS LC may populate a conformation or state with rapid transverse relaxation (see below) (i.e. lifetime line broadening) (Fawzi et al., 2011) or intermediate chemical shift timescale conformational exchange either within the phase separated state or at the phase interface, in analogy to the “dark state” of α-synuclein at the lipid membrane surface (Bodner et al., 2009). In either case, resonance positions of dispersed CTD are not significantly affected by FUS LC droplets, supporting a model where CTD binds FUS LC primarily in its phase separated state and not as dispersed monomers.
Diffusion kinetics within FUS LC liquid phase separated state
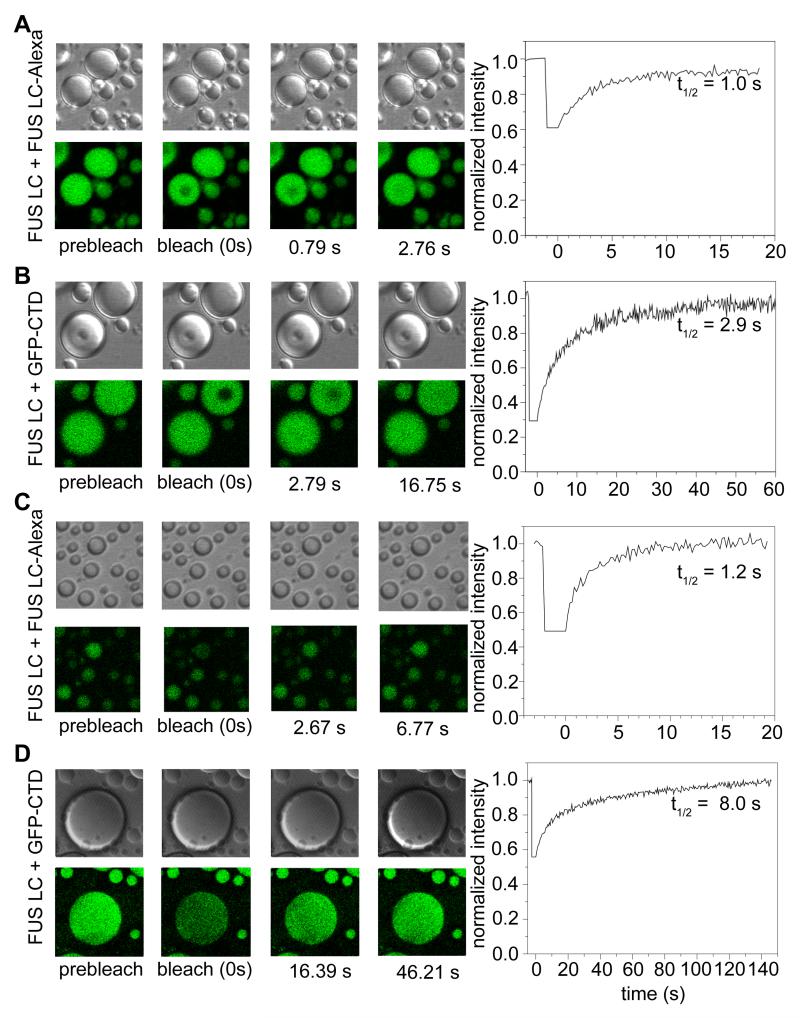
Flow and fusing of liquid droplets implies rapid rearrangement and diffusion of the structural components within the liquid phase-separated state. To determine the diffusion kinetics within the FUS LC liquid phase-separated state, we used fluorescence recovery after photobleaching (FRAP) of small portions of droplets formed in 150 mM NaCl doped with 0.01% FUS LC labeled with Alexa-488 at a central engineered cysteine position (S86C). When we bleached a 2.5 μm region of ~8 μm droplets, we observed rapid signal recovery with a halftime of 1.1 ± 0.1 s (Figure 4A). These values can be used to estimate the translational diffusion coefficient (Poitevin and Wahl, 1988) for FUS LC within the phase separated state at 0.4 μm2/s. This value is similar to that observed for protein components within germline P granules in live cells, implying the presence of intermolecular interactions that slow translational diffusion with an effective apparent viscosity three orders of magnitude higher than that of water (Brangwynne et al., 2009). Interestingly, although the qualitative behaviors are similar in FUS LC phase separated states formed in salt-free buffer, diffusion appears faster by nearly an order of magnitude (Figure S4).
Figure 4.
Rapid diffusion and turnover of FUS LC droplet contents. Example differential interference contrast and fluorescence images (left panels) and FRAP timecourses (right) of partial droplet photobleaching experiments used to measure diffusion constants within the droplets of FUS LC + FUS LC-Alexa (A) and FUS LC + GFP-CTD (B). Half times for the selected timecourse are labeled in the recovery curves. Example differential interference contrast and fluorescence images (left panels) and FRAP timecourses (right) of full droplet photobleaching experiments used to measure rate of exchange between droplet and dispersed states of FUS LC + FUS LC-Alexa (C) and FUS LC + GFP-CTD (D). See also Figure S4.
We also confirmed that the slow translational diffusion within the FUS LC phase separated state applies to the RNA polymerase II C-terminal domain (CTD) partitioned into the FUS LC phase separated state. Using GFP-tagged CTD, FRAP measurements of recovery halftime using the same parameters as above result in a slower halftime of 3.5 ± 0.5 s. Therefore, GFP-CTD diffuses at a rate of 0.1 μm2/s, slower than that of phase separated FUS LC and consistent with the larger size of the GFP-tagged construct (Figure 4B). Taken together, the slow diffusion rates of both FUS LC and GFP-CTD demonstrate hindered translational motion.
Binding and turnover kinetics of FUS LC liquid droplet contents
For liquid phase separated assemblies to be reversible structures and functional reaction centers, molecules must be able to cross the phase boundary to enter and exit. Therefore, we elucidated the molecular kinetics of the incorporation of FUS into and out of the phase separated state. We used FRAP experiments to measure the rate of turnover of droplet contents. Photobleaching of entire smaller (~5 μm) FUS LC droplets results in >95% signal recovery and follows an exponential profile with 1.1 ± 0.1 s half time (Figure 4C). We also observed rapid and complete fluorescence recovery after full droplet photobleach for the binding of GFP-CTD to FUS LC droplets (Figure 4D) with a half time of 7 ± 0.4 s for large ~50 μm droplets. These data suggest that monomers of both FUS LC and GFP-CTD incorporate into the phase separated state and that the droplet contents are completely turned over. Importantly, although the analyzed droplets remained effectively static by DIC microscopy on this timescale (Figure 4A-D, upper panels), the contents are entirely replaced. Therefore, we find no evidence for stable aggregates or static structure on the 10 s or slower timescale.
Visualizing the structural details of liquid phase separated FUS
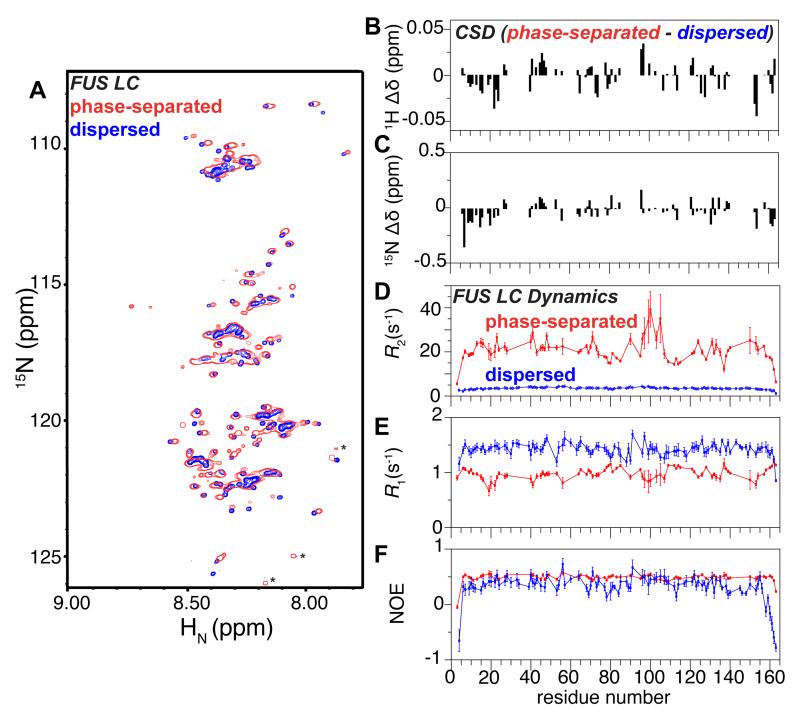
As for CTD (Figure S3), we did not observe significant NMR chemical shift deviations for FUS LC in the presence of suspended FUS LC droplets. However, given that we observed FUS LC droplet fusion in our microscopy experiments, we hypothesized that it would be possible to collapse the spontaneously formed droplets into a single macroscopic phase-separated FUS LC phase. Using incubation on ice to increase the driving force for droplet formation followed by centrifugation to fuse the droplets due to their higher density, our 15 ml samples of 1 mM FUS LC phase separated to form a ~400 μl viscous, protein-dense phase stable for weeks at room temperature. FUS LC concentration in the phase is approximately 7 mM (120 mg/ml FUS LC) as determined by spectrophotometry. We then obtained a residue-specific NMR characterization of FUS LC structure and local motions within the phase separated state. Despite the high viscosity, FUS LC yielded observable but broad resonances in the 1H 15N HSQC, suggesting that some local motions are retained in the phase separated state (Fig S5A). Importantly, the high concentration and hence excellent NMR signal-to-noise enabled aggressive resonance line sharpening of the 1H-15N HSQC spectra, resulting in many resolved peaks (Figure 5A, S5A-C). Surprisingly, overlays of these spectra with those of the monomeric, dispersed FUS LC (50 μM) show high similarity, demonstrating that the global disordered structure formed by monomeric FUS LC is retained in the phase-separated state. We assigned 77 resonances by transfer from the known dispersed phase assignments and by three-dimensional NMR experiments (see Experimental Procedures). The available chemical shift deviations, backbone 1H and 15N, are distributed across the entire FUS LC domain (Figure 5B, C). As anticipated from similar 1H 15N HSQC spectra, phase-separated FUS LC gives rise to 1H 13C HSQC spectra highly similar to dispersed FUS LC (Fig S5D-F), although broad resonances preclude atomic level analysis of these 1H and 13C shifts. Taken together, the lack of large chemical shift differences between the phase-separated and dispersed phases that would be expected for significant conformational changes indicate that the local structure of the protein remains disordered in the phase-separated state.
Figure 5.
Phase separated FUS LC remains disordered and dynamic. (A) 1H 15N HSQC spectrum of the phase separated FUS LC is highly similar to the monomer in the dispersed state solution, suggesting a similar overall conformation. Asterisks denote presumed C-termini of small quantities of impurities or truncations where favorable relaxation properties amplify their signal compared to FUS LC resonances. 1H (B) and 15N (C) chemical shift differences between FUS LC in the phase-separated and dispersed states indicate that small chemical shift differences are distributed across the chain. R2, R1, and (1H)-15N nuclear Overhauser effect (NOE) values (D, E, F, respectively) for phase separated FUS LC (red) are consistent with slowed motion compared to the dispersed phase (blue, repeated from Figure 1) but demonstrate that FUS LC retains reorientational mobility. Data are represented as mean +/− st dev. See also Figure S5.
Despite this structural similarity between FUS LC in dispersed and liquid phase-separated states, local motions of the peptide backbone as measured by 15N spin relaxation experiments show evidence for restricted mobility compared to that of the monomeric, dispersed protein. R2 values range from 15 to 35 s−1 away from the termini (compared to 3 to 5 s−1 in the dispersed state) (Figure 5D), R1 values from 0.75 to 1.2 s−1 (compared to 1.3 to 1.7 s−1) (Figure 5E), and heteronuclear NOE values of ~0.5 (compared to 0.2 to 0.6) (Figure 5F). Although the complex motions of an intrinsically disordered domain undergoing transient intermolecular interactions complicate interpretation of relaxation parameters using simple motional models, the differences in all three spin relaxation parameters between dispersed and phase-separated states all point to slower average reorientational motion in the protein-dense liquid phase-separated state. However, the wide range of 15N R2, particularly the sharply peaked values between residues 95 to 105, suggest that conformational exchange on the intermediate chemical shift timescale intermediate or line broadening due to transient population of a high molecular weight state (Fawzi et al., 2011) may contribute to the observed transverse relaxation. However, backbone Carr-Purcell-Meiboom-Gill 15N relaxation dispersion experiments did not show significant dispersion >3 s−1 at any resolved positions over the range from 50 Hz to 1000 Hz (data not shown), though these experiments are primarily sensitive to exchange only on the ~100 μs to ms timescale. Therefore, further work is necessary to understand the detailed contacts and conformational exchange for phase separated FUS.
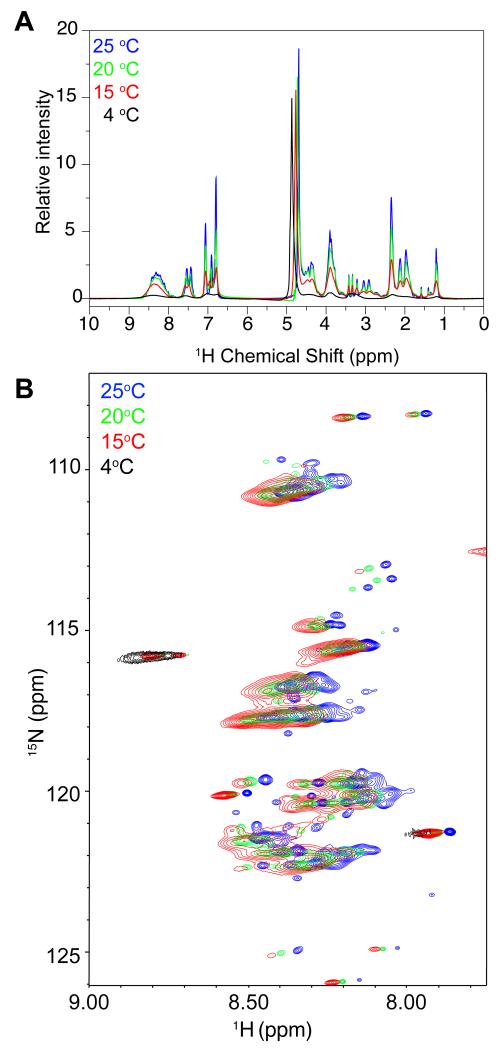
Intriguingly, the local dynamics of FUS LC in the liquid phase-separated state appear to be highly temperature sensitive. We observed a dramatic decrease in signal intensity as temperature decreases to 4 °C (Figure 6). Although this broadening may arise from chemical exchange on the intermediate chemical shift timescale, this line broadening is also concomitant with an apparent solid/liquid phase transition in the protein-dense phase itself observed by a reversible white/opaque transition. These data suggest that liquid-like granules can have temperature-dependent changes in physical properties.
Figure 6.
NMR resonance intensities arising from phase separated FUS LC are highly temperature dependent. One dimensional 1H (A) and 1H 15N HSQC (B) experiments show decreased signal intensity and increased line broadening as temperature decreases.
DISCUSSION
FUS droplets: an in vitro model of self-assembled FUS granules and puncta
Liquid phase-separated states of biomolecules represent one strategy a cell can take to create sharp local concentration differences in order to spatially confine cellular components and control reaction rates without a membrane compartment (Brangwynne, 2013). Here we show that the low complexity domain of FUS (FUS LC) is structurally disordered both as a monomer and within a liquid phase-separated state that directly recruits the C-terminal repeat domain of RNA polymerase II.
FUS LC contains the features necessary for liquid phase separation: highly valent but individually weak interactions. In the case of FUS LC, the degenerate tyrosine/glutamine containing repeats provide the high valency interaction sites needed to create a macroscopic protein-dense/water phase (Kato et al., 2012). On their own, each interaction between tyrosine repeats is weak, but by tethering multiple sites together on a single chain, a dynamic contact network formed. Our data suggest that hydrophobic interactions stabilize FUS LC phase separation, as would be predicted by the requirement for tyrosine residues for FUS LC hydrogel formation (Kato et al., 2012). FUS LC contacts are distinct from the opposite charge and cation-pi (RG/RGG to FG) interactions that stabilize the phase separation of the nuage/chromatoid body associated protein Ddx4 (Nott et al., 2015). Liquid phase separation of full-length FUS may also be mediated in part by additional interactions, possibly involving RGG motifs present in full-length FUS which may also bind RNA quadruplex junctions (Phan et al., 2011) and Tudor domains (Chen et al., 2011). However, our data suggest that electrostatic interactions are not the primary drivers for phase separation in full-length FUS.
The LC domain is required for punctate distribution and RNA polymerase II colocalization in the nucleus (Yang et al., 2014). However, nucleic acid species are known components of FUS granules and nuclear foci (Bentmann et al., 2012; Daigle et al., 2013; Schwartz et al., 2014; Yang et al., 2014). In parallel to reports on the formation of FUS fibrous assembly (Schwartz et al., 2013), RNA species can increase full-length FUS phase separation extent (Figure 3) likely via interactions outside the LC domain (Figure S3).
The liquid-liquid phase-separated state of FUS LC shares some characteristics with the FUS LC hydrogels described by McKnight and coworkers. First, the phase-separated form is stabilized by low temperature and high salt. Second, these liquid phase-separated states also concentrate the CTD of RNA polymerase II. However, liquid phase separated FUS is distinct from FUS hydrogels. Although the contacts stabilizing the two forms may share tyrosine repeat interactions, the persistence time of the contacts in the liquid phase separated state are much shorter, consistent with disorder observed in liquids. FUS hydrogels are fibrillar in structure with stable molecular contacts while the liquid phase separated states show fast rearrangement as observed by FRAP. Phase-separated droplets of FUS form and disperse rapidly (seconds to minutes) upon change of temperature. In addition, the distribution of intermolecular contacts is different in the fibrils and the droplets. Fibrillar structure in the hydrogels implies a regular and repeating pattern of contacts, most likely in-register parallel β-sheet structure common to amyloid fibrils of disordered protein domains including yeast prions (Engel et al., 2011; Shewmaker et al., 2009; Shewmaker et al., 2006). In FUS LC liquid phase separated states, dynamic reorganization is suggested by local mobility observed in NMR relaxation experiments. Therefore, the protein-protein contacts within the phase separated state do not have fibrillar order but rather form non-specific contacts where each repeat interacts with any of the other tyrosine repeats, in analogy to the disordered combinatorial binding leading to phase separation by chains of folded protein interaction domains (Li et al., 2012). Future work testing the effect of FUS LC ALS-associated mutants on phase separation and the propensity to convert from the liquid states characterized here into static inclusions could lead to insight into the mechanism of formation of FUS protein inclusions in ALS.
Phase separated FUS LC structure and dynamics
The direct observation of a liquid phase-separated state by NMR offers the opportunity to directly determine the structure and motions of the protein architecture underlying granules or membraneless organelles. We observed that the constituent proteins in the FUS LC liquid phase separated state remain disordered with only small changes in the 1HN, 15N, and 13C chemical shifts, suggesting that the structure in the phase-separated state is largely the same as in the dispersed monomer. However, changes in all three spin relaxation experiments point to slowed motions at the backbone positions compared to the monomer in the dispersed state, reflecting direct intermolecular interaction or crowding from high protein concentration. The distribution of these slower motions across the FUS LC chain suggests broad involvement of the whole LC domain in phase separation.
Together, our data suggest that while FUS LC global motion (translational diffusion as monitored by FRAP) is dramatically slowed, local structure and motions are largely preserved. These data are consistent with a model where transient (“flickering”) intermolecular contacts are formed along FUS LC chain, analogous to those identified for intramolecular interactions in yeast prion proteins of similar sequence composition (Mukhopadhyay et al., 2007). These interactions are sufficiently short-lived and of low enough population that the individual segments of the chain are on average free to reorient. Therefore, this work supports a view of membraneless organelles as fundamentally dynamic structures made up of intrinsically disordered proteins, exhibiting both diffusional motion of protein components and local motion of the protein chain. We find no evidence for static assemblies within the phase-separated state nor significant structural or conformational changes upon incorporation into this phase. Hence, we suggest FUS LC self assembly as well as recruitment of RNA polymerase II CTD are mediated by the same weak, multivalent interactions, rather than by adoption of specific conformers upon phase separation. Similar phase separation has been observed for a variety of sequences, including highly charged proteins such as Ddx4 which bear little sequence composition resemblance to FUS. Therefore, further work is necessary to determine if retention of disordered structure within liquid phase-separated states is a general feature of membraneless organelles.
Phase separated FUS LC avidly recruits RNA polymerase II CTD. Phosphorylation of CTD correlates with its ability to bind FUS and localize in nuclear FUS granules (Kwon et al., 2013; Schwartz et al., 2012; Schwartz et al., 2014), but the mechanisms that control the association and the rules governing cross-incorporation of proteins into membraneless organelles have yet to be determined. Therefore, future studies aimed at cracking the code connecting the sequence of amino acids in low complexity repeats to their interactions may enable prediction of the molecular assembly of heterogeneous mixtures of low complexity domain proteins.
These studies using NMR spectroscopy enable a residue-by-residue view of a model of FUS granules. However, the studies described here were performed in vitro on purified FUS and lack the complexity of components and regulation present for granules formed in vivo. First, the liquid phase-separated state characterized with residue-level resolution here is formed by the isolated FUS LC. Although the domain readily phase separates with the same features as the full-length protein, additional interactions (e.g. via RGG domains or with RNA, Figure 2D) may play roles in full-length FUS phase separation. Second, the recombinant proteins explored here are unmodified. Posttranslational modification of FUS may alter the critical concentration for self-assembly or change its affinity for phase-separated states. The in vitro phase-separation mediated by the highly charged disordered domains of Ddx4 is inhibited by asymmetric dimethylation (aDMA) of arginine residues modified by protein arginine methyltransferase 1 (PRMT1) in vivo (Nott et al., 2015). Similar regulation of liquid phase separation by arginine methylation may also occur for FUS, whose stress granule localization and inclusion formation is altered by PRMT1 activity (Tradewell et al., 2012; Yamaguchi and Kitajo, 2012). Additionally, FUS LC phosphorylation by the phosphatidylinositol 3-kinase-related kinases is associated with shuttling FUS from the nucleus to the cytoplasm (Deng et al., 2014; Gardiner et al., 2008). Therefore experiments examining the ability of FUS phosphorylation to act as the on/off switch for liquid phase separation are ongoing in our laboratory.
EXPERIMENTAL PROCEDURES
Protein expression of FUS and RNA polymerase II CTD were performed in E. coli cultures from constructs described in detail in Supplemental Experimental Procedures Section 1, including an MBP fused form of FUS (MBP-FUS) created in a previously described vector (Peti and Page, 2007). Samples for NMR spectroscopy were produced in M9 minimal media with 15N and 13C precursors as appropriate for the experiment.
Resonance assignment for FUS LC (conditions: 50 μM U-13C/U-15N FUS LC in 20mM MES pH 5.5, 10% 2H2O, 25 °C) was achieved by a combination of three dimensional 1H/13C/15N experiments with parameters appropriate for disordered proteins as described in the Supplemental Experimental Procedures Section 2. pH 5.5 was selected for optimal spectra with reduced amide proton water exchange. Assignment of phase separated FUS LC (see Supplemental Experimental Procedures, Section 3 for sample preparation) was achieved with HSQC-NOESY-HSQC incorporating two 15N dimensions appropriate for a high concentration phase containing 1H/15N labeled protein (see details in Supplemental Experimental Procedures, Section 4). 1H 13C (real time) HSQC spectra of phase separated FUS LC were measured at - natural 13C isotopic abundance (U-15N only) and compared to constant time (T=2/JCC=54 ms) HSQC spectra of U-13C/15N dispersed phase FUS LC samples. Dynamics of samples of U-15N FUS LC were monitored by 15N R1, 15N R2, and heteronuclear NOE experiments (details in Supplemental Experimental Procedures). Secondary structure propensity (SSP) was calculated using SSP v1.0 (The Hospital for Sick Children) with default parameters including automated chemical shift re-referencing (Marsh et al., 2006). δ2D secondary structure decomposition predictions were performed with default parameters (Camilloni et al., 2012). Far UV CD experiments for global structure of FUS LC were performed using default parameters on a Jasco J-815 spectropolarimeter.
The phase diagram for FUS LC phase separation was determined using 750 μl samples of FUS LC at concentrations from 50 to 250 μM (conditions: 0 to 300 mM NaCl, 50mM MES pH 5.5) using absorbance at 600 nm in 1 cm plastic cuvette. Full-length FUS phase separation was monitored using 50 μl samples of MBP-FUS at 5 μM in triplicate. RNA preparations were made from torula yeast RNA type VI (Sigma #R6625). See supplemental experimental procedures for detailed sample preparation.
Imaging of FUS phase separation was performed using 20 μl samples on glass coverslips with differential interference contrast (DIC) on an Axiovert 200M microscope (Zeiss) with a 40× NA objective to examine the formation and fusion of liquid-liquid phase-separated states of FUS LC domain using 200 μM and 50 μM (control) FUS LC (conditions: 150 mM NaCl, 50mM MES pH 5.5) or MBP-FUS at 10 μM in the presence of TEV protease or control (protease buffer only). Fluorescence recovery after photobleaching (FRAP) experiments were performed on an LSM 710 confocal microscope (Zeiss) with a 40× W NA objective using a 488-nm laser line in conjunction with differential interference contrast microscopy. The FUS LC S86C-Alexa 488 or GFP-CTD were doped into 200 μM FUS LC samples and subsequently bleached using ten iterative pulses (total time ~2s) with full laser power. The circular bleached spots were 2.5 μm diameter of ~8-10 μm droplets for partial droplet photobleach or the entire size for full droplet bleaching. Fluorescence recovery was imaged at 2% laser intensity appropriate for both GFP and Alexa-488 fluorophores. Images were processed using ImageJ. The recovery curve, halftime recovery, and immobile species were calculated using the FRAP_Calculator v3 macro for ImageJ (National Institutes of Health). Translational diffusion coefficients were calculated from partial droplet FRAP experiments using the relationship for circular bleach regions (Poitevin and Wahl, 1988; Yguerabide et al., 1982).
Supplementary Material
Highlights.
N-terminal prion-like low complexity domain of FUS (FUS LC) is disordered in solution
FUS LC phase separates to recruit RNA polymerase II C-terminal domain
Droplet granules models are viscous but dynamic assemblies lacking static structures
Local structure of phase-separated FUS LC remains disordered
ACKNOWLEDGMENTS
We thank Jamie Fried, Kohana Leuba, Rute Silva, and Louis Taylor for early work on FUS self-association and Steven McKnight and Masato Kato for helpful conversations and kindly providing GFP-CTD. We thank Frank Shewmaker, Michael Clarkson, Alex Conicella, and Jinfa Ying for helpful suggestions and Julie Forman-Kay, Cliff Brangwynne, Mike Rosen, and Tony Hyman for stimulating conversations. We thank Geoff Williams, the Leduc Bioimaging Facility at Brown University, Gary Wessel, Mamiko Yajima, and Eric Gustafson for assistance in microscopy. Research reported here was supported in part by Medical Research Grant #20133966 from the Rhode Island Foundation (to N.L.F.), a Richard B. Salomon Faculty Research Award from Brown University (to N.L.F.), and pilot project awards (to N.L.F.) as part of Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P20 GM 103430-13 and P20 GM 104937-06. This research is based in part upon work conducted using the Rhode Island NSF/EPSCoR Proteomics Shared Resource Facility, which is supported in part by the National Science Foundation EPSCoR Grant No. 1004057, National Institutes of Health Grant No. 1S10RR020923, a Rhode Island Science and Technology Advisory Council grant and the Division of Biology and Medicine, Brown University. This research is based in part on data obtained at the Brown University Structural Biology Core Facility, which is supported by the Division of Biology and Medicine, Brown University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014;3 doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Valdmanis PN, Dion PA, Daoud H, Kabashi E, Noreau A, Gauthier J, Hince P, Desjarlais A, Bouchard JP, et al. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73:1176–1179. doi: 10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, Haass C. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) The Journal of biological chemistry. 2012;287:23079–23094. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodner CR, Dobson CM, Bax A. Multiple tight phospholipid-binding modes of alpha-synuclein revealed by solution NMR spectroscopy. Journal of molecular biology. 2009;390:775–790. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol. 2013;203:875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilloni C, De Simone A, Vranken WF, Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51:2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle JG, Lanson N.a., Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Human Molecular Genetics. 2013;22:1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, Ito D, Murray ME, Dickson DW, Seyfried NT, et al. FUS is Phosphorylated by DNA-PK and Accumulates in the Cytoplasm after DNA Damage. J Neurosci. 2014;34:7802–7813. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drepper C, Herrmann T, Wessig C, Beck M, Sendtner M. C-terminal FUS/TLS mutations in familial and sporadic ALS in Germany. Neurobiol Aging. 2011;32:548, e541–544. doi: 10.1016/j.neurobiolaging.2009.11.017. [DOI] [PubMed] [Google Scholar]
- Engel A, Shewmaker F, Edskes HK, Dyda F, Wickner RB. Amyloid of the Candida albicans Ure2p prion domain is infectious and has an in-register parallel beta-sheet structure. Biochemistry. 2011;50:5971–5978. doi: 10.1021/bi200142x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Atomic-resolution dynamics on the surface of amyloid protofibrils probed by solution NMR. Nature. 2011;480:268–272. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner M, Toth R, Vandermoere F, Morrice N.a., Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. The Biochemical Journal. 2008;415:297–307. doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hotta K, Ranganathan S, Liu R, Wu F, Machiyama H, Gao R, Hirata H, Soni N, Ohe T, Hogue CW, et al. Biophysical properties of intrinsically disordered p130Cas substrate domain--implication in mechanosensing. PLoS Comput Biol. 2014;10:e1003532. doi: 10.1371/journal.pcbi.1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S-C, Polymenidou M, Cleveland DW. Converging Mechanisms in ALS and FTD: Disrupted RNA and Protein Homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci U S A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nomura T, Watanabe S, Kaneko K, Yamanaka K, Nukina N, Furukawa Y. Intranuclear aggregation of mutant FUS/TLS as a molecular pathomechanism of amyotrophic lateral sclerosis. J Biol Chem. 2014;289:1192–1202. doi: 10.1074/jbc.M113.516492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peti W, Page R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein expression and purification. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitevin E, Wahl P. Study of the translational diffusion of macromolecules in beads of gel chromatography by the FRAP method. Biophys Chem. 1988;31:247–258. doi: 10.1016/0301-4622(88)80030-9. [DOI] [PubMed] [Google Scholar]
- Riggi N, Cironi L, Suva ML, Stamenkovic I. Sarcomas: genetics, signalling, and cellular origins. Part 1: The fellowship of TET. J Pathol. 2007;213:4–20. doi: 10.1002/path.2209. [DOI] [PubMed] [Google Scholar]
- Ryu HH, Jun MH, Min KJ, Jang DJ, Lee YS, Kim HK, Lee JA. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging. 2014;35:2822–2831. doi: 10.1016/j.neurobiolaging.2014.07.026. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes & Development. 2012;26:2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Podell ER, Han SS, Berry JD, Eggan KC, Cech TR. FUS is sequestered in nuclear aggregates in ALS patient fibroblasts. Mol Biol Cell. 2014;25:2571–2578. doi: 10.1091/mbc.E14-05-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–925. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two prion variants of Sup35p have in-register parallel beta-sheet structures, independent of hydration. Biochemistry. 2009;48:5074–5082. doi: 10.1021/bi900345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci U S A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS biology. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N, Silani V, LeClerc AL, Keagle P, Gellera C, Ratti A, Taroni F, Kwiatkowski TJ, Jr., McKenna-Yasek DM, Sapp PC, et al. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology. 2009;73:1180–1185. doi: 10.1212/WNL.0b013e3181bbff05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky JA, Wright PE. Assemblages: functional units formed by cellular phase separation. J Cell Biol. 2014;206:579–588. doi: 10.1083/jcb.201404124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tradewell ML, Yu Z, Tibshirani M, Boulanger MC, Durham HD, Richard S. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21:136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- Wang X, Schwartz JC, Cech TR. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Kitajo K. The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS One. 2012;7:e49267. doi: 10.1371/journal.pone.0049267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Gal J, Chen J, Zhu H. Self-assembled FUS binds active chromatin and regulates gene transcription. Proc Natl Acad Sci U S A. 2014;111:17809–17814. doi: 10.1073/pnas.1414004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yguerabide J, Schmidt JA, Yguerabide EE. Lateral mobility in membranes as detected by fluorescence recovery after photobleaching. Biophys J. 1982;40:69–75. doi: 10.1016/S0006-3495(82)84459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- Zou ZY, Cui LY, Sun Q, Li XG, Liu MS, Xu Y, Zhou Y, Yang XZ. De novo FUS gene mutations are associated with juvenile-onset sporadic amyotrophic lateral sclerosis in China. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.