Disease-causing microbes have become rapidly resistant to antibiotic drug therapies and diseases that were thought to be eradicated, are re-emerging. Tuberculosis for example is reappearing even in the developed world causing more than 1.1 million deaths a year worldwide. The methylerythritol phosphate (MEP) pathway,[1] an alternative to the mevalonate pathway,[2] is used for the biosynthesis of isopentenyl diphosphate (IPP, 1) and dimethylallyl diphosphate (DMAPP, 2), the crucial building blocks involved in the formation of essential terpenoids of most pathogenic bacteria (including Mycobacterium tuberculosis) and in plant plastids. Therefore the MEP pathway is a target for the development of new antimicrobial agents as it is essential for microorganisms, and absent in humans.[3] In the last step of this biosynthetic route, (E)-4-hydroxy-2-methylbut-2-en-1-yl diphosphate (HMBPP, 3) is converted into a mixture of IPP 1 and DMAPP 2 (Scheme 1).
Scheme 1.
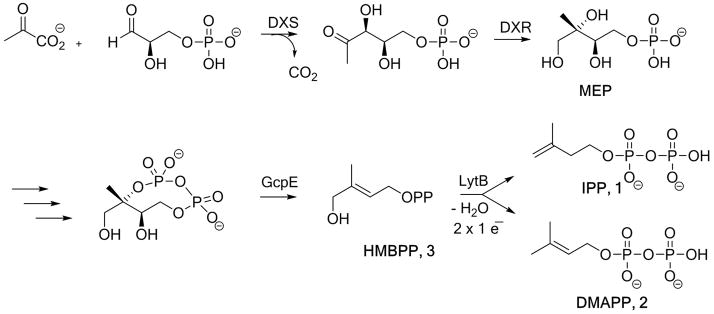
Methylerythritol phosphate (MEP) pathway. DXS: 1-deoxy-D-xylulose 5-phosphate synthase; DXR: 1-deoxy-D-xylulose 5-phosphate reductoisomerase; GcpE: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate reductase; LytB: 4-hydroxy-3-methylbut-2-enyl 1-phosphate reductase.
This reaction is catalyzed by a peculiar [4Fe-4S] center of the LytB/IspH protein.[4] The substrate-free LytB protein contains a special diamagnetic [4Fe-4S]2+ cluster, which is EPR silent. Two Mössbauer spectroscopic studies reported independently that one of the four iron sites of the substrate-free LytB protein has an unusual high isomer shift (δ = 0.89 mms−1).[4, 5] This value is identical within experimental error to that of an unusual fourth iron site in the citrate bound form of aconitase.[6] Therefore it has been proposed that the coordination sphere of this special iron site comprises three inorganic sulfurs from the iron sulfur cluster and additional 3 or 2 non-sulfur ligands (O and/or N) in a binding motif similar to those of substrate-bound aconitase.[4, 6]
First X-ray structures of substrate-free LytB from Aquifex aeolicus[7] and Escherichia coli[8] report the presence of a [3Fe-4S]+ cluster, but in later published work the structure of HMBPP bound LytB from E. coli has been refined to a structure with a [4Fe-4S] cluster.[9] The structures of LytB containing the [4Fe-4S] cluster bound to (E)-4-amino-3-methylbut-2-en-1-yl diphosphate 4 or (E)-4-mercapto-3-methylbut-2-en-1-yl diphosphate 5, two analogues of HMBPP, were also reported.[10] These amino and thiol analogues of HMBPP have been shown to be extremely potent LytB inhibitors displaying Ki = 20 nM and Ki = 54 nM respectively.[11] The crystal structure of the substrate-free LytB in its [4Fe-4S]2+ state has not been solved yet. The lack of a structure for the substrate-free LytB motivated us to perform a spectroscopic study using synchrotron based nuclear resonance vibrational spectroscopy (NRVS) also called nuclear inelastic scattering (NIS) in combination with density functional theory based quantum chemical-molecular mechanical (QM/MM) calculations[12] in order to get more insight into the structure of substrate-free LytB. In the present work we additionally present NRVS data of LytB bound to its natural substrate 3 and of LytB bound to the inhibitors 4 or 5. NRVS[13, 14] detects iron-involved molecular vibrations and is sensitive to the movement of the 57Fe Mössbauer nucleus. Therefore it is complementary to other vibrational methods such as IR or Raman spectroscopy. Since no optical selection rules apply for NRVS, it represents all individual modes of the iron, which in turn are sensitive to iron-ligand distances. Combined with simulations based on quantum mechanical calculations of the iron center[15] NRVS can be used to make structural predictions of the iron center and its environment. This has been shown recently for the CO-inhibited Mo-nitrogenase[16] as well as for the active site of [NiFe] hydrogenase.[17]
The E. coli His6-tagged LytB protein was produced on LB containing 57FeCl3 and purified under anaerobic conditions as described in [4]. As high concentrations of the Mössbauer isotope 57Fe (> 3 mM) are required for NRVS, the integrity of the enzyme was verified by measuring its activity.[11] The activity was similar before and after concentration (800 nmol·min−1·mg−1).
The present study shows that the [4Fe-4S] center of the substrate-free LytB is coordinated to three bridging sulfurs, and to three water molecules, but not to an additional amino acid from the protein backbone.
Figure 1a displays the experimental NRVS data (the energy dependence of nuclear inelastic scattering) of the substrate-free form of LytB. The obtained NRVS data-set contains three separate regions typical for [4Fe-4S] clusters.[16, 17] The first region extends to ca. 240 cm−1 and contains several distinct bands with strong maxima at 136 cm−1, 161 cm−1 and 180 cm−1. The second region ranges from 240 cm−1 to 340 cm−1 with an especially characteristic band at 277 cm−1. The third region from 340 cm−1 to 440 cm−1 shows no distinct peaks.
Figure 1.
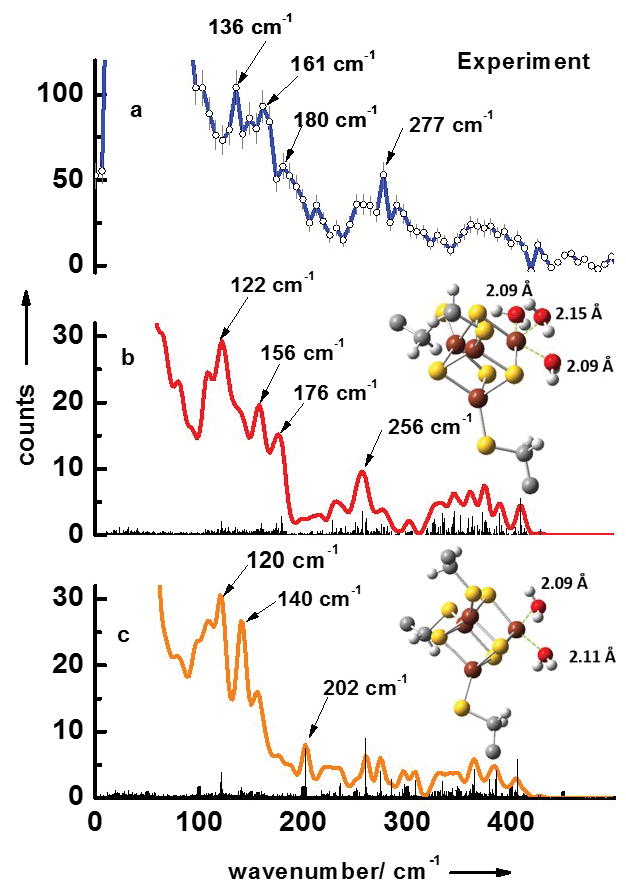
NRVS data of the substrate-free form of LytB (a). Simulated NRVS data are obtained via combined quantum chemical and molecular mechanics (QM/MM) calculations based on the pdb entry of the [3Fe-4S] substrate-free LytB cluster[8] (3F7T.pdb) assuming model structures of the active site complexes with (b) three water ligands and (c) with two water ligands.
In order to pin down the ligand structure of the unusual 4th iron site of the substrate-free LytB, we have performed simulations of NRVS data by quantum mechanical (QM) DFT-calculations on the [4Fe-4S] unit with different ligands combined with molecular mechanics (MM) calculations of the whole protein shell. Since inserting the missing iron in the substrate-free LytB structure harbouring an incomplete Fe/S cluster (pdb 3F7T) already indicated the presence of three water molecules (see supporting information of ref. [4]), we have performed a QM/MM simulation with this structural model. The results are shown in Fig. 1b. The simulated data set also predicted signals in the same three separate regions found in the NRVS experiments. The first region ranges up to 220 cm−1, with also three distinct bands at 122 cm−1, 156 cm−1 and 176 cm−1, and contains mostly pure S-Fe-S′ bending modes as well as mixed bending and stretching modes. Below 50 cm−1, protein modes exist that show movements of the protein backbone coupled to the whole [4Fe-4S] entity but without any contributions from stretching or bending iron ligand modes. The second region from 240 cm−1 to 340 cm−1 contains mostly pure Fe-S stretching modes and a mixed bending and stretching mode with a very distinct band at 256 cm−1. This band is caused by several almost degenerate modes, the most intense occur at 250.4, 256.6, 256.7 and 257.0 cm−1. All these modes are characteristic for the unusual 4th iron site and have considerable S-Fe-O stretching character coupled to a rotation of the water ligands around the Fe-O bond. This very distinct band is also visible in the experimental data at 277 cm−1. The third region extends above 360 cm−1 and contains modes with mixed stretching and bending character as well as pure Fe-S stretching modes. Figure 1c shows the results of the QM/MM simulations of the active site complex simulated with two water ligands. This simulation does not reproduce the experimental observation of the three regions discussed above, which also has been reported as a NRVS signature for synthetic [4Fe-4S]2+ model complexes[18] as well as ferredoxins.[19] Moreover, simulation with two water ligands shown in Fig. 1c does not reproduce the very characteristic band experimentally observed at 277 cm−1. This supports our conclusion that the unusual 4th iron of the [4Fe-4S] center of substrate-free LytB is coordinated to three water molecules and three bridging sulfurs of the cluster.
We have also measured the NRVS data of LytB bound to its natural substrate 3 as well as to the inhibitors (E)-4-amino-3-methylbut-2-en-1-yl diphosphate 4 and (E)-4-mercapto-3-methylbut-2-en-1-yl diphosphate 5 (Fig 2a–c). Like for the substrate-free form of LytB all NRVS data sets display separate three regions. Fig. 3 contains the results of calculated NRVS data based on X-ray structure data.[9, 10, 20] There is reasonable agreement between calculated and experimental data and we attribute the deviations to the presence of slightly different protein conformations in protein crystals and in solution. For LytB in complex with 4 a NRVS data set has been calculated based on the structure recently published by the groups of Groll and Oldfield (see Figure 4Sc in the supplement), which for no obvious reasons, show significantly less agreement with the experimental data displayed in Fig. 2b than the calculation based on 3ZGL.pdb.[20]
Figure 2.
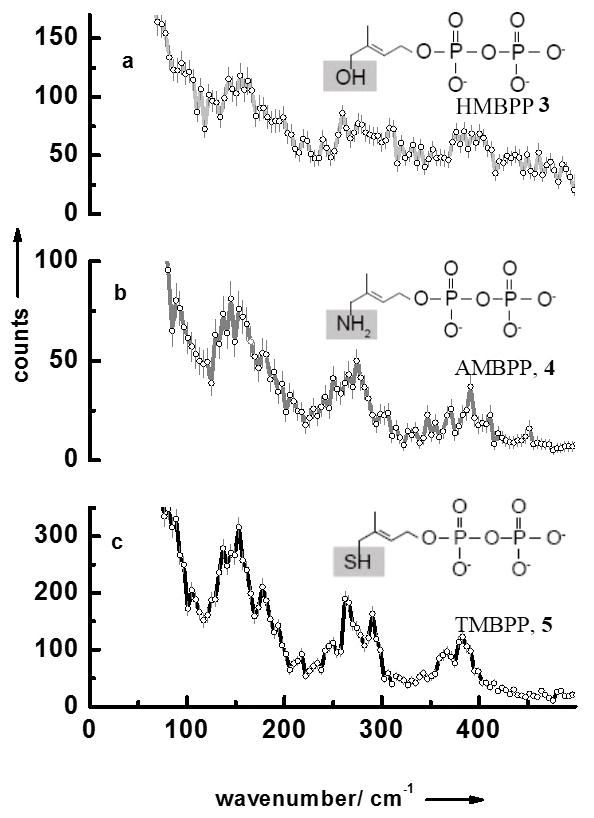
NRVS data of (a) LytB bound to its substrate 3, (b) LytB in complex with inhibitor 4, and (c) LytB in complex with inhibitor 5.
Figure 3.
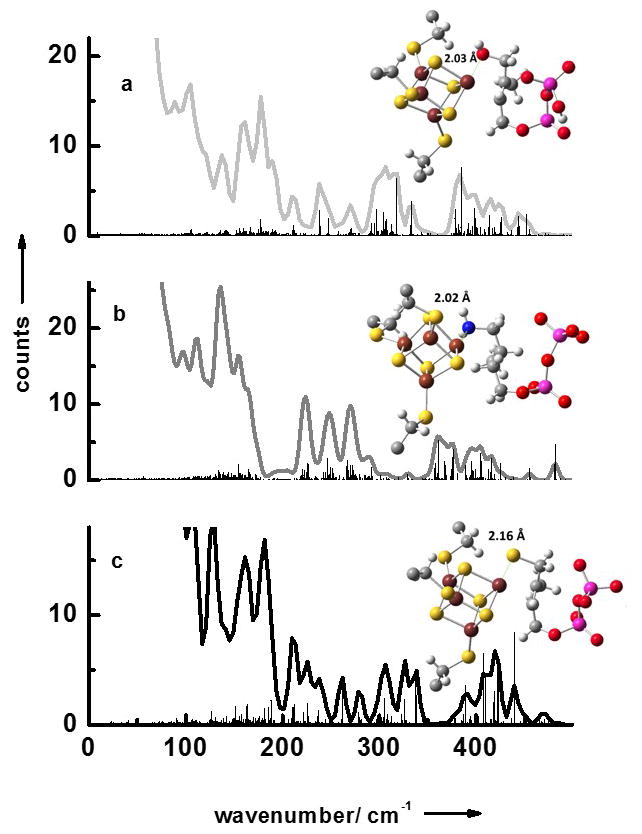
Simulated NRVS data obtained via combined quantum chemical and molecular mechanics (QM/MM) calculations based on crystal structures of the active site/inhibitor complexes: (a) LytB with substrate 3 (3KE8.pdb),[9] (b) LytB in complex with inhibitor 4 (3ZGL.pdb),[20] and (c) LytB in complex with inhibitor 5 (4H4E.pdb).[10]
The corresponding partial density of states (pDOS) obtained from the experimental as well as from the simulated NRVS data displayed in Figs. 1, 2 and 3 are shown in the supplement (figure 2S and 3S). From pDOS, it is possible to determine the Lamb-Mössbauer factor (fLM) and several thermodynamic parameters, including the total vibration amplitude (|x|), the mean internal energy (U), the specific heat capacity (cV), the entropy (Svib), the free enthalpy (Gvib=U-TSvib) as well as the normalized mean force constant (D) of the protein sample. Table S1 shows parameters that are calculated from the experimental pDOS (Fig. 2S, 3S) for LytB bound to its substrate 3 and for substrate-free LytB. The mean force constant for the substrate-bound LytB is 216 N/m and therefore smaller than for substrate-free LytB with 260 N/m. This could indicate that the substrate-free conformation of LytB has a [4Fe-4S] cluster that is bound to the protein in a stiffer state than in the substrate-bound form, which exhibits a smaller mean force constant.
Additional evidence for coordination of water molecules at the apical iron site of the [4Fe-4S] cluster can be found in the X-ray structure of LytB in complex with propynyl diphosphate, an inhibitor with an IC50 of 6.7 μM. In this latter structure, evidence was found for a water molecule (or a hydroxide ion) bound to the apical ion with a Fe-O bound length of 2.1 Å.[21]
The presence of three labile water ligands on the apical iron would also explain the instability of the [4Fe-4S]2+ in the crystallisation experiments. The inability to obtain crystals with a complete [4Fe-4S] can be attributed to the oxygen sensitivity of the enzyme. However, the oxygen sensitivity of GcpE, another enzyme of the MEP pathway, was reported to be higher than that of LytB[22] and surprisingly the X-ray structures of substrate-free GcpE from A. aeolicus[23] and T. thermophiles[24] revealed an unique iron in the [4Fe-4S]2+ with a tetrahedral coordination and linked to a glutamate of the protein. This coordination by a protein residue and not adventitious water makes the protein more stable and the reported oxygen sensitivity might be due to a higher exposition of the Fe/S cluster to the solvent.
In addition to substrate-bound aconitase,[6] a coordinating water/hydroxide ion was also observed in the X-ray structure of other [4Fe-4S] enzymes catalyzing a dehydration step, including the (R)-2-hydroxyisocaproyl-CoA dehydrase (α-cluster) of Clostridium difficile[25] and the quinolinate synthase of Thermotoga maritima[26]. To the best of our knowledge, the coordination of the unique site of a [4Fe-4S] cluster by three water molecules, as seen here for LytB, is unprecedented.
The mechanism of the reaction catalysed by LytB is still under investigation but it is now well established that the first step of this mechanism is the binding of the OH group of the substrate to the apical iron of the [4Fe-4S]. Previous Mössbauer investigations evidenced a change from octahedral to tetrahedral in the coordination geometry of this iron upon substrate binding.[27] According to this NRVS study, this binding is most probably accompanied by the release of three water molecules.
Supplementary Material
Footnotes
We are grateful to Prof. M. Rohmer for helpful discussion. We thank Prof. A. Boronat (University of Barcelona, Spain) and his group for providing us with the E. coli strain overexpressing LytB. We acknowledge M. Parisse for technical assistance. This work was supported by the ‘Agence Nationale de la Recherche’ (ANR-2011-BSV5-028) and COST Action 1201 to M.S., and by NIH grant GM25521 to C.D.P as well as by the collaborative research center SFB/TRR 88 “Cooperative effects in homo- and hetero-metallic complexes (3MET)”, by NANOKAT and by the German Federal Ministry of Education and Research under 05K13UK2 to V.S..
Contributor Information
Isabelle Faus, Fachbereich Physik, TU Kaiserslautern, Erwin-Schrödinger-Strasse 46, D-67653 Kaiserslautern (Germany), Fax: ++49 631 205 4958.
Dr. Annegret Reinhard, Fachbereich Physik, TU Kaiserslautern, Erwin-Schrödinger-Strasse 46, D-67653 Kaiserslautern (Germany), Fax: ++49 631 205 4958
Dr. Sergej Rackwitz, Fachbereich Physik, TU Kaiserslautern, Erwin-Schrödinger-Strasse 46, D-67653 Kaiserslautern (Germany), Fax: ++49 631 205 4958
Dr. Juliusz A. Wolny, Fachbereich Physik, TU Kaiserslautern, Erwin-Schrödinger-Strasse 46, D-67653 Kaiserslautern (Germany), Fax: ++49 631 205 4958
Dr. Kai Schlage, P01, Petra III, DESY, Notkestraße 85, D-22607 Hamburg (Germany)
Dr. Hans-Christian Wille, P01, Petra III, DESY, Notkestraße 85, D-22607 Hamburg (Germany)
Dr. Aleksandre Chumakov, ESRF - The European Synchrotron, CS40220, 38043, Grenoble Cedex 9 (France)
Dr. Sergiy Krasutsky, Department of Chemistry, University of Utah, 315 South 1400 East RM 2020, Salt Lake City, Utah 84112 (USA)
Dr. Philippe Chaignon, Université de Strasbourg, UMR 7177 CNRS, Institut Le Bel, 4 rue Blaise Pascal, CS 90032, 67081 Strasbourg Cedex (France)
Prof. C. Dale Poulter, Department of Chemistry, University of Utah, 315 South 1400 East RM 2020, Salt Lake City, Utah 84112 (USA)
Dr. Myriam Seemann, Email: mseemann@unistra.fr, Université de Strasbourg, UMR 7177 CNRS, Institut Le Bel, 4 rue Blaise Pascal, CS 90032, 67081 Strasbourg Cedex (France)
Prof. Volker Schünemann, Email: schuene@physik.uni-kl.de, Fachbereich Physik, TU Kaiserslautern, Erwin-Schrödinger-Strasse 46, D-67653 Kaiserslautern (Germany), Fax: ++49 631 205 4958
References
- 1.a) Rohmer M. Nat Prod Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]; b) Eisenreich W, Rohdich F, Bacher A. Trends Plant Sci. 2001;6:78–84. doi: 10.1016/s1360-1385(00)01812-4. [DOI] [PubMed] [Google Scholar]
- 2.Bloch K. Steroids. 1992;57:378–383. doi: 10.1016/0039-128x(92)90081-j. [DOI] [PubMed] [Google Scholar]
- 3.Rohmer M, Grosdemange-Billiard C, Seemann M, Tritsch D. Curr Opin Invest Drugs. 2004;5:154–162. [PubMed] [Google Scholar]
- 4.Seemann M, Janthawornpong K, Schweizer J, Böttger LH, Janoschka A, Ahrens-Botzong A, Tambou EN, Rotthaus O, Trautwein AX, Rohmer M, Schünemann V. J Am Chem Soc. 2009;131:13184–13185. doi: 10.1021/ja9012408. [DOI] [PubMed] [Google Scholar]
- 5.Xiao Y, Chu L, Sanakis Y, Liu P. J Am Chem Soc. 2009;131:9931–9933. doi: 10.1021/ja903778d. [DOI] [PubMed] [Google Scholar]
- 6.Beinert H. J Biol Inorg Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 7.Rekittke I, Wiesner J, Röhrich R, Demmer U, Warkentin E, Xu W, Troschke K, Hintz M, No JH, Duin EC, Oldfield E, Jomaa H, Ermler U. J Am Chem Soc. 2008;130:17206–17207. doi: 10.1021/ja806668q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gräwert T, Rohdich F, Span I, Bacher A, Eisenreich W, Eppinger J, Groll M. Angew Chem Int Ed. 2009;48:5756–5759. doi: 10.1002/anie.200900548. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2009;121:5867–5870. [Google Scholar]
- 9.Gräwert T, Span I, Eisenreich W, Rohdich F, Eppinger J, Bacher A, Groll M. Proc Natl Acad Sci. 2010;107:1077–1081. doi: 10.1073/pnas.0913045107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Span I, Wang K, Wang W, Jauch J, Eisenreich W, Bacher A, Oldfield E, Groll M. Angew Chem Int Ed. 2013;52:2118–2121. doi: 10.1002/anie.201208469. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2013;125:2172–2175. [Google Scholar]
- 11.Janthawornpong K, Krasutsky S, Chaignon P, Rohmer M, Poulter CD, Seemann M. J Am Chem Soc. 2013;135:1816–1822. doi: 10.1021/ja309557s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senn HM, Thiel W. Angew Chem Int Ed. 2009;48:1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2009;121:1220–1254. [Google Scholar]
- 13.Seto M, Yoda Y, Kikuta S, Zhang XW, Ando M. Phys Rev Lett. 1995;74:3828–3831. doi: 10.1103/PhysRevLett.74.3828. [DOI] [PubMed] [Google Scholar]
- 14.Sturhahn W, Toellner TS, Alp EE, Zhang S, Ando M, Yoda Y, Kikuta S, Seto M, Kimball CW, Dabrowski B. Phys Rev Lett. 1995;74:3832–3835. doi: 10.1103/PhysRevLett.74.3832. [DOI] [PubMed] [Google Scholar]
- 15.Maylis Orio M, Pantazis DA, Neese F. Photosynth Res. 2009;102:443–453. doi: 10.1007/s11120-009-9404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott AD, Pelmenschikov V, Guo Y, Yan L, Wang H, George SJ, Dapper CH, Newton WE, Yoda Y, Tanaka Y, Cramer SP. J Am Chem Soc. 2014;136:15942–15954. doi: 10.1021/ja505720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamali S, Wang H, Mitra D, Ogata H, Lubitz W, Manor BC, Rauchfuss TB, Byrne D, Bonnefoy V, Jenney FE, Jr, Adams MWW, Yoda Y, Alp E, Zhao J, Cramer SP. Angew Chem Int Ed. 2013;52:724–728. doi: 10.1002/anie.201204616. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2013;125:752–756. [Google Scholar]
- 18.Xiao Y, Koutmos M, Case DA, Coucouvanis D, Wang H, Cramer SP. Dalton Trans. 2006;18:2192–2201. doi: 10.1039/b513331a. [DOI] [PubMed] [Google Scholar]
- 19.Mitra D, Pelmenschikov V, Guo Y, Case DA, Wang H, Dong W, Tan M-L, Ichiye T, Jenney FE, Jr, Adams MWW, Yoda Y, Zhao J, Cramer SP. Biochemistry. 2011;50(23):5220–5235. doi: 10.1021/bi200046p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borel F, Barbier E, Kratsutsky S, Janthawornpong K, Rohmer M, Dale Poulter C, Ferrer JL, Seemann M. to be published. [Google Scholar]
- 21.Span I, Wang K, Wang W, Zhang Y, Bacher A, Eisenreich W, Li K, Schultz C, Oldfield E, Groll M. Nat Commun. 2012;3:1042. doi: 10.1038/ncomms2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolff M, Seemann M, Tse Sum Bui B, Frapart Y, Tritsch D, Garcia-Estrabot A, Rodriguez-Concepción M, Boronat A, Marquet A, Rohmer M. FEBS Lett. 2003;1:115–120. doi: 10.1016/s0014-5793(03)00317-x. [DOI] [PubMed] [Google Scholar]
- 23.Lee M, Gräwert T, Quitterer F, Rohdich F, Eppinger J, Eisenreich W, Bacher A, Groll M. J Mol Biol. 2010;404:600–610. doi: 10.1016/j.jmb.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 24.Rekittke I, Nonaka T, Wiesner J, Demmer U, Warkentin E, Jomaa H, Ermler U. FEBS Lett. 2011;585:447–451. doi: 10.1016/j.febslet.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Knauer SH, Buckel W, Dobbek H. J Am Chem Soc. 2011;133:4342–4347. doi: 10.1021/ja1076537. [DOI] [PubMed] [Google Scholar]
- 26.Cherrier MV, Chan A, Darnault C, Reichmann D, Amara P, Ollagnier de Choudens S, Fontecilla-Camps JC. J Am Chem Soc. 2014;136:5253–5256. doi: 10.1021/ja501431b. [DOI] [PubMed] [Google Scholar]
- 27.Ahrens-Botzong A, Janthawornpong K, Wolny JA, Tambou EN, Rohmer M, Krasutsky S, Poulter CD, Schünemann V, Seemann M. Angew Chem Int Ed. 2011;50:11976–11979. doi: 10.1002/anie.201104562. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2011;123:12182–12185. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


