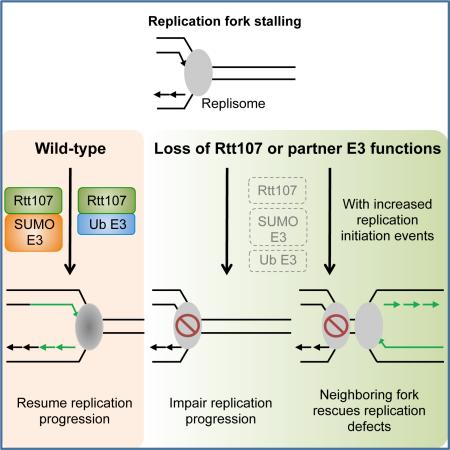
Summary
Elucidating the individual and collaborative functions of genome maintenance factors is critical for understanding how genome duplication is achieved. Here we investigate a conserved scaffold in budding yeast, Rtt107, and its three partners: a SUMO E3 complex, a ubiquitin E3 complex, and Slx4. Biochemical and genetic findings show that Rtt107 interacts separately with these partners and contributes to their individual functions, including a role in replisome sumoylation. We also provide evidence that Rtt107 associates with replisome components, and both itself and its associated E3s are important for replicating regions far from initiation sites. Corroborating these results, replication defects due to Rtt107 loss and genotoxic sensitivities in mutants of Rtt107 and its associated E3s are rescued by increasing replication initiation events through mutating two master repressors of late origins, Mrc1 and Mec1. These findings suggest that Rtt107 functions as a multi-functional platform to support replication progression with its partner E3 enzymes.
Keywords: Replication stress, Homologous recombination, Genome replication, Protein sumoylation, Histone modification
Graphical Abstract
Introduction
Exogenous and endogenous sources of stress generate many obstacles to the progress of DNA replication. To cope with this, cells have evolved networks of proteins that facilitate genome duplication. These include DNA metabolizing enzymes that remove impediments or enable repair synthesis, protein modifying enzymes that change substrate properties, and scaffolds that establish specific associations favorable for overcoming replication obstacles. Among them, some key factors play roles in multiple processes through collaborations with diverse regulatory proteins. Unraveling how these multi-tasking factors function in specific contexts during replication has been challenging due to the complexities of their interactions, but is vital to our understanding of genome maintenance and associated diseases.
Here we examine a group of such factors in the model system budding yeast, focusing on the central member Rtt107 (or Esc4). A key feature of this conserved scaffold protein is its six BRCT domains, a structure known to support multiple protein-protein interactions (Leung and Glover, 2011). The first four BRCTs of Rtt107, which are devoid of phospho-binding residues, associate with three genome maintenance factors: the Rtt101 cullin ubiquitin ligase complex, the Smc5/6 SUMO ligase complex, and another scaffold Slx4 (Chin et al., 2006; Ohouo et al., 2010; Roberts et al., 2006). Rtt107 and its three partners are individually required for genome duplication and cell viability under replication stress. Rtt107 is implicated in histone dynamics, with the Rtt101 ligase (or E3) composed of cullin Rtt101, linker Mms1, and substrate adaptor Mms22 (referred to as Rtt101Mms22) (Collins et al., 2007; Zaidi et al., 2008). Rtt101Mms22 is implicated in ubiquitinating acetylated histone H3 to facilitate nucleosome assembly during replication (Han et al., 2013). Both Rtt107 and Rtt101Mms22 mutants relieve growth inhibition caused by persistent histone H3 acetylation, suggesting a common role in histone regulation (Collins et al., 2007). Additionally, Rtt107 is implicated with Slx4 in reducing the Rad53 kinase-mediated DNA damage checkpoint response by counteracting the function of a Rad53-activator, Rad9; a regulation dubbed ‘checkpoint dampening’ (Ohouo et al., 2013; Cussiol et al., 2015).
Currently, it is unclear whether the checkpoint regulatory function of Rtt107 is shared by Rtt101Mms22, or how the two known Rtt107 functions are linked to those of the other Rtt107 interactor, the octameric Smc5/6 SUMO E3 complex that is implicated in replication fork regulation using both SUMO-dependent and -independent means (Ampatzidou et al., 2006; Branzei et al., 2006; Chen et al., 2009; Xue et al., 2014). The lack of understanding of these issues is partly due to insufficient information on how these proteins associate with each other; specifically, whether Rtt107 and its partner proteins form a mega-complex or exist as several independent complexes in cells. Additional gaps in our understanding of this group of proteins include their exact roles in DNA replication and how these roles are linked with other regulatory elements in the replication program.
Insights into these issues will be invaluable for understanding how Rtt107 and its partner proteins facilitate replication. Here, we address multiple facets of this question using a combination of biochemical and genetic approaches, and physical measurements of DNA replication through two-dimensional agarose gel (2D gel) electrophoresis, replication-coupled single-stranded DNA (ssDNA) profiling, and pulsed field gel electrophoresis (PFGE). Our results show that Rtt107 forms three distinct complexes in which the specific Rtt107 partner defines the function of each complex. We reveal a role for Rtt107 in collaborating with the Smc5/6 SUMO ligase to influence the sumoylation of specific replisome components, and that Rtt107 physically interacts with these sumoylation substrates. We also demonstrate that a main function of Rtt107 and its E3 partners during S phase lies in promoting replication progression. Interestingly, the lack of Rtt107 and its E3 partners can be partially compensated for by mutating two master repressors of late origins. Additional lines of evidence suggest that this compensation stems from improving chromosomal duplication through increased replication initiation events. These results support an intriguing model whereby Rtt107 collaborates with its partner E3 enzymes to assist replisomes in the synthesis of large replicons.
Results
Rtt107 interacts separately with Slx4, the Rtt101Mms22 complex, and the Smc5/6 complex
To understand how Rtt107 and its associated proteins affect replication, we first clarified if Rtt107 and its three binding partners formed distinct complexes or one mega-complex. These two interaction modes make different predictions. The ‘distinct complexes’ model predicts that: 1) the three Rtt107 partners do not interact amongst themselves, though each associates with Rtt107; 2) the interaction between Rtt107 and each of its partners is independent of the other two partners; 3) each complex exerts specific functions not shared by the others. Opposite observations would support the ‘mega-complex’ model. We tested these predictions to clarify how these proteins interact in vivo.
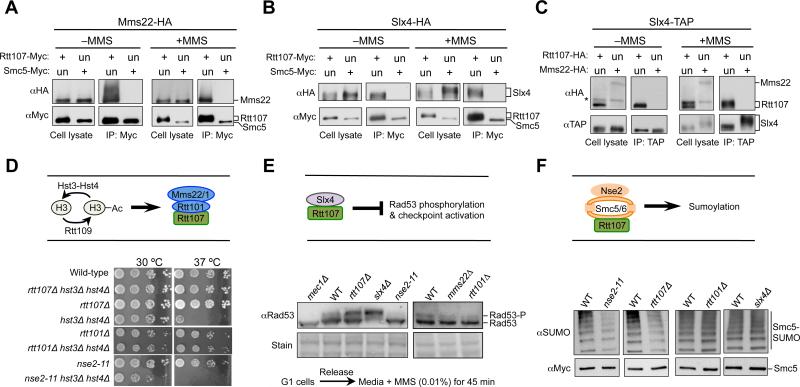
First, we used co-immunoprecipitation (co-IP) assays to query the association among the Rtt107 partners in the absence and presence of the replication stress agent methyl methanesulfonate (MMS). To this end, each of the examined proteins was endogenously tagged with an epitope at its C-terminal end. While Rtt107 co-purified with Mms22 either in the presence or absence of MMS, Smc5 did not (Figure 1A). Similarly, Rtt107, but not Smc5, associated with Slx4 in both conditions (Figure 1B). These results suggest that Smc5 is not part of a complex containing Mms22 or Slx4. In addition, the latter two proteins did not interact with each other, as Slx4 co-purified with Rtt107, but not Mms22 in both conditions (Figure 1C). Similarly, yeast two hybrid assay (Y2H) assays showed that while Slx4 and subunits of the Smc5/6 and the Rtt101Mms22 complexes interacted with Rtt107, no interactions among these Rtt107 binding partners were detected (Figure S1A-S1B). Thus, both co-IP and Y2H findings show that the three Rtt107 partners do not interact amongst themselves.
Figure 1. Rtt107 forms separate complexes with its three partners.
(A-C) The three Rtt107 binding partners do not associate among themselves. Cells were grown in the presence or absence of 0.03% MMS (2 h). Immunoprecipitation of Myc-tagged Rtt107, but not Smc5, co-purifies Mms22 (A) or Slx4 (B). Immunoprecipitation of TAP-tagged Slx4 co-purifies with Rtt107, but not Mms22 (C). An upward shift of Rtt107 and Slx4 bands in MMS-treated samples is due to phosphorylation (Flott and Rouse, 2005; Roberts et al., 2006; Rouse, 2004). The asterisk denotes a background band. Un: untagged; +: tagged versions of proteins. Note that longer exposures of western blots confirm that Smc5 and Mms22, which are less abundant than Rtt107, do not associate with Slx4 nor between themselves.
(D) Mutants of Rtt101 or Rtt107, but not Nse2, rescue the temperature sensitivity of hst3Δ hst4Δ cells. Ten-fold serial dilutions of cells were spotted. H3: newly synthesized histone H3.
(E) Mutations of Slx4 or Rtt107, but not Rtt101Mms22 and Nse2, increase the levels of Rad53 phosphorylation. -P: phosphorylated form. Equal loading is shown by stain.
(F) nse2-11 or rtt107Δ, but not slx4Δ or rtt101Δ, reduces the sumoylation levels of Smc5. Cells were treated with 0.03% MMS for 2 h, and Smc5-Myc was immunoprecipitated and analyzed by Western blot using a SUMO-specific antibody (top) that recognizes the sumoylated forms, and a Myc antibody that detects the unmodified forms (bottom).
See also Figure S1.
Rtt107 interaction with each partner is independent of the other two partners
To test the second prediction described above, we examined whether Rtt107 associates with each partner in the absence of the other two partners using co-IP before and after MMS treatment. We found that the Rtt107-Smc5/6 interaction was sustained when MMS22 or SLX4 was deleted (Figure S1C-S1D). Similarly, the Rtt107-Slx4 interaction was not diminished when either the Mms1 subunit of Rtt101Mms22 or the Nse5 subunit of Smc5/6 was absent (Figure S1E-S1F), and the Rtt107-Mms22 interaction was sustained in the absence of Slx4 or Nse5 (Figure S1G-S1H). These results show that the interaction between Rtt107 and each of its partners is independent of the other two partners.
Each Rtt107-containing complex affects a unique protein modification pathway
We moved on to examine the third prediction of the ‘distinct complexes’ model by using functional readouts. Rtt107 and Rtt101Mms22 act downstream of Rtt109, which acetylates newly synthesized histone H3 during S phase (schematized in Figure 1D, top). This H3 marker has to be deacetylated by Hst3 and Hst4 later in the cell cycle, otherwise cells endure persistent DNA damage that is particularly deleterious at higher temperatures (e.g. Maas et al., 2006, Celic et al., 2006). Such defects caused by Hst3 and Hst4 loss are suppressed by preventing the modified form of H3 from being incorporated into chromatin through removal of Rtt101Mms22 or Rtt107 (Collins et al., 2007). We confirmed that rtt107Δ and rtt101Δ suppressed the temperature sensitivity of cells lacking Hst3 and Hst4, but found that mutants of the Smc5/6 complex did not confer suppression (Figure 1D, bottom). In fact, mutation of the SUMO ligase subunit of the complex, Nse2/Mms21 (Nse2 hereafter), sensitized hst3Δ hst4Δ (Figure 1D, bottom), and a similar effect has also been reported for slx4Δ (Celic et al., 2008). These findings suggest that Smc5/6 and Slx4 do not collaborate with Rtt107-Rtt101Mms22 in histone acetylation-related regulation.
Rtt107 and Slx4 reduce Rad53 checkpoint activation by counteracting the Rad53 activator Rad9 (schematized in Figure 1E, top). We recapitulated the finding that removal of Rtt107 or Slx4 increases Rad53 phosphorylation levels, but found that cells lacking Nse2 SUMO ligase activity (nse2-11), Mms22 or Rtt101 all exhibited lower Rad53 phosphorylation levels than wild-type cells (Figure 1E, bottom). Thus, Rtt101Mms22 and Smc5/6 likely do not share functions with Rtt107-Slx4 in DNA damage checkpoint dampening.
As indicated above, the biological effects linking Rtt107 to Smc5/6 during replication stress remain unclear. As the latter is a SUMO E3 (schematized in Figure 1F, top), we examined the effect of Rtt107 on sumoylation of an Nse2 substrate, Smc5 (Zhao and Blobel, 2005). Both rtt107Δ and nse2 SUMO ligase mutants exhibited lower levels of Smc5 sumoylation, while rtt101Δ and slx4Δ sustained wild-type levels of Smc5 sumoylation (Figure 1F, bottom). These results suggest that Rtt107-Smc5/6, but not the other two Rtt107 partners, influence Smc5 sumoylation.
In summary, our biochemical, Y2H, and genetic results support the ‘distinct complexes’ model and strongly suggest that Rtt107 serves as a scaffold for three independent complexes, each with a unique partner that influences a different protein modification pathway.
Smc5/6 acts independently of Rtt107 in regulating recombination intermediate metabolism
After clarifying the manner by which Rtt107 interacts with its partners, we delved deeper to address the functional relationship between Rtt107 and its least understood partner, Smc5/6. Our findings presented above suggested a role for Rtt107 in the SUMO-dependent functions of Smc5/6 (Figure 1F). We asked whether Rtt107 is also involved in the SUMO-independent functions of Smc5/6, such as its interaction with and inhibition of the Mph1 DNA helicase that promotes recombination (Chen et al., 2009; Xue et al., 2014). Below, we present biochemical and genetic evidence showing that Rtt107 is not involved in this function of Smc5/6.
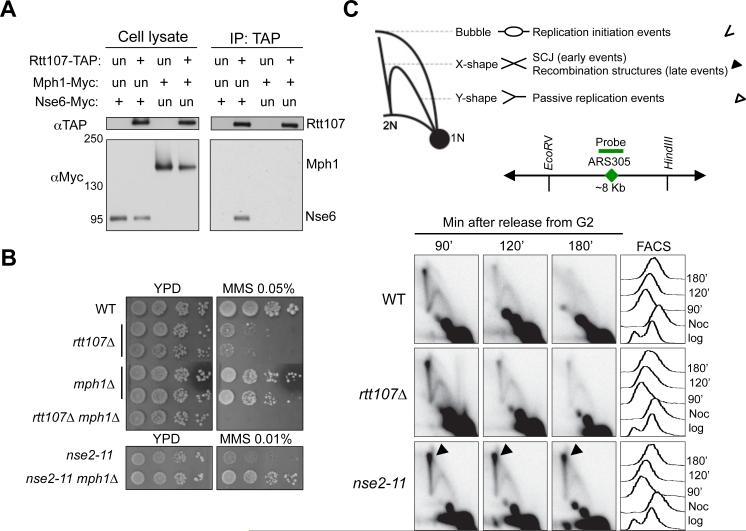
First, Rtt107 co-IPed with Nse6, but not Mph1, suggesting that Rtt107 does not associate with Mph1 (Figure 2A). Second, mph1Δ did not reduce the Rtt107-Smc5/6 interaction; conversely, rtt107Δ did not alter the Mph1-Smc5/6 interaction (Figure S2A-S2B). Thus, Rtt107 and Mph1 independently associate with Smc5/6, and do not affect each other's interaction with Smc5/6. Third, while mph1Δ suppressed the MMS sensitivity of nse2 mutants by reducing toxic recombination structures (Chen et al., 2009; Xue et al., 2014), it exacerbated that of rtt107Δ cells (Figure 2B). Fourth, while nse2 mutants increased recombination intermediate levels compared to wild-type cells as expected (Chen et al., 2009), rtt107Δ did not, as shown by 2D gel analysis of the region surrounding early origin ARS305 (Figure 2C).
Figure 2. Smc5/6 regulates Mph1 and recombination independently of Rtt107.
(A) Rtt107 interacts with the Nse6 subunit of the Smc5/6 complex, but not the DNA helicase Mph1. Experiments were carried out and results are presented as in Figure 1A.
(B) mph1Δ suppresses the MMS sensitivity of the nse2 mutant but not that of rtt107Δ. Experiments were done as in Figure 1D, except that cells were spotted on normal media (YPD) or YPD containing the indicated amount of MMS.
(C) Unlike nse2 mutants, rtt107Δ cells do not accumulate recombination intermediates. Top, schematic that depicts DNA structures visualized by 2D gel. Symbols for each structure are indicated at the right. Bubble DNA represents replication initiation structures; Y-shaped structures represent passive replication intermediates; X-shaped DNA are either sister chromatin junctions (SCJs) that form at an early time point, or other recombination structures that appear later in S phase. Middle, schematic for ARS305, probe position, and the restriction enzyme sites used for 2D gel analysis. Bottom, 2D gel and FACS results. Cells were arrested in nocodazole and released into medium containing 0.03% MMS, and samples were examined at the indicated time points.
See also Figure S2.
These findings suggest that Rtt107 is not involved in this SUMO-independent function of Smc5/6. We conclude that while Smc5/6 and Rtt107 interact and similarly influence Smc5 sumoylation, each also associates with other partners to perform additional functions. Partially overlapping functions have also been reported for Rtt107 and Rtt101Mms22, as well as for Rtt107 and Slx4. For example, Rtt101Mms22 interacts with the replication protein Ctf4 independently of Rtt107 (Mimura et al., 2010). Thus, both Rtt107 and its three partners are multi-functional proteins collaborating with different factors, and their functions overlap only partially (schematized in Figure S2C).
Rtt107 promotes replisome sumoylation and interacts with replisome components
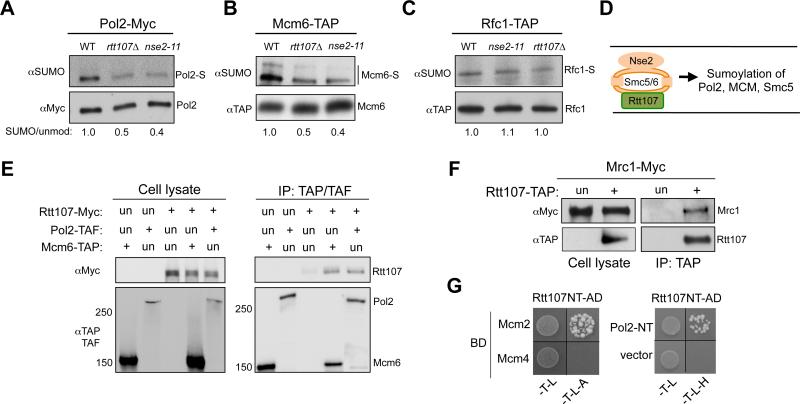
As our data showed that Rtt107 and Smc5/6 shared SUMO-dependent, but not SUMO-independent roles, we asked whether Rtt107 influences Smc5/6-mediated sumoylation events directly relevant to replication. As both Rtt107 and Smc5/6 are present at stalled replication forks (Lindroos et al., 2006; Roberts et al., 2008), we examined replisome components known to be sumoylated under replication stress (Cremona et al., 2012). We found that sumoylation of both DNA polymerase epsilon subunit Pol2 and replicative helicase subunit Mcm6 was reduced in nse2 SUMO ligase mutants (Figure 3A-3B). Importantly, a similar reduction was detected in rtt107Δ cells (Figure 3A-3B). These effects were specific, as sumoylation of another replication protein, Rfc1, was unaffected in either rtt107Δ or nse2 mutants (Figure 3C). In addition, rtt107Δ did not alter the sumoylation of other Nse2 substrates, such as cohesin and condensin subunits (Figure S2D). Moreover, the other Smc5/6 binding partner, Mph1, did not affect Pol2 and Smc5 sumoylation (Figure S2E). These findings suggest that Rtt107 is a unique Smc5/6 interactor that promotes its sumoylation of specific substrates, including replisome components and Smc5 (schematized in Figure 3D).
Figure 3. Rtt107 associates with replisome components and affects the sumoylation of Pol2 and Mcm6.
(A-C) Rtt107 and Nse2 mutations reduce the sumoylation levels of specific replisome components. rtt107Δ and nse2-11 reduced the sumoylation levels of Pol2 (A) and Mcm6 (B), but not Rfc1 (C). Experiments were done as in Figure 1F. The relative ratios of sumoylated forms to the unmodified protein forms are indicated.
(D) Schematic of the shared function of Smc5/6 and Rtt107 suggested by this study.
(E-F) Rtt107 associates with replisome components Mcm6, Pol2, and Mrc1. G1 cells were released into medium containing MMS for 90 min. Experimental procedures and labels are the same as those in Figure 1A. (E) Immunoprecipitation of Pol2 or Mcm6 co-purifies Rtt107. (F) Immunoprecipitation of Rtt107 co-purifies Mrc1.
(G) Rtt107 associates with Mcm2 and Pol2 in Y2H assay. Reporter strains contained the DNA binding domain (BD) and DNA activation domain (AD) constructs. Cells were spotted onto SC-Trp-Leu (-T-L) media for plasmid selection and on SC-Trp-Leu-Ade (-T-L-A) or SC –Trp-Leu-His (-T-L-H) media to detect reporter activation. Rtt107 NT: 1-660 amino acids; Pol2 NT: 1-1264 amino acids.
Considering that Rtt107 is a scaffold protein, we asked whether its observed effects on Pol2 or Mcm6 sumoylation are mediated by interactions with these proteins. Indeed, immunoprecipitation of Pol2 or Mcm6 co-purified Rtt107 (Figure 3E). Rtt107 also co-immunoprecipitated with another replisome component, Mrc1 (Figure 3F). Finally, Y2H studies using Rtt107 as prey against replisome components showed that Rtt107 interacted with Mcm2 and the N-terminal domain of Pol2 (Figure 3G). These results suggest a potential mechanism for Rtt107 in promoting sumoylation of replisome components by bridging the interaction between these substrates and the Smc5/6 E3 ligase complex.
Rtt107 loss impairs replication progression without affecting the origin firing program
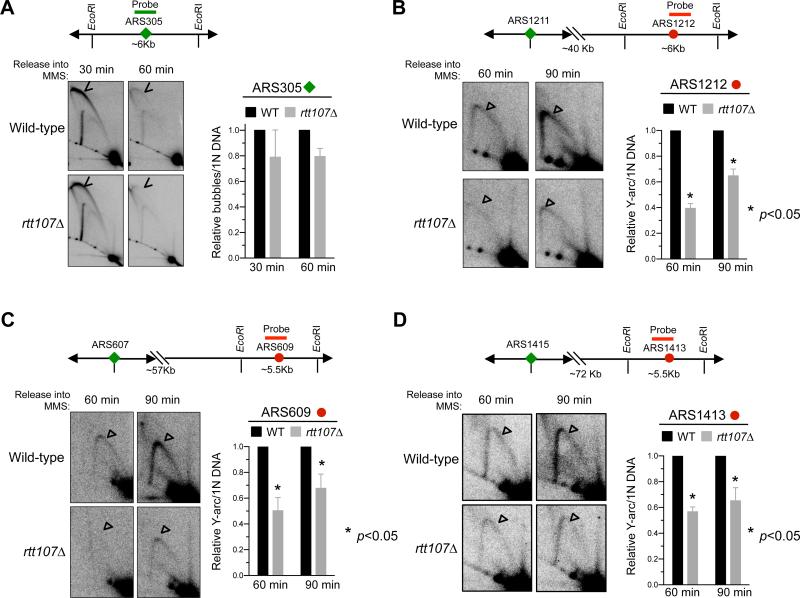
Our results hitherto show that Rtt107 supports multiple functions defined by each of its three partners, including the sumoylation of Smc5 and replisome subunits. We then addressed how these functions of Rtt107 collectively influence replication under stress. After exposure to MMS, rtt107Δ cells are known to reduce chromosome entry into pulsed field gels, an indicator of incomplete replication (Rouse, 2004). Here we used 2D gel and genome-wide methods to show that rtt107Δ defects lie in replication progression, but not initiation.
We synchronized cells in G1 and released them into S phase in the presence of MMS (Figure S3A). Consistent with previous reports, wild-type cells showed robust firing at early origin ARS305 at 30 minutes after release, and no firing at three late origins, namely ARS1212, ARS609, and ARS1413 (Figure 4A, S3B). This is indicated by the presence and absence of bubble replication initiation structures, respectively. The origin firing patterns of rtt107Δ cells closely resembled that of wild-type cells at this time point (Figure 4A, S3B), suggesting that Rtt107 is not required for regulating initiation at these examined origins. We extended this finding genome-wide by measuring replication fork-associated ssDNA formation (Feng et al., 2006). As shown by a representative 550 kb region of chromosome XV, the profiles of wild-type and rtt107Δ cells closely resembled each other (Figure S3C). This result and our 2D gel findings indicate that Rtt107 is dispensable for the origin firing program.
Figure 4. 2D gel analyses reveal differential effects of rtt107Δ in early versus late replicated regions.
Samples were taken after G1 cells were released into media containing MMS. Representative 2D gel results for examining a ~6 kb region containing the early origin ARS305 (A) or later origins as indicated (B-D). Graph in (A) depicts the relative ratio of bubble DNA (arrowheads) versus 1N DNA and those in (B-D) depict those of Y-shaped DNA (unfilled triangles) versus 1N DNA. In each case, two different spore clones were tested, and means and SEM are plotted.
See also Figure S3.
Next, we examined the above-mentioned loci in mid to late S phase. At both 60 and 90 minutes, rtt107Δ cells showed reduced levels of Y-shaped replication intermediates compared with wild-type cells at the three late origins, located > 40 kb from the nearest activated early origins (Figure 4B-4D). The finding of a normal firing program but defective replication at regions far from early origins in rtt107Δ cells points to the importance of Rtt107 in fork progression under MMS conditions. As the three examined late origins are located on chromosomes of different lengths, this role likely reflects a general function not restricted to chromosome size. We note that the replication progression defect of rtt107Δ differs from that of DNA repair mutants, which show defective replication within 5-10 kb regions from fired origins (Vazquez et al. 2008). Consistent with this, rtt107Δ strongly sensitized mutants of DNA repair pathways in replication stress conditions (Figure S4). Both observations support a DNA repair-independent role for Rtt107 in replication progression.
Removal of a potent late origin inhibitor Mrc1 alters the replication program in rtt107Δ cells
After establishing that Rtt107 promotes replication progression, we examined how rtt107 mutants respond to global de-repression of late origin firing. In principle, de-repression of late origin firing reduces average replicon size, and thus should benefit cells defective in progression. Though evidence supporting this concept is seen in higher eukaryotes (reviewed in McIntosh and Blow 2012), none has been found in budding yeast. Such evidence in yeast would support a conserved relationship between replication progression and late origin firing, and would further define the roles for Rtt107.
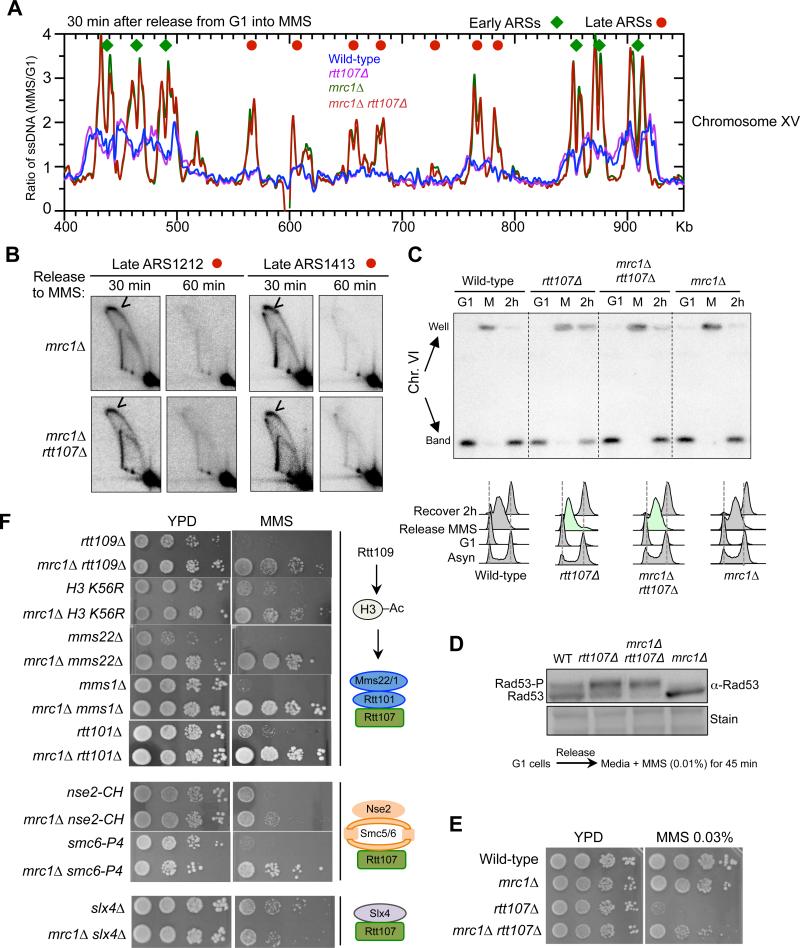
We first queried mrc1Δ, as it causes the largest increase in the numbers of fired late origins amongst a collection of mutants in hydroxyurea (HU) conditions (Crabbe et al., 2010). Our replication fork-associated ssDNA analyses showed a similar effect for mrc1Δ in MMS conditions, with 174 late origins activated (Figure 5A, representative profile). Average profiles were generated by meta-analysis in areas spanning 20 kb on both sides of early or late origins (Figure S5A). As seen earlier, global increases in origin firing correlate with slower fork movement, likely because cellular levels of certain replication factors and DNA precursors are limited (e.g. Poli et al., 2012; Zhong et al., 2013). This replication profiling of MMS-treated mrc1Δ cells shows that Mrc1 loss results in potent late origin firing, accompanied by reduced fork movement.
Figure 5. mrc1Δ increases global late origin firing and rescues defects due to the loss of Rtt107 and its interacting E3s.
(A) Replication fork-associated ssDNA profiles show that mrc1Δ increases late origin firing. The profiles of a segment of Chromosome XV are shown as representative images of the genome-wide data. G1 cells were released into media containing MMS for 30 min. Relative amount of ssDNA was determined as the ratio of fluorescent signal from the 30 min S phase sample to that from the G1 sample. The early and late origins are indicated by green diamonds and red dots, respectively.
(B) 2D gel results verify that mrc1Δ leads to firing at two late origins, ARS1212 and ARS1413, in MMS. Experimental procedures and labels are identical to those in Figure 4.
(C) mrc1Δ increases chromosomal replication in rtt107Δ cells. G1 cells were released into media containing 0.03% MMS for 1 h (Release MMS; M), then recovered in normal media for 2 h (Recover 2 h; 2h). FACS shows the bulk DNA synthesis of each strain, with mrc1Δ rtt107Δ exhibiting faster progression than rtt107Δ cells (green). Chromosomes separated by PFGE were examined by Southern blot using a probe specific to Chr. VI. Quantification of the results seen Figure. S5B.
(D) mrc1Δ does not reduce Rad53 phosphorylation levels in rtt107Δ cells. Experiments were done as in Figure 1E.
(E-F) mrc1Δ suppresses the MMS sensitivity of cells defective in Rtt107 or its interacting E3s, but not Slx4. MMS sensitivities were examined as in Figure 2B. (E) Diagrams on the right summarize the relevant complexes and proteins examined. MMS concentrations for panels containing the following mutants are rtt109Δ: 0.02%; H3 K56R and mms22Δ: 0.01%; rtt101Δ, mms1Δ and nse2-CH: 0.03%; smc6-P4: 0.003%, and slx4Δ: 0.05%.
See also Figure S5.
The profiles of mrc1Δ rtt107Δ resembled that of mrc1Δ (Figure 5A, S5A). De-repression of two late origins in these mutants were verified by 2D gel analyses, as bubble-shaped replication firing structures were detected 30 minutes after release from G1 and largely diminished by 60 minutes (Figure 5B). Taken together, these results show that the effect of mrc1Δ on the replication program is maintained in rtt107Δ cells under MMS conditions.
Mrc1 loss rescues replication defects of rtt107Δ cells without correcting checkpoint levels
We proceeded to examine whether mrc1Δ-mediated increase of late origin firing benefits chromosomal replication in rtt107Δ cells. G1 arrested cells were released into MMS-containing media followed by recovery in normal media. PFGE and Southern blotting were performed to examine the replication of specific chromosomes. As shown in Figure 5C and S5B, nearly 95% of chromosome VI entered the gel (indicative of replication completion) in wild-type and mrc1Δ cells, but only ~60% did so in rtt107Δ cells after a 2 hr recovery. This number increased to ~80% in mrc1Δ rtt107Δ cells (Figure 5C, S5B). Similar findings were made for a large chromosome (Figure S5C). Thus, mrc1Δ improves chromosomal replication in rtt107Δ cells.
We excluded the possibility that the mrc1Δ suppression was due to resetting Rad53 phosphorylation levels, as Mrc1 removal in rtt107Δ cells did not reduce Rad53 phosphorylation levels when G1 cells were released into media containing MMS, though mrc1Δ alone exhibitedlower levels of Rad53 phosphorylation as expected (Figure 5D, S5D) (Alcasabas et al, 2001). This finding is consistent with the notion that although Mrc1 is a Rad53 activator, only the other Rad53 activator, Rad9, is involved in Rad53 hyper-activation caused by rtt107Δ and slx4Δ (Ohouo et al., 2013; Cussiol et al., 2015).
Mrc1 loss in rtt107Δ cells increases survival of replication stress
Correlating with improved chromosomal replication in rtt107Δ cells, mrc1Δ increased rtt107Δ cell survival of MMS on plates or during acute treatment (Figure 5E, S5E). Similar suppression was seen toward the replication stress agent, Camptothecin (CPT) (Figure S5F). Incomplete mrc1Δ-mediated suppression is consistent with its partial suppression of chromosomal replication in rtt107Δ cells. Regardless, that the mrc1Δ-mediated changes to rtt107Δ replication dynamics correlate with improved cell viability contrasts with observations that mrc1Δ exacerbates the phenotypes of many other replication mutants (e.g. Collins et al., 2007; Costanzo et al., 2010). In addition, it differs from the negative interactions observed between rtt107Δ and DNA repair mutants (Figure S4). Moreover, although mrc1Δ slows replication fork speed, other mutants with slower fork movement either negatively affected rtt107Δ or showed no suppression, suggesting that this feature unlikely accounts for the suppression of rtt107Δ defects(Figure S5G). These controls highlight the unique positive interaction between mrc1Δ and rtt107Δ.
Mrc1 loss benefits mutants of Rtt107-associated E3s that impair replication progression
Considering that Rtt107 functions with its three partners during replication, we examined which Rtt107 partner(s) shares a similar genetic relationship with Mrc1 and affects replication of regions far from origins. We found that mrc1Δ suppressed the MMS sensitivities of cells lacking any of the subunits of the Rtt101Mms22 complex, and mutants of Rtt109 and H3K56 acetylation site (Figure 5F). Additionally, mrc1Δ suppressed the MMS sensitivity of two mutants of the Smc5/6 complex (Figure 5F). In contrast, mrc1Δ enhanced the MMS sensitivity of slx4Δ cells (Figure 5F). We note that the differential genetic interactions of rtt107Δ and slx4Δ with mrc1Δ agrees with a recent report (Leung et al., 2014) and is consistent with our conclusion that the mrc1Δ suppression is not due to resetting of Rad53 activation levels (Figure 5D, S5D). In line with these genetic interactions, 2D gel analyses of three regions located > 40 kb away from origins showed that, like rtt107Δ, smc6 mutation or rtt101Δ also diminished replication at these regions (Figure S5H). These results suggest that Rtt107 and associated E3 complexes support replication progression, and that the loss of their functions can be compensated for by increased global origin firing.
A separation-of-function allele of Mec1 de-represses late origins in rtt107Δ cells
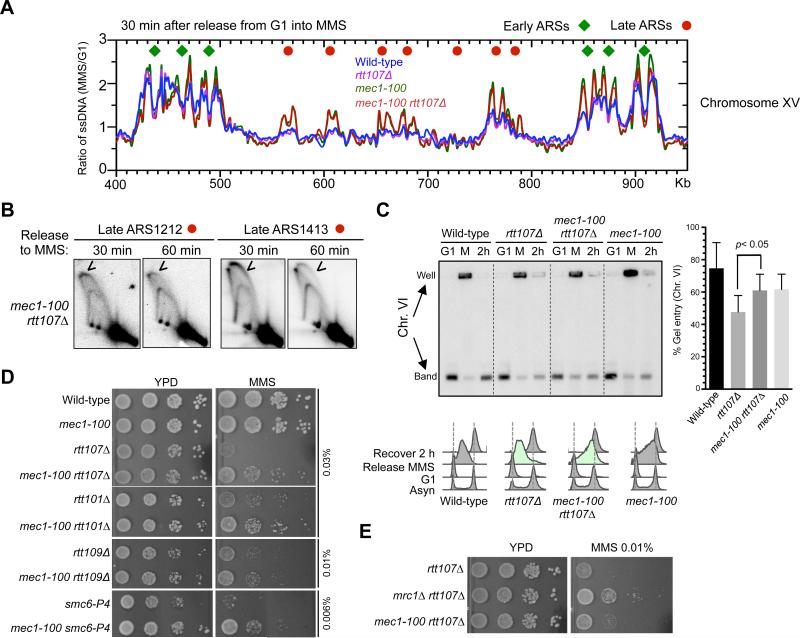
To further test the above notion, we asked if increasing late origin firing by another means can benefit cells lacking Rtt107 and its associated E3s. Among the mutants known to de-repress late origins, mrc1Δ and mec1-100 have the strongest effects (Crabbe et al., 2010). mec1-100 is a separation-of-function allele of the Mec1 checkpoint kinase, and under MMS conditions, it is known to specifically defective in repressing late origin firing (Paciotti et al., 2001; Zhong et al., 2013, Tercero et al., 2003), thus it is a useful allele for testing the above idea.
mec1-100 indeed exhibited a similar, though less potent, effect on the de-repression of late origins compared to mrc1Δ (Figure 6A, S6A). It has a slight reduction in fork movement, consistent with a previous study (Zhong et al., 2013). These features of mec1-100 were retained in rtt107Δ cells (Figure 6A, S6A). 2D gel analyses at two late origins, ARS1212 and ARS1413, confirmed origin firing in the rtt107Δ mec1-100 double mutant (Figure 6B). Thus, mec1-100 has similar, though less potent, effects as mrc1Δ on origin firing in rtt107Δ cells.
Figure 6. mec1-100 suppresses defects of cells lacking Rtt107 or its associated E3s.
(A) mec1-100 de-represses late origins in rtt107Δ cells. Replication fork-associated ssDNA profiles of a segment of Chromosome XV are shown as in Figure 5A.
(B) 2D gel analyses confirm that mec1-100 leads to firing of late origins, ARS1212 and ARS1413, in rtt107Δ background. Experimental procedures and labels are identical to those in Figure 5B.
(C) mec1-100 increases chromosomal replication in rtt107Δ cells. Experimental scheme and results are presented as in Figure 5C and S5B. Quantification of results from 3 trials is plotted with means and SD.
(D) mec1-100 rescues the MMS sensitivity of rtt107Δ and related E3 mutants.
(E) Side-by-side comparison shows that mec1-100 is less effective than mrc1Δ in suppressing the MMS sensitivity of rtt107Δ cells. Experiments were done as in Figure 5E, except that plates were grown for a shorter time to better reveal differences between mec1-100 and mrc1Δ suppression.
See also Figure S6.
mec1-100 rescues defects in cells lacking Rtt107 and its partner E3s
In a similar experimental scheme as described above (Figure 5C), PFGE analyses of a medium- and a large-sized chromosome showed that mec1-100 enhanced replication in rtt107Δ cells, albeit to a lesser degree than mrc1Δ (Figure 6C, S6B). Consistently, mec1-100 partially rescued the MMS and CPT sensitivities of rtt107Δ cells (Figure 6D, S6C). Like mrc1Δ, mec1-100 also improved the MMS resistance of Rtt101Mms22, Rtt109, and Smc5/6 mutants (Figure 6D). The magnitude of suppression by mec1-100 was lower than that of mrc1Δ (Figure 6E), commensurate with their effects on the replication program. The fact that phenotypic suppression correlates in degree with changes in replication dynamics supports a model that additional origin firing benefits cells defective for Rtt107 or its partner E3s by providing an alternative means for completing replication.
Increased number of fired origins on a single chromosome improves its replication in rtt107Δ cells
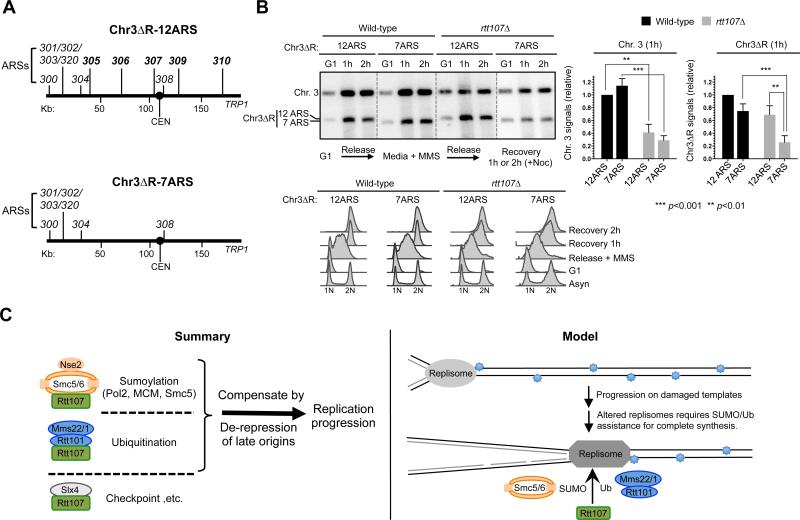
As strong increases in origin firing at global level occurs concurrently with reduced fork speeds, we used an alternative strategy to provide direct evidence that increased origin firing per se improves chromosomal replication in rtt107Δ cells. This strategy alters a small number of origins on an artificial chromosome to avoid affecting the global replication program and fork speed. If the proposed model were correct, a chromosome with more origins should be replicated more efficiently than one with fewer in rtt107Δ cells, but not in wild-type cells. Two variants of chromosome III (or Chr. 3) lacking a portion of the right arm (referred to as Chr3ΔR, 170 kb) can be maintained in cells containing a normal Chr. 3 (Theis et al., 2007, 2010) (Figure 7A). One variant has 12 ARSs and the other has 7 ARSs, and are referred to as Chr3ΔR-12ARS and Chr3ΔR-7ARS, respectively. The extra five ARSs in the former are known to fire, and thus greatly reduce replicon sizes (Theis et al., 2007, 2010). Using PFGE coupled with Southern blot analysis, fully replicated Chr. 3 and Chr3ΔRs can be monitored as distinct bands.
Figure 7. Increasing the number of origins alone improves chromosomal replication in rtt107Δ mutants.
(A) Schematic of Chr3ΔR constructs. The right arm of Chromosome III (Chr. 3) is deleted in both constructs. Chr3ΔR-12ARS contains 12 ARSs on the remaining region, while Chr3ΔR-7ARS contains 7 ARSs.
(B) PFGE and FACS analyses of cells containing Chr3ΔR. Experiments were similar to Figure 5C. A representative result and quantification of 6 trials are shown with means and SD plotted; the values were normalized to wild-type cells containing Chr3ΔR-12ARS in both plots. The left plot shows that Chr. 3 is replicated to a lesser extent in rtt107Δ cells than in wild-type cells. The right plot shows that Chr3ΔR-7ARS replicated more poorly than Chr3ΔR-12ARS in rtt107Δ cells, but not in wild-type cells. FACS shows that the two versions of Chr3ΔR did not affect S phase progression.
(C) Summary of results and a working model. See Discussion for details.
Similar to three other chromosomes (Figure 5C, S5C, 6C, S6B), Chr. 3 replication in rtt107Δ cells was reduced compared with wild-type cells, as evidenced by the lower levels of Chr. 3 signals in gels, regardless of Chr3ΔR-12ARS or Chr3ΔR-7ARS (Figure 7B). When examining Chr3ΔR signals in gels, wild-type cells showed no statistically significant differences between -12ARS and -7ARS, suggesting that reduced number of origins was tolerated (Figure 7B). However, in rtt107Δ cells, the signal levels of Chr3ΔR-12ARS were ~3-fold higher than those of Chr3ΔR-7ARS (Figure 7B), indicating improved replication. These findings provide direct evidence that increasing origin firing alone can improve chromosomal replication in rtt107Δ cells.
Discussion
Replication stress has serious implications for cell survival and human disease. A large number of proteins in multiple cellular processes are employed to cope with such stress. Some factors have taken on multiple functions and lie at the nexus of several processes. These features make them critical for cell survival upon replication stress, but also more challenging to study their individual functions. Here we elucidate several important attributes of the multi-functional scaffold Rtt107 and its associated proteins (summarized in Figure 7C, left). Our data suggest that Rtt107 serves as the common scaffold for three distinct complexes, with each binding partner affecting a unique protein modification pathway. In particular, we propose a function for Rtt107 in promoting sumoylation together with Smc5/6. We also show that Rtt107 and its partner E3s support replication progression under replication stress. Strikingly, modulation of the replication program by increasing late origin firing correlated with better replication and increased survival in rtt107Δ and mutants of its associated E3s. We further provide several lines of evidence which indicate that increased replication initiation events account for the suppression. These results suggest that Rtt107 supports replication via multiple mechanisms, a subset of which can be compensated for by modifying regulatory circuitries controlling origin firing. Our model for the specific roles of Rtt107 and its associated E3s and the biological implication is discussed below.
Rtt107 interacts separately with three partners, one of which affects replisome sumoylation
Unlike scaffolds that simultaneously bind to several partners, our co-IP and Y2H results support the conclusion that Rtt107 forms mutually exclusive complexes with its partners (Figure 1A-1C, S1). This notion is strengthened by genetic and functional analyses (Figure 1D-1F). For example, examination of the shared functions between Rtt107 and Rtt101Mms22 (histone pathway) or Slx4 (checkpoint dampening pathway) suggests that Smc5/6 is likely not involved in these. Rather, we provide evidence that both Rtt107 and Smc5/6 affect the sumoylation of Pol2 and Mcm6, as well as Smc5 (Figure 1F, 3A-3B). That Rtt107 physically associates with replisome components (Figure 3E-3G) suggests that it may bridge the substrates and their E3 ligase. As both Rtt107 and Smc5/6 are recruited to stalled replication forks (Lindroos et al., 2006; Roberts et al., 2008), such sumoylation may influence replication restart by altering DNA polymerase or helicase properties. This idea is consistent with previous findings that ubc9 SUMO E2 and nse2 SUMO ligase mutants impair replication in MMS (Branzei et al., 2006; Cremona et al., 2012). We thus propose a plausible model for the role of Rtt107-Smc5/6 in replication, one that will serve as an important basis for investigation of replisome sumoylation.
It is worth noting that Rtt107's partners do not interact solely with Rtt107 (Figure S2C). In particular, we show that Smc5/6 plays a role in recombinational repair independently of Rtt107 via another binding partner, Mph1 (Figure 2; S2A-S2B). Therefore, Rtt107 and Mph1 each dictates an Smc5/6 function - in replisome sumoylation vs. SUMO-independent recombination regulation, respectively. This is an example wherein a small set of proteins can form multiple modular complexes to execute a larger, diverse set of functions.
Unique features of Rtt107-facilitated replication progression suggest a model for replisome progression
Our work reveals several unique features of Rtt107-based regulation. Both site-specific 2D gel and genome-wide ssDNA analyses suggest that rtt107Δ cells are proficient for the origin firing program, but are defective in replication progression, especially in regions far (> 40 kb) from fired origins (Figure 4, S3), suggesting that Rtt107 is particularly important for completing replication at the distal regions of a large replicon. Rtt101Mms22 and Smc5/6 mutants shared the same defects (Figure S5H). These features differentiate them from those of DNA repair and checkpoint proteins, which affect proximal replisome progression (Tercero and Diffley, 2001; Vazquez et al., 2008). In line with this, our genetic findings suggest that Rtt107 unlikely affects replication through a DNA repair function (Figure S4).
Another unique feature is that mutating two master repressors of late origins suppresses rtt107 defects. It is known that mrc1Δ and/or mec1-100 exacerbates mutants of many proteins aiding replication progression (e.g. Collins et al., 2007; Costanzo et al., 2010). That they globally increased late origin firing in rtt107Δ cells in a manner commensurate with their suppression of replication stress sensitivity and chromosomal replication defects (Figures 5A-5C, 5E, 6, S5A-S5F, S6) suggests that increased origin firing compensates for the impaired replication progression caused by Rtt107 loss.
Additional findings support this conclusion. First, mrc1Δ did not rescue Rad53 hyper-activation in rtt107Δ cells, suggesting that its suppression is not through Rad53 inactivation (Figure 5D, S5D). Consistent with this notion, mrc1Δ did not suppress the phenotype of slx4Δ (with higher Rad53 activation), rather suppressed the E3 mutants (with lower Rad53 activation)(Figure 1E; 5F). Second, genetic data suggest that reducing replication fork movement per se did not suppress the rtt107Δ phenotype (Figure S5G). Third, increasing the number of fired origins on a small chromosome improves its replication in rtt107Δ cells (Figure 7A-B), indicating that increasing replication initiation can compensate for Rtt107 loss.
Our findings suggest an intriguing model of how Rtt107 and its partner E3 enzymes promote replication progression (Figure 7C, right). We propose that the replisome is subjected to many forms of regulations as it traverses damaged templates, such as changes in protein-protein interactions and recruitment of additional proteins; as such, replisome behavior and properties are likely altered as it progresses along the replicon. Rtt107 and the E3 enzymes can be particularly important for assisting specifically altered forms of replisomes during the later part of synthesis. SUMO and ubiquitin modifications are likely important contributors to such Rtt107-mediated regulation: Smc5/6 acts through replisome sumoylation, while Rtt101 regulates histone dynamics, which is tightly linked to replisome functions (Burgess and Zhang, 2013).
In summary, our work brings to light Rtt107's distinct collaborations with three partners to achieve a versatile and multi-faceted cellular response to replicative stress. We have revealed unique features of this response and its implications for replication regulation under stress. Our findings also highlight the innate flexibilities of the replication program. While late origin inhibition can be important for cell survival under certain conditions, removal of this regulation can provide a backup mechanism when specific replication defects arise. As the proteins and regulatory circuitries examined in this study are evolutionarily conserved, these findings should also contribute to a greater understanding of replication stress response circuitries in human cells and how these may be altered during pathogenesis.
Experimental Procedures
Yeast strains, plasmids and general manipulation
Yeast strains are derivatives of W1588-4C, a RAD5 variety of W303 (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1 rad5-535) (Zhao and Blobel, 2005). At least two strains per genotype were examined in each experiment. Strains and plasmids are included in Supplemental Table (one strain is listed for each genotype). Standard yeast methods were used for strain construction. All fusion proteins supported cellular resistance to MMS, suggesting that tagging did not affect protein function (data not shown). Fusion proteins also supported known protein-protein interactions as seen in Figure 1. In addition, the functionality of some fusion constructs has been reported previously (e.g. Chen et al., 2009; Zaidi et al., 2008). Y2H and DNA damage sensitivity assays were performed using standard procedures with the plates incubated for 2-4 days at 30 °C, unless indicated otherwise. For Y2H, AD and BD constructs were introduced into reporter strains and cells were grown on SC-Trp-Leu plates. Positive interactions were assessed by growth after spotting or replica plating cells onto SC-Trp-Leu-His, SC-Trp-Leu-His+3mM 3AT or SC-Trp-Leu-Ade containing plates, as described previously (Duan et al., 2009). Cell viability was assessed by treating asynchronous cells with 0.015% MMS for 2 hrs. MMS was quenched by adding 5% Sodium Thiosulfate. Equal numbers of untreated and MMS-treated cells were sonicated and spread onto YPD media in triplicate and grown at 30 °C for 2 days. Colonies were counted and percentage of viability was calculated as: 100 × (colony numbers of MMS-treated cells / colony numbers of untreated cells). Means and SDs are shown unless otherwise indicated, and student's t-test was used for statistical analysis.
Co-IP assays
Co-IP assays were performed as in Chen et al., 2009. In brief, mid-log phase cells growing in the presence or absence of 0.03% MMS (2 hrs) were collected and lysed by glass bead beating, and proteins were immunoprecipitated using IgG-sepharose beads (for TAP tagged proteins) or with Protein G beads and antibodies. Benzonase (Novagen) was included during cell lysis to remove DNA. Samples were washed and eluted in SDS-containing buffer. In Figure 3E-3F, cells were arrested in G1 by alpha factor, released into YPD containing 0.03% MMS, and harvested 90 minutes after release, when cells were in S phase as assayed by FACS analyses. In other figures panels, +MMS samples were treated for 0.03% MMS (2 hrs). Mms22 is phosphorylated and the different appearance of its band may be due to this modification. Rtt107 interaction with Mms22, Rtt101 and Mms1 has been shown by Mimura et al., 2010 using co-IP and other methods. Nse5 degradation was achieved by adding 1mM IAA to media for 30 minutes followed by a second dose of 0.5 mM IAA added for 2 hrs.
Sumoylated protein detection
Sumoylated proteins were detected as in Hang et al., 2011. In brief, mid-log phase cells were treated with 0.03% or 0.3% MMS for 2 hrs, and lysed in denaturing conditions followed by immunoprecipitation. Proteins were incubated with Protein G agarose beads and Myc antibody or with IgG beads (for TAP tagged proteins), washed and eluted. Samples were separated by SDS-PAGE and analyzed by Western blot. An antibody specific to SUMO (Zhao and Blobel, 2005) was used to detect SUMO forms, while an antibody to Myc (9E10, MSKCC Monoclonal Antibody facility) or TAP (Sigma) was used to detect unmodified forms of proteins.
Rad53 phosphorylation analysis
TCA extraction was performed as in Chen et al., 2013. In brief, G1 arrested cells were released into 0.01% MMS containing media for 45 minutes and collected for Figure 1E and 5D. For Figure S5D, cells were arrested in G1 and released into 0.03% MMS containing media and samples were collected at 30, 60, and 90 minutes. Cell pellets were resuspended in 20% TCA, and lysed by glass bead beating. Lysates were washed with 5% TCA and centrifuged to collect protein precipitants. The pellets were then resuspended in Laemmli buffer, neutralized by 2M Tris, and boiled prior to separation by an SDS-PAGE. A Rad53-specific antibody (yC-19, Santa Cruz) was used to detect unmodified and phosphorylated forms of Rad53 by Western blotting.
2D agarose gel electrophoresis
2D gel analyses were performed as previously described (Friedman and Brewer, 1995). To examine recombination intermediates, cells were arrested in G2 phase by nocodazole and released into YPD media containing 0.03% MMS. Samples were taken at 90, 120, and 180 minutes to examine the ARS305 locus, since at these times recombination intermediates are abundant while replication intermediates are diminished. For other figure panels in which replication intermediates were examined, cells were arrested by alpha-factor in G1, released into media containing 0.03% MMS, and collected at 30, 60 and 90 minutes. DNA was extracted and digested by enzymes indicated in the figures and separated by agarose gel electrophoresis in two dimensions. DNA was transferred to Hybond-XL membrane (GE Healthcare) and analyzed by Southern blot hybridization using probes specific for the loci. Primers used for amplification of the probes are available upon request. For quantification, the signals of 1N DNA were obtained from shorter exposures while those of replication intermediates from longer exposures to ensure both types of signals are within the linear detection range of the PhosphorImager. Note that crosslinking agents were used to preserve recombination intermediates only in Figure 2 (Branzei et al., 2006). Our detection of replication structures at examined loci in wild-type cells is similar to previous reports (e.g. Shirahige et al., 1998; Tourriere et al., 2005).
PFGE analysis
G1 arrested cells were released into 0.03% MMS containing media for 90 minutes. 5% sodium thiosulfate was added to quench MMS and cells were released into media containing nocodazole (15 ug/ml) to prevent 2nd cell cycle entry. Samples from the indicated time points were embedded into agarose plugs and analyzed on a Bio-Rad CHEF apparatus as previously described (Cremona et al., 2012). The percentage of gel entry at recovery 2 hr for a particular chromosome was calculated by dividing the chromosomal band signals with the total chromosomal signals (band and well) after adjusting background signals. For Figure 7B, signals at recovery 1 hr were compared for Chr. 3 and Chr3ΔR variants in the plots shown. Relative ratio of Chr3ΔR/Chr. 3 yielded the same conclusion that more origins on Chr3ΔR improved the replication of this mini-chromosome in rtt107Δ cells but not in wild-type cells.
Chromoduction
Chr3ΔR variants were introduced to cells using chromoduction as described in Theis et al., 2007, 2010. In brief, donor and recipient strains were grown to late log phase in SC-Leu-Trp media and YPD, respectively. 5×106 cells of each strain were mixed in liquid YPD and incubated at 30° C for 4-8 hrs to achieve mating. Cells were collected by centrifugation and washed twice with sterile water and then plated out on SC-Leu-Trp media containing 60 μg Canavanine and 10 μg cyclohexamide for selecting recipient cells that receive Chr3ΔR without chromosomal transfer. Successful chromoduction of Chr3ΔR fragments were confirmed by PFGE and Southern blot.
ssDNA mapping
ssDNA mapping was performed as described in Peng et al 2014 and detailed in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
A genome maintenance scaffold Rtt107 interacts separately with three partners.
Partner interactions determine distinct functions, including replisome sumoylation.
Rtt107 supports replication progression without affecting initiation.
Increasing numbers of fired origins compensates for Rtt107 and its associated E3s.
Acknowledgements
We thank Iestyn Whitehouse for assistance with meta-analysis; David Toczyski, Maria Pia Longhese, Judy Campbell, Hannah Klein, Steve Brill, Lorraine Symington, and Carol Newlon for providing strains and plasmids; Zhao, Hurwitz and Pan lab members, particularly Prabha Sarangi and Xiao Peng, for discussion; Xiao Peng for the Nse5 degron, Koyi Choi and Lei Wei for subset of 2H analysis. This study was supported by US National Institutes of Health grants GM080670, American Cancer Society grant RSG-12-013-01-CCG, and a Leukemia and Lymphoma Society Scholar Award to X.Z., a Pathway to Independence Award (4R00GM08137804) from the National Institutes of Health to W.F., the Italian Association for Cancer Research (AIRC IG 14171), Fondazione Telethon (GGP12160), and European Research Council (REPSUBREP 242928) grants to D.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Ampatzidou E, Irmisch A, O'Connell MJ, Murray JM. Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Celic I, Verreault A, Boeke JD. Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics. 2008;179:1769–1784. doi: 10.1534/genetics.108.088914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci U S A. 2009;106:21252–21257. doi: 10.1073/pnas.0908258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Szakal B, Castellucci F, Branzei D, Zhao X. DNA damage checkpoint and recombinational repair differentially affect the replication stress tolerance of Smc6 mutants. Mol Biol Cell. 2013;24:2431–2441. doi: 10.1091/mbc.E12-11-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JK, Bashkirov VI, Heyer WD, Romesberg FE. Esc4/Rtt107 and the control of recombination during replication. DNA Repair. 2006;5:618–628. doi: 10.1016/j.dnarep.2006.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Thomas A, Pantesco V, De Vos J, Pasero P, Lengronne A. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol. 2010;17:1391–1397. doi: 10.1038/nsmb.1932. [DOI] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol Cell. 2012;45:422–432. doi: 10.1016/j.molcel.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cussiol JR, Jablonowski CM, Yimit A, Brown GW, Smolka MB. Dampening DNA damage checkpoint signalling via coordinated BRCT domain interactions. EMBO J. 2015;34:1704–1717. doi: 10.15252/embj.201490834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Yang Y, Chen YH, Arenz J, Rangi GK, Zhao X, Ye H. Architecture of the Smc5/6 Complex of Saccharomyces cerevisiae Reveals a Unique Interaction between the Nse5-6 Subcomplex and the Hinge Regions of Smc5 and Smc6. J Biol Chem. 2009;284:8507–8515. doi: 10.1074/jbc.M809139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol. 2006;8:148–155. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Rouse J. Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem J. 2005;391:325–333. doi: 10.1042/BJ20050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- Han J, Zhang H, Zhang H, Wang Z, Zhou H, Zhang Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 2013;155:817–829. doi: 10.1016/j.cell.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang LE, Liu X, Cheung I, Yang Y, Zhao X. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol. 2011;18:920–926. doi: 10.1038/nsmb.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CC, Glover JN. BRCT domains: easy as one, two, three. Cell Cycle. 2011;10:2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung GP, Aristizabal MJ, Krogan NJ, Kobor MS. Conditional genetic interactions of RTT107, SLX4, and HRQ1 reveal dynamic networks upon dna damage in Saccharomyces cerevisiae. G3. 2014;4:1059–1069. doi: 10.1534/g3.114.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, Sjogren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- McIntosh D, Blow JJ. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb Perspect Biol 4. 2012:a012955. doi: 10.1101/cshperspect.a012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Yamaguchi T, Ishii S, Noro E, Katsura T, Obuse C, Kamura T. Cul8/Rtt101 forms a variety of protein complexes that regulate DNA damage response and transcriptional silencing. J Biol Chem. 2010;285:9858–9867. doi: 10.1074/jbc.M109.082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohouo PY, Bastos de Oliveira FM, Almeida BS, Smolka MB. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell. 2010;39:300–306. doi: 10.1016/j.molcel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Ohouo PY, Bastos de Oliveira FM, Liu Y, Ma CJ, Smolka MB. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature. 2013;493:120–124. doi: 10.1038/nature11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol. 2001;21:3913–3925. doi: 10.1128/MCB.21.12.3913-3925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Raghuraman MK, Feng W. Analysis of ssDNA gaps and DSBs in genetically unstable yeast cultures. Methods Mol Biol. 2014;1170:501–515. doi: 10.1007/978-1-4939-0888-2_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 2012;31:883–894. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Kobor MS, Bastin-Shanower SA, Ii M, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell. 2006;17:539–548. doi: 10.1091/mbc.E05-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Zaidi IW, Vaisica JA, Peter M, Brown GW. Regulation of Rtt107 recruitment to stalled DNA replication forks by the cullin Rtt101 and the Rtt109 acetyltransferase. Mol Biol Cell. 2008;19:171–180. doi: 10.1091/mbc.E07-09-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J. Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J. 2004;23:1188–1197. doi: 10.1038/sj.emboj.7600129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- Theis JF, Dershowitz A, Irene C, Maciariello C, Tobin ML, Liberi G, Tabrizifard S, Korus M, Fabiani L, Newlon CS. Identification of mutations that decrease the stability of a fragment of Saccharomyces cerevisiae Chromosome III lacking efficient replicators. Genetics. 2007;177:1445–1458. doi: 10.1534/genetics.107.074690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis JF, Irene C, Dershowitz A, Brost RL, Tobin ML, di Sanzo FM, Wang JY, Boone C, Newlon CS. The DNA damage response pathway contributes to the stability of Chromosome III derivatives lacking efficient replicators. PLoS Genet. 2010;6:e1001227. doi: 10.1371/journal.pgen.1001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Vazquez MV, Rojas V, Tercero JA. Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair. 2008;7:1693–1704. doi: 10.1016/j.dnarep.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Xue X, Choi K, Bonner J, Chiba T, Kwon Y, Xu Y, Sanchez H, Wyman C, Niu H, Zhao X, et al. Restriction of replication fork regression activities by a conserved SMC complex. Mol Cell. 2014;56:436–445. doi: 10.1016/j.molcel.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmstrom J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008;9:1034–1040. doi: 10.1038/embor.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci U S A. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Nellimoottil T, Peace JM, Knott SR, Villwock SK, Yee JM, Jancuska JM, Rege S, Tecklenburg M, Sclafani RA, et al. The level of origin firing inversely affects the rate of replication fork progression. J Cell Biol. 2013;201:373–383. doi: 10.1083/jcb.201208060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.