Abstract
Folding may be described conceptually in terms of trajectories over a landscape of free energies corresponding to different molecular configurations. In practice, energy landscapes can be difficult to measure. Single-molecule force spectroscopy (SMFS), whereby structural changes are monitored in molecules subjected to controlled forces, has emerged as a powerful tool for probing energy landscapes. We summarize methods for reconstructing landscapes from force spectroscopy measurements under both equilibrium and nonequilibrium conditions. Other complementary, but technically less demanding, methods provide a model-dependent characterization of key features of the landscape. Once reconstructed, energy landscapes can be used to study critical folding parameters, such as the characteristic transition times required for structural changes and the effective diffusion coefficient setting the timescale for motions over the landscape. We also discuss issues that complicate measurement and interpretation, including the possibility of multiple states or pathways and the effects of projecting multiple dimensions onto a single coordinate.
Keywords: protein folding, nucleic acid folding, reaction coordinate, statistical mechanics, optical trapping, AFM
INTRODUCTION
Energy landscape theory provides the fundamental biophysical framework for understanding structure formation in biological macromolecules, such as proteins and nucleic acids (33, 92, 94, 104). In landscape theory, folding is understood in terms of the statistical mechanics of the ensemble of allowed conformational microstates: The landscape consists of a hyperdimensional surface representing the free energy of each structure of the molecule in a configuration space that spans all possible conformations. Because the three-dimensional conformation of linear biopolymers is minimally specified by the bond angles in the polymer chain, this landscape is inherently multidimensional. In this picture, the folding process may be viewed as a diffusive search over the landscape hypersurface—biased by the conformational energy changes implicit in the landscape—to arrive at the native structure, which is typically the minimum-energy configuration, in accordance with Anfinsen’s hypothesis (2).
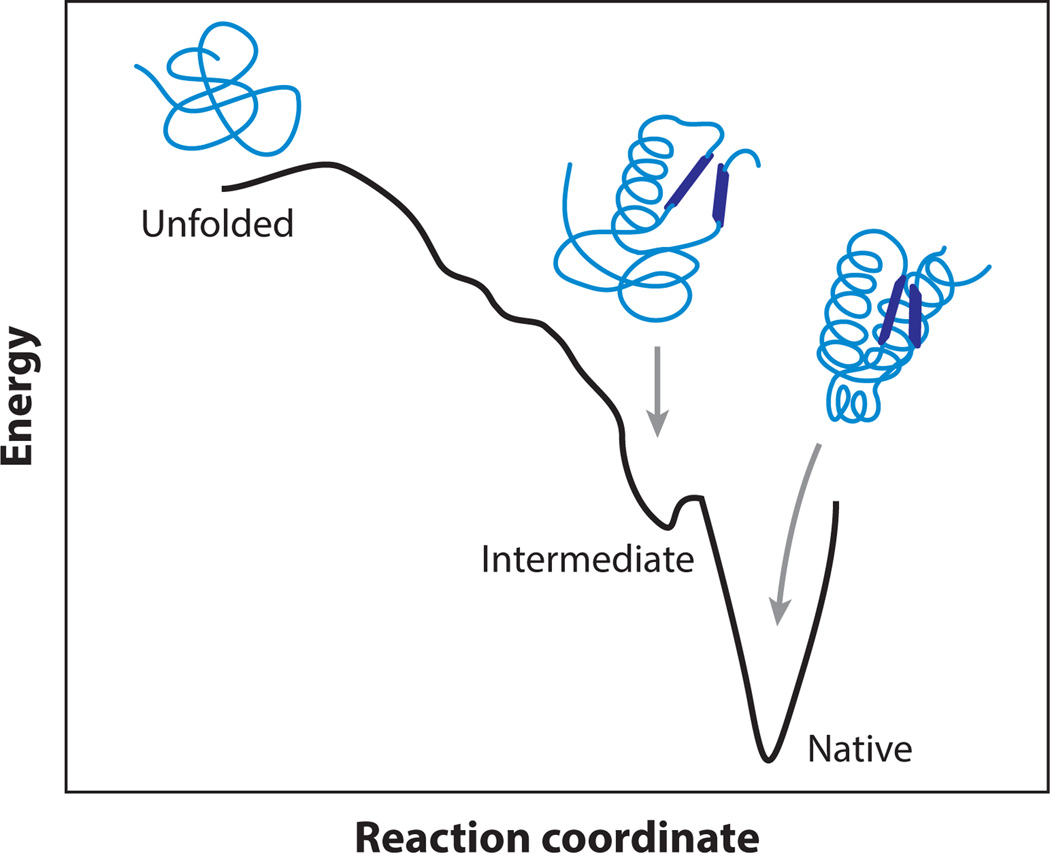
A generic folding landscape, reduced to a single-dimensional coordinate for simplicity, is illustrated schematically in Figure 1, which plots the free energy against a measure of the molecular configuration. Unfolded molecules tend to have high enthalpy and high entropy, whereas folded molecules tend to have low enthalpy and low entropy. Many biomolecules have evolved to fold rapidly and efficiently; these are generally thought to have funnel-shaped landscapes, as illustrated, minimizing the time needed to arrive at the native structure (33, 94). Nevertheless, landscapes usually contain energy barriers within the funnel that slow down the folding, owing to the fact that the decreases in enthalpy and entropy during folding are not synchronized. Because intramolecular interactions vary with environmental conditions (e.g., temperature, pH, ionic strength, solutes, etc.), the folding landscape is not unique. Instead, it is a sensitive function of the ambient conditions and can be tuned to stabilize nonnative structures, such as partially folded states, unfolded states, or even polymer aggregates (18). We note that one-dimensional (1D) representations of energy landscapes are more than just notional cartoons: In these, the full, hyperdimensional landscapes are projected onto a single “reaction coordinate” that (hopefully) encapsulates the essence of the folding trajectory. In practice, this reaction coordinate often represents an experimental observable used to follow the progress of the folding transition. Projections onto a judiciously chosen coordinate often describe the essential features of folding surprisingly well despite their simplicity. That said, multidimensional effects cannot always be ignored, as discussed below.
Figure 1.
Notional cartoon of an energy landscape in one dimension. An unfolded molecule has high energy and high entropy, whereas a folded molecule has low energy and low entropy. The funnel-like shape of the landscape leading to the native state may be punctuated with barriers and metastable intermediates.
The utility of the landscape picture also lies in the fact that one can, in principle, predict the folding behavior of a molecule given its energy landscape. For example, metastable states, such as partially folded intermediates, are formed at local minima in the landscape. The abundance of different structures at equilibrium can be computed from their relative energies using the Boltzmann relation. Specific folding pathways can be identified as low-energy channels, formed in the landscape, that connect different states. The rate for crossing a barrier can also be predicted using Kramers’ theory (55, 75). Comparing rates over different barriers then allows the kinetic partitioning between pathways on the landscape to be determined (104). Even the “internal friction” of a molecule is captured, from changes in the effective diffusion coefficient owing to the local roughness of the landscape (12, 52, 120).
Although specific features of an energy landscape, such as the presence of metastable intermediates and the heights of barriers, can be characterized by various approaches (18), the full shape of the landscape is hard to determine. Ideally, to make such a determination, one would observe a single molecule folding at atomic resolution on the timescale of bond rotations. This goal cannot currently be achieved experimentally, although it can be done computationally in certain large-scale simulations (10). In recent years, however, advances in single-molecule experiments have led to the development of new methods for measuring 1D profiles of energy landscapes directly from the folding trajectories of individual molecules. In particular, several complementary methods have been developed for reconstructing such landscape profiles using force spectroscopy, where the molecule is unfolded (and, optionally, refolded) under controlled mechanical tension. Here, we review methods for reconstructing landscapes in model-independent ways, based both on equilibrium and nonequilibrium measurements of molecular conformations. We also discuss some model-dependent approaches that are necessarily more approximate but easier to implement experimentally. Finally, we review how experimentally derived landscape profiles can be applied to identify key parameters of the folding, and we critically discuss issues that complicate interpretation of the data.
MEASURING LANDSCAPE PROFILES USING FORCE SPECTROSCOPY
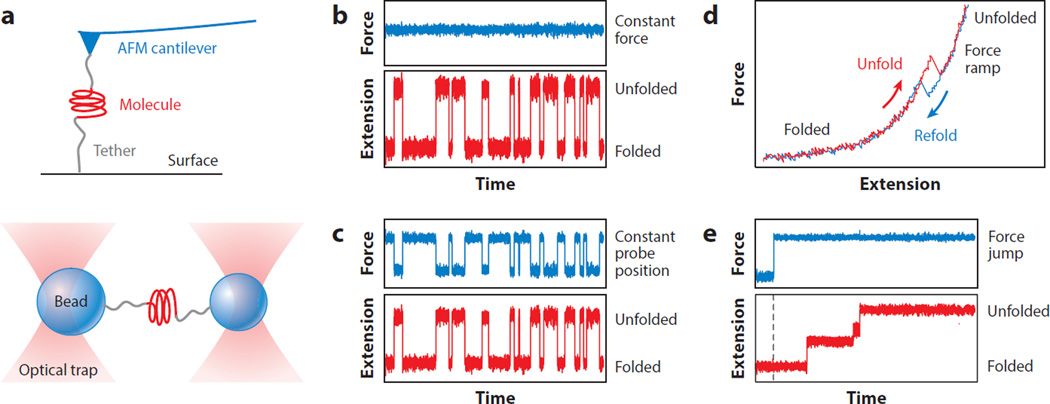
In single-molecule force spectroscopy (SMFS) experiments, mechanical tension is applied across a single molecule so as to perturb its structure, and the extension of the molecule is measured as its structure changes under the applied load (49, 111, 119). Various types of force probes have been used for SMFS, most commonly atomic force microscopes (AFMs), optical tweezers, and magnetic tweezers (49, 86). The molecule of interest is attached at one point to a force probe and at another point to a fixed surface or a second force probe (Figure 2a); often, the attachment points are at the ends of the polymer. The response of the molecule to applied force is subsequently measured in one of several modalities: (a) constant force, where fluctuations in extension are recorded as the load is held constant with a force clamp (Figure 2b); (b) constant position, where fluctuations in both molecular extension and force are recorded as the probe is held at an unchanging position (Figure 2c); (c) force ramp, where the molecular extension is recorded as the force is ramped up or down by moving the probe at a constant speed (Figure 2d); and (d) force jump, where the molecular extension is recorded after the force is changed abruptly to a different value (Figure 2e). The first two modalities record equilibrium fluctuations, whereas the last two are out of equilibrium, owing to the rapid force changes. In all cases, the effect of the external force is to stretch out the molecule as it unfolds. The natural reaction coordinate to use is thus the molecular extension, which increases as the unfolding reaction proceeds.
Figure 2.
Single-molecule force spectroscopy (SMFS) measurement modalities. (a) SMFS measurements can involve a molecule tethered between a surface and a force probe, as illustrated for an atomic force microscope (top) or between two probes, as illustrated for optical tweezers (bottom). (b) In constant-force mode, the extension fluctuates as the molecule unfolds and refolds while the force is clamped. (c) In constant-position mode, both the extension and the force fluctuate as the structure changes. (d) In force-ramp mode, the elastic stretching of the molecule is interrupted by a “rip” when the molecule unfolds or refolds; hysteresis between unfolding and refolding curves indicates a nonequilibrium process. (e) In force-jump mode, the force is abruptly jumped and then clamped at the new value. The extension then increases in steps as the molecule changes structure; here, unfolding is shown. Abbreviation: AFM, atomic force microscope.
SMFS offers one of the few means available to probe the full folding energy landscape experimentally. The key to this success is that SMFS allows the trajectory of a single molecule to be observed directly at high resolution along the entire length of the folding reaction coordinate. Numerous trajectories can be observed for a given molecule, providing sufficient statistics to determine the energy of the molecule throughout the folding pathway. Landscape profiles have been reconstructed, with varying degrees of success, using each of the measurement modalities described above, but the specific characteristics of the different modalities require somewhat different analytical methods.
Landscape Reconstructions from Equilibrium Measurements of Extension Trajectories
Conceptually, the simplest method for reconstructing energy landscape profiles is based on obtaining the probability distribution of the molecular extension x, P(x), from equilibrium constant-force measurements (109). The desired landscape profile is the equilibrium free-energy function G(x), which is found directly from P(x) by inverting the Boltzmann relation. To within a constant,
| 1. |
where kBT is the thermal energy. Woodside et al. (109) demonstrated this approach using high-resolution optical trapping measurements of the folding of DNA hairpins as a model system. The extension record shown in Figure 3a yielded P(x) (Figure 3b), and hence G(x) (Figure 3c). An essential requirement of this type of reconstruction is that the applied load be held constant over the full temporal bandwidth of the folding process. This requirement is not easily met by instruments using an active force clamp, where the feedback loop closure time (that is, the time required to measure the position and adjust the force in response) tends to be slow compared with the timescale of the structural transitions of interest. However, it can be met by the use of a passive force clamp instead, where the force probe has zero effective stiffness (48).
Figure 3.
Landscape reconstruction from equilibrium extension measurements. (a) The extension trajectory for a DNA hairpin held at constant force fluctuates between folded (low extension) and unfolded (high extension). (b) The distribution of extension values before (black) and after (red) deconvolution to remove the effects of the instrumental compliance. (c) Landscape reconstructed from Equation 1 before (black) and after (red) deconvolution. (d) A DNA hairpin subjected to a harmonic constraint samples a small range of extensions for a given probe position (different colors indicate different probe positions). Metastable intermediates can be seen in the extension histograms (right). Reprinted from Reference 30, copyright 2011, with permission from Elsevier. (e) Landscape reconstructed from panel d, displayed at a constant force of 15 pN.
Reconstructing energy landscapes using the inverse Boltzmann transform (Equation 1) poses some serious technical challenges. The molecule spends the least time of all exploring the tops of any high-energy barriers, making it difficult to accumulate adequate statistics near these significant features of the landscape. The system must have the requisite temporal response to record such brief and rare molecular excursions. Furthermore, a large number of folding trajectories must be measured, placing extraordinary demands on instrumental stability to avoid blurring of the results caused by noise. It is also essential that certain effects of instrumentation and noise on the measurement be understood, because the dynamics of the molecule are convolved with the dynamics of the force probe. If the true distribution of the molecular extension is p(x), and the point-spread function (PSF) of the probe is S(x), then the observed extension distribution is P(x) = S(x) ⊗ p(x). The confounding effect of the force probe may be removed by deconvolution to recover the intrinsic properties of the molecule. In the example above, this was achieved by first measuring the PSF directly for a nonfolding DNA molecule of fixed length tethered between two beads and then applying an iterative nonlinear deconvolution algorithm (Figure 3b) (72). The resulting landscape (Figure 3c) agreed well with the profile anticipated from a model for the hairpin landscape, with no free fitting parameters (109, 110). More recently, Thirumalai and coworkers (57) derived a theoretical approach that models the mechanics of each element in the measurement, permitting deconvolutions to be performed even in cases where it may not be practical to measure the instrumental PSF directly.
A similar approach to landscape reconstruction can be applied to measurements where the position of the force probe is held constant, rather than the force. The system is again in equilibrium, but force varies linearly with the molecular extension owing to the finite probe stiffness. The changing force complicates interpretation because both the landscape and the PSF are load dependent. However, these effects can be taken into account in the deconvolution, as demonstrated in optical trapping studies of the GCN4 leucine zipper by Rief and colleagues (44).The landscape was reconstructed empirically by performing the deconvolution point wise, using a position-dependent PSF (44), and by applying the model for measurement mechanics of Thirumalai and coworkers (57).
The necessity to deconvolve instrumental effects places practical limits on the accuracy of any reconstructed landscapes. For example, if the probes are compliant and broaden P(x) excessively, it may be difficult to remove instrumental effects fully: As a rule of thumb, sharpening features by more than a factor of 3 or 4 is difficult to achieve reliably by deconvolution (72). To overcome this limitation, stiffer probes may be used. One important source of compliance broadening is the elasticity of the molecular “handles” used to apply loads to the molecule (Figure 2a). Although compliant handles are desirable for accurate measurements of rates (22, 67, 107), stiff handles are better for landscape reconstructions. One way to increase handle stiffness is to use shorter handles (42). An alternative is to replace the handle material with something stiffer, as done by Pfitzner et al. (96), who showed that using rigid DNA origami beams in place of the double-stranded DNA handles typically used in optical trapping experiments sharpened the distributions and improved the reconstruction.
The other primary source of series compliance lies within the force probe itself, namely, the laser trap (for optical tweezers) or the cantilever (for AFM). Here, measurements based on a constant probe position provide an advantage over constant-force measurements with a zero-stiffness probe. For sufficiently stiff probes, the motion of the molecule over its energy landscape is constrained by the harmonic potential imposed, such that only a portion of the range of the reaction coordinate is explored in a given measurement, analogous to umbrella sampling techniques used in simulations (105). This constraint allows the molecule to sample the states near the energy barrier more frequently than would otherwise occur. These are precisely the states that are the most influential for the folding kinetics, but also—normally—the least occupied. La Porta and colleagues (30) showed that harmonically constrained folding trajectories of a DNA hairpin (Figure 3d) led to reliable landscape reconstructions (Figure 3e) and did so significantly faster than constant-force trajectories. Harmonically constrained trajectories should allow reconstruction techniques to be extended to molecules with higher energy barriers, hence slower rates, than might be practical otherwise.
Landscape Reconstructions from Nonequilibrium Force-Ramp Measurements
One limitation of equilibrium-based approaches is that the necessary folding equilibrium is not always attained in experiments. For example, when the energy barrier is high, the folding rate may be so slow that the only practical approach is to observe transitions well away from equilibrium, by ramping or jumping the force. Many such examples have been reported for both proteins (32, 39, 91, 99) and nucleic acids (24, 31, 93, 98). To address such cases, alternative methods for reconstructing landscapes have been developed on the basis of nonequilibrium statistical mechanics.
One of these methods makes use of fluctuation theorems to recover equilibrium free energies from measurements of the nonequilibrium mechanical work performed during force-ramp measurements by the force probe as the molecular structure is perturbed. The essential concept underlying fluctuation theorems, like Jarzynski’s equality (73), is that, even in the presence of dissipation, free energies can be computed from the distribution of values of the mechanical work (19). Hummer & Szabo (64) extended this idea to determine the free energy along the reaction coordinate using data from trajectories where the force probe is displaced at a constant speed to ramp the force and induce the structural transition. Assuming a time-dependent perturbation from the force probe of V = −½ ks[x−z(t)]2, where ks is the effective probe stiffness and z(t) = z0 + υt is the position of the probe (trap or cantilever) moving at a velocity υ, the external work at time t along a trajectory is given by . The free-energy profile at zero force is then
| 2. |
where β = (kBT)−1. Typically, only a small window of the landscape around the equilibrium position is sampled at any given time. Hence, an average over several time slices is used, and the full profile is reconstructed from a weighted histogram average, as
| 3. |
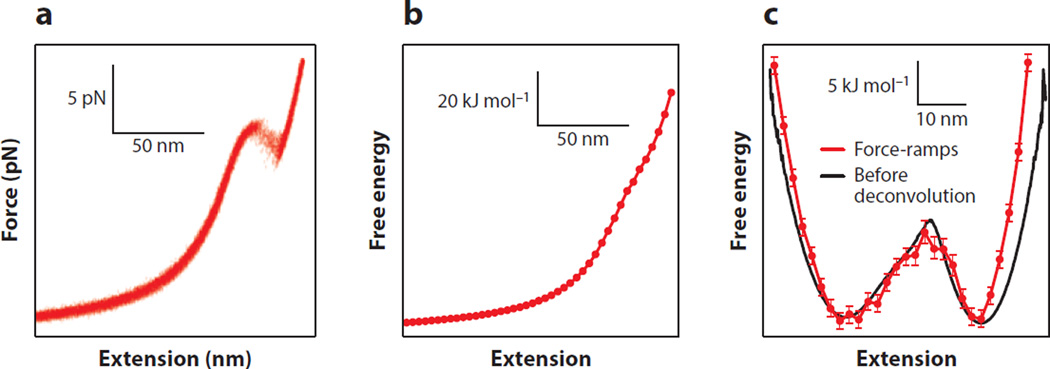
This approach was tested experimentally by measuring DNA hairpins with optical tweezers (51). From a set of force-extension curves (Figure 4a), the profile was reconstructed at zero force (Figure 4b), then tilted by the application of a constant force, which adds energy to the landscape in the amount of ΔG(x) = −F·x, for comparison to a constant-force reconstruction (Figure 4c). This reconstruction agrees well with the constant-force reconstruction obtained before force probe deconvolution (that is, instrumental effects must still be removed). However, the spatial resolution of the reconstruction is somewhat lower, owing to the coarse binning needed to produce well-defined work distributions, particularly near the barrier position. As a general rule, nonequilibrium methods typically require ever more data to recover equilibrium energies as the amount of dissipated work increases (45). In particularly difficult cases, when the transition is far from equilibrium or when the extension change for unfolding is large, the reconstruction may become unreliable or incomplete for some values of the reaction coordinate. Illustrating this challenge, a reconstruction of the landscape for the protein titin from AFM unfolding curves reported that the profile was unreliable in the region immediately beyond the barrier (56).
Figure 4.
Landscape reconstruction from force-ramp measurements. (a) Multiple force-extension curves of a DNA hairpin showing unfolding at ~14 pN. (b) The free-energy landscape at zero force reconstructed from the force ramps using Equation 3 is dominated by the stretching energy of the handles. (c) The landscape from the force ramps (red), tilted to 14 pN, agrees well with the profile from a constant-force measurement of the same hairpin (black) before deconvolution. Adapted from Reference 51.
An alternate strategy for reconstructing landscape profiles from force-ramp data that may alleviate the foregoing problem was proposed by Hummer & Szabo (65). Instead of proceeding directly from force-extension curves to the energy landscape as above, they first found the energy as a function of the experimental control parameter, G(z), and then used an inverse Weierstrass transform to recover the desired profile as a function of the reaction coordinate, G(x). This approach eliminates the need to bin the data, as described above. However, to obtain reliable results, the probe stiffness must be high compared to the effective stiffness imposed by the local curvature of any landscape features measured. The method was tested successfully using simulated data, but it has not yet been validated experimentally.
Landscape Reconstructions from Nonequilibrium Force-Jump Measurements
Fluctuation theorems based on continuous changes in the control parameter (19) cannot be used for force-jump measurements, where the changes are discontinuous. Zhang et al. (116) developed an alternative approach applicable to such measurements, under the assumption of overdamped Langevin dynamics, which was based on inverting the nonequilibrium stationary probability density. Folding trajectories, starting in the unfolded state at xU and ending at xF, sample the probability density ρNE(x), obtained from integrating the Fokker–Planck equation associated with the Langevin dynamics, according to
| 4. |
for xF < x < xU, and ρNE(x) = ρNE(xU) for x ≥ xU. Here, α is a normalization constant, and D(x) is the coefficient for diffusion along the landscape profile. The landscape is then given by
| 5. |
for xF < x < xU. Comparing to Equation 1, the second term in Equation 5 corrects the free energy for the bias in the probability density arising from being out of equilibrium.
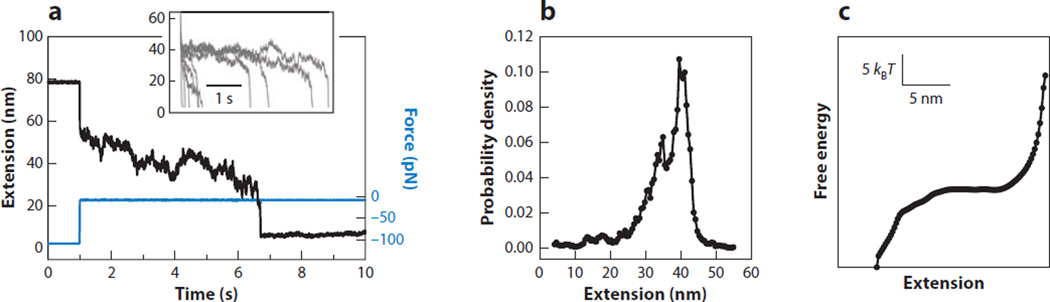
Brujić and colleagues (77) demonstrated this method using force-jump refolding trajectories of the protein ubiquitin measured with an AFM (Figure 5a). The reconstructed landscape (Figure 5b) was found to disagree with a previous model for polyubiquitin (8), but it matched better to a type of model originally proposed for RNA folding (66).Note that Equation 5 requires knowledge of D(x), which is difficult to obtain experimentally. However, it becomes independent of D whenever this diffusion coefficient is spatially invariant, an assumption often made in the literature for simplicity, despite being generally incorrect (12). Moreover, the properties of the force probe almost certainly affect any reconstruction and need to be removed (e.g., via deconvolution), although this issue has not yet been confronted. Another potential complication is that qualitatively different folding pathways may be probed during rapid force quenches, when compared to measurements performed closer to equilibrium (66), and therefore the reconstructed landscapes may not be directly comparable.
Figure 5.
Landscape reconstruction from force-jump measurements. (a) Upon a decrease of the force to near zero (blue), the extension of a polyubiquitin molecule (black) decreases as it folds. Different folding curves at 10 pN show folding over different times (inset). (b) Extension distribution of the nonequilibrium trajectories from Equation 4. (c) Free-energy profile from Equation 5. Reprinted with permission from Reference 77. Copyright 2013 by the American Physics Society. Available online at http://link.aps.org/abstract/PRL/v110/e128301.
MODEL-DEPENDENT APPROXIMATIONS FOR KEY FEATURES OF LANDSCAPES
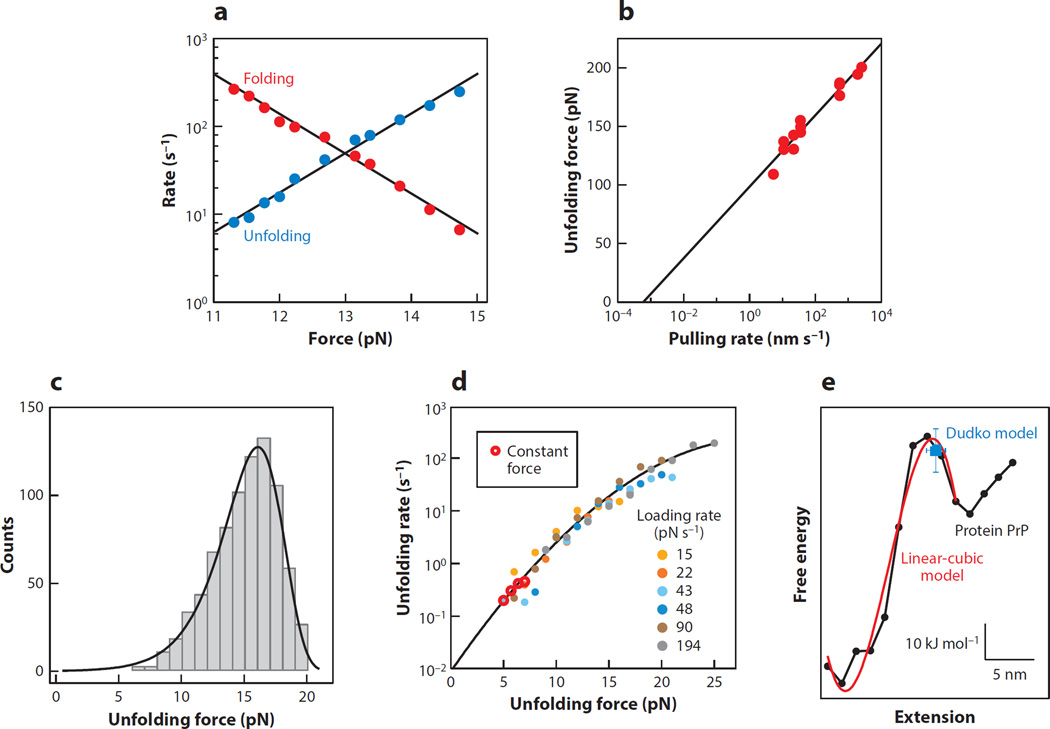
In addition to reconstructing landscapes using model-free approaches, various methods have been developed for characterizing the key features of landscapes using model-dependent approximations. A classic approach is the Bell–Zhurkov model for a force-dependent barrier height. In this model, proposed by Bell (6) and based on earlier work by Zhurkov (118), the effect of the force is simply to change the height of the barrier by an amount F·Δx‡, where Δx‡ is the distance to the barrier along the reaction coordinate from the initial state of the molecule. The rate for barrier crossing thus varies exponentially with load, as k(F) = k0exp[−β(ΔG0‡−F·Δx‡)], where k0 and ΔG0‡ are the rate and barrier height at zero force, respectively, allowing k0 and Δx‡ to be found from the force dependence of the rates. Purely exponential behavior is often seen in rates measured at constant force (Figure 6a). However, the model neglects the fact that the barrier actually moves with force, thereby changing Δx‡. As a consequence, it is generally only appropriate for determining Δx‡ from a comparatively narrow range of forces (where any small motions of the barrier are negligible), and the rate extrapolated to zero force, k0, tends to be somewhat unreliable.
Figure 6.
Model-dependent approximations of the landscape profile. (a) Over a narrow force range, the rates for an RNA hairpin from constant-force measurements are described well by the Bell–Zhurkov model with an exponential force dependence. (b) The unfolding force of titin found from force ramps varies logarithmically with the pulling speed, according to the Evans model (Equation 7). Adapted from Reference 97. Reprinted with permission from AAAS. (c) The unfolding force distribution of a riboswitch aptamer from force ramps is well described by the Dudko model (Equation 8). Adapted from Reference 47. (d) The force-dependent unfolding rates of the riboswitch aptamer, found from force ramps at different loading rates, lie on a single curve, agreeing with constant-force results but covering a larger range of forces. The results are well described by the Dudko model (Equation 9). Data from Reference 47, figure courtesy of O. Dudko. (e) The landscape reconstructed from force ramps of the protein PrP (black) is well described by a linear-cubic model (red), and the Dudko model (blue) recovers the barrier height and location. Adapted from Reference 112.
Evans&Ritchie (41) improved the simple Bell–Zhurkovmodel, applying Kramers’ theory (75) to derive an expression for the distribution of forces for unfolding a molecule using a force ramp. Their unfolding force distribution, p(F), is
| 6. |
where r is the ramp (loading) rate. Typically, the barrier distance determines the width of the distribution (wider for smaller Δx‡), whereas k0 determines the forces needed for a given loading rate. Of particular interest is the expression for the most probable unfolding force, Fmp,
| 7. |
which increases logarithmically with the loading rate. This model has been applied widely to dynamic force spectroscopy measurements. One example, for the unfolding of the protein titin (97), is shown in Figure 6b. Nevertheless, the Evans–Ritchie model also assumes that the energy barrier does not move under load.
This formalism was extended by Dudko et al. (36) to include a barrier that moves with force, deriving closed-form analytical expressions for the unfolding force distribution for two distinct landscape profiles:
| 8. |
where
| 9. |
Here, ν parameterizes the shape of the landscape, with ν = 2/3 for a linear-cubic potential, and ν = 1/2 for a cusp; ν = 1 recovers Equation 6. An application of the Dudko model is illustrated in Figure 6c for force-ramp measurements of the energy landscape for a riboswitch aptamer (47).
Dudko et al. (37) subsequently showed how the force distributions from force-ramp measurements may be transformed, in a model-independent fashion, into force-dependent rates, and thereby mapped onto the rates obtained in constant-force measurements. The rates from measurements acquired at different pulling speeds all collapse onto a single master curve, the same curve as for the rates from constant-force measurements, as illustrated for the riboswitch aptamer in Figure 6d (47).Notably, load-dependent rates can typically be determined over a wider range of forces by using force ramps than from constant-force trajectories because different pulling speeds can be used. As illustrated in Figure 6d, the rates are no longer simply exponential with load (compare to Figure 6a), but they instead display curvature in the semilogarithmic plot, reflecting a reduction of Δx‡ as force increases.
The methods described in this section are typically easier to implement than full-up reconstruction approaches and therefore have enjoyed much wider application in practice, but they do involve model-dependent approximations. To assess the validity of certain of these approximations (e.g., the assumption of a particular barrier shape in the Dudko model), one can compare model-dependent results to full profile reconstructions. Such a comparison was recently made for the protein PrP (Figure 6e), which has a two-state native folding pathway (113). The full landscape, reconstructed from force-ramp measurements, was found to be well approximated by a linear-cubic barrier profile and agreed (within error) with the barrier position and height found from the Dudko model in this limit (112).
The methods described above involve characterizing the landscape mainly by identifying the barrier height and position and, optionally, using the general barrier shape. However, finer-scale features of the landscape can also play significant roles in folding. One notable example is the roughness of the landscape surface, which can reflect so-called internal friction in the polymer (3, 12, 52, 54). Such friction affects the effective diffusion coefficient for trajectories over landscapes and hence the rates of structural changes. Extending earlier work by Zwanzig (120), Hyeon & Thirumalai (68) showed that roughness of the landscape can lead to non-Arrhenius mechanical unfolding rates. In the context of force-ramp measurements, the most probable unfolding force increases with the roughness (for a given loading rate, r) and also varies with temperature, T, allowing the roughness to be estimated from measurements of Fmp at different values of r and T. After accounting for temperature-induced changes in Δx‡ (89), this method was used to measure a roughness factor of 4–6 kBT in the folding of bacteriorhodopsin helices (71) and titin (103). In some cases, however, a Gaussian-distributed roughness does not appear to explain the experimental observations. For example, force-jump measurements of polyubiquitin unfolding found a power-law distribution for the unfolding rates, suggesting a more complex, “glassy” landscape with a distribution of barrier heights (17).
MULTIPLE PATHWAYS AND MULTIPLE STATES
Many of the examples presented above involve simple two-state systems, where the molecule is mainly either folded or unfolded. However, many biomolecules exhibit multistate folding. There may be multiple metastable intermediates found along a single folding pathway, as has been reported for some riboswitch aptamers (4, 47), the GCN4 leucine zipper (44), and a membrane fusion protein (43). In other cases, there may be multiple folding pathways, whether leading to the native state or to misfolded states, as found for some riboswitch aptamers (88), RNA pseudoknots (24, 98) and hairpins (1, 79), calmodulin (101), and the prion protein PrP (113). The presence of multiple states and/or multiple pathways raises important issues that must be considered when reconstructing folding landscapes.
First, it is essential to determine the overall kinetic scheme, i.e., what transitions exist and how the different states are connected by these to form pathways. Such an analysis is best undertaken from measurements made close to equilibrium, because nonequilibrium measurements tend to select against pathways with slower rates, and they may obscure sequential states protected by high barriers. The kinetic scheme can be found by identifying different states present in the trajectories (e.g., via their extensions and lifetimes) and analyzing the transitions between the different states. A variety of methods have been applied to identify states and transitions in single-molecule records, including thresholding and step-finding algorithms (21, 106) and hidden Markov models (5, 14). Hidden Markov analysis, in particular, is predicated on the assumption of a particular kinetic scheme; the scheme that best fits the data must therefore be identified. A new approach based on signal-pair correlation analysis has also been demonstrated (58); it considers the correlations between specific ranges of the signal, producing multiple correlation functions that are fit simultaneously to functional forms expected from different schemes to determine the most appropriate one in a way that is robust against experimental noise (115).
If all the states happen to lie along a single folding pathway, then the landscape can be reconstructed using one of the methods outlined in the first section. Such an approach has already been demonstrated, for example, for a multistate DNA hairpin (109) and the GCN4 leucine zipper (44, 57). However, the sequential application of two-state models (36, 37, 41) to build up a piecewise picture of the landscape is not guaranteed to produce correct results, particularly when using force ramps, because the unfolding forces associated with the first (lower-force) transitions typically affect those of the later (higher-force) transitions. Put another way, only when there is no correlation between the unfolding forces of sequential transitions, that is, when such transitions can be treated as independent two-state processes, is a piecewise reconstruction valid (117). To treat the more general case, Zhang & Dudko (117) recently introduced a method for transforming the transitions in multistate force-ramp records into a map of the microscopic rates for each possible transition. The force dependence of each of the microscopic rates can then, in principle, be fitted to a model (such as Equation 9) to characterize each of the barriers in the landscape.
For the case of systems with multiple folding pathways, the contributions from the various pathways must somehow be separated from one another before the landscape for a given pathway can be reconstructed because all pathways get projected onto the identical reaction coordinate. This separation is nontrivial, and no general solution has been identified. One specific approach to the problem was demonstrated in a recent study of the prion protein, PrP, which can fold into nonnative states, but only from its unfolded state (113). Force ramps starting in the native state induce the molecule to unfold along the native pathway, whereas misfolding subsequent to native unfolding can be suppressed by choosing a high pulling speed, thereby isolating the native folding pathway experimentally (112). It should be noted, however, that, when measurements start too far from equilibrium (e.g., with large force jumps), the molecule may sample pathways that would normally be very improbable, owing to their high energy, complicating any interpretations.
ENERGY-LANDSCAPE ANALYSIS OF FOLDING
Once armed with experimentally determined landscapes, items such as folding/misfolding events, structure/function relationships, and connections among the various features of the landscape can be probed with greater understanding. Here, we highlight some applications that relate to key biophysical aspects of the folding process. Perhaps most fundamental of all is an ability to test the premise that folding is well described as diffusion over an energy landscape by demonstrating that the observed kinetics can be correctly recovered from the profile using Kramers’ theory. Yu et al. (112) did just that for the native folding pathway of the protein PrP. According to Kramers (75), the rate of diffusive escape over an energy barrier is
| 10. |
where κw is the stiffness (curvature) of the potential well, κb the stiffness of the barrier, and D the diffusion coefficient over the barrier. Using values for κw, κb, and ΔG‡ as a function of force found by tilting the reconstructed landscape, the measured rates for both folding and unfolding could be fit globally over several orders of magnitude with the single parameter, D.
The ability to determine D over a barrier is a very useful feature of SMFS measurements. Most studies have been limited to determining the value of D in the unfolded potential well (25, 53, 85), but both theory (12) and experiment (15, 85) suggest that D should decrease substantially from the unfolded well to the barrier because of restrictions in conformational flexibility caused by intrachain interactions as the transition state forms. D can be estimated not only from landscape reconstructions, via Equation 10, but also from approximations, such as in Equation 9, which implies (for the case of a linear-cubic potential):
| 11. |
Such approaches have been used to estimate D for the folding of proteins (112) as well as for nucleic acids, such as DNA hairpins, RNA pseudoknots, and a riboswitch aptamer (87), with values in the range of 10−14 to 10−12 m2/s. When using landscape reconstructions to determine D, however, it is essential to remove first the effects of the force probe by deconvolution; otherwise, Equation 10 yields an effective D that is dominated by the properties of the probe and that may be too low by orders of magnitude. For example, for data corresponding to the DNA hairpin folding presented in Figure 3, the landscape before deconvolution suggestsD = 5 × 10−15, but afterward, D = 5 × 10−13. A similar effect likely led to an underestimate of D (reported as 10−16–10−15 m2/s) in AFM force-jump measurements of protein relaxation times (9) and refolding landscapes (77).
An additional property of folding that can be explored via landscapes is the transition path time. Transition paths are the microscopic paths taken across a barrier during a given structural transition; these are of special interest because they contain information about the folding mechanism. However, they tend to be extremely brief, hence difficult to measure (27, 28). In contrast to the transition rate, which is dominated by ΔG‡, the transition path time, τtp, is insensitive to ΔG‡ and is dominated instead by D. In the high-barrier limit,
| 12. |
where γ is Euler’s constant (23, 27, 63). Using landscape reconstructions for the protein PrP, τtp was found to be ~2 µs (112), consistent with values found in the handful of experimental (28) and computational (100) studies that have been done. Slightly longer transition times were found for nucleic acids, ~10 µs, with a linear dependence on length for the zippering of a double helix (87). It should be noted, however, that such indirect determinations of τtp assume the validity of Equation 12, which has yet to be tested experimentally.
INTERPRETATION OF RECONSTRUCTED LANDSCAPES
Energy landscapes reconstructed from SMFS have enjoyed notable success in explaining folding phenomena using the simple physical picture of diffusion over 1D profiles. However, the best way to interpret such landscapes remains an important question. One issue that has attracted controversy concerns the folding energy barrier associated with the reconstructed profile. The physical distance to this barrier, Δx‡, contains important clues about the structural basis for the folding transition and has been the focus of several studies. For proteins, Δx‡ has been found to range from just a few angstroms (32, 78, 97, 99) to several nanometers (43, 44, 101", 112), with the longer distances tending to correspond to helical proteins. For nucleic acids, a similarly wide range has been reported (4, 24, 31, 47, 81, 82, 88, 93, 98, 109", 110), with the long distances tending to occur when there is no appreciable tertiary structure. In certain cases, Δx‡ can change abruptly as a function of load, owing to a change in the nature of the barrier (82). Structural evolution from a molten globule to native state can also affect Δx‡, with the molten globule being significantly more compliant (39). The barrier distance also depends on the experimental geometry of the pulling force, with shearing geometries yielding smaller values than unzipping geometries for both proteins (16, 70) and nucleic acids (76).
It is tempting to interpret Δx‡ in terms of the physical structure of the transition state, but this interpretation assumes that the reaction coordinate (here, the molecular extension, x) is “good” in that the dynamics along this coordinate capture, in some physical sense, the dynamics of the many conformational degrees of freedom that are projected onto it. The suitability of x as a reaction coordinate can be tested by means of the splitting probability, pfold(x), which represents the fractional chance that a trajectory starting at a given x value will reach the folded state before the unfolded state (34). For a good reaction coordinate, pfold ~ 0.5 for trajectories that start at the very top of the barrier, indicating that this barrier represents a proper separatrix between states. Morrison et al. (84) found that x is indeed a good reaction coordinate for certain DNA hairpins with two-state folding behavior, but the outcome is less clear-cut for a DNA hairpin with three states or for a three-state protein. In the case of the three-state protein, x is better as a reaction coordinate for one of the two structural transitions than for the other, highlighting the fact that the quality of the projection may differ not only among molecules but also within a given molecule. A complementary test is to compare values of pfold(x) computed from the measured trajectories to those derived from the landscape; agreement between these is a necessary condition for a good coordinate. One such comparison for a DNA hairpin (26) did not find particularly good agreement, although the agreement improved considerably with a position-dependent diffusion coefficient. Instrumental effects that alter the apparent landscape were also not taken into account.
Fernández and colleagues (7) challenged the relevance of landscapes reconstructed from equilibrium measurements of “hopping” between states (Figure 2b,c) by claiming that these do not properly probe the position of the folding barrier but reflect instead a downhill collapse of the unfolded polymer. In their representation, the folding energy landscape is originally barrier free at zero force. A so-called entropic barrier then appears as the entire landscape is tilted by the application of an external load, but the appearance of this particular form of barrier is a consequence of the applied force and not some intrinsic property of the folding molecule. On this basis, they concluded that the folding barriers—and the barrier positions—reported in earlier SMFS work were simply experimental artifacts. Although barrier-free folding (analogous to a system that has crossed the spinodal curve in phase space) may indeed occur in special cases (38), most folding is thought to involve a true barrier crossing. An alternative explanation for their controversial conclusion is that an apparently barrier-free profile at zero force can arise as a consequence of a “bad” projection onto the reaction coordinate, namely, one where the true barrier overlaps with the low-energy states. By expanding the picture to include information from a second, “good” reaction coordinate in a two-dimensional (2D) landscape projection, Dudko et al. (35) refuted the conclusions of Fernández and coworkers, showing that the force-dependent kinetics can indeed still probe the true zero-force barrier even when it is hidden because of such a bad projection. Compliant transition states that stretch easily along x (those with large Δx‡) were predicted to result in the 1D projection being trustworthy over a range of external forces, whereas the barrier itself may remain hidden at low loads, particularly for certain brittle barriers (small Δx‡).
The inclusion of a second dimension in the landscape projection can also account for kinetics where the force dependence does not match the expected pattern of Equation 9. Stepwise changes in the semilogarithmic slopes of force-dependent unfolding rates have often been interpreted in terms of probing different structural barriers under varying force regimes, implying a switch with force in the position of the barrier, Δx‡ (70, 82). Perhaps even more striking are rates that vary nonmonotonically with force (50, 83), which have been attributed to the existence of competing pathways (40, 95), including so-called catch bonds. However, Suzuki & Dudko (102) showed that such complex force dependence could result instead from the presence of a single barrier, provided that the rates involved are governed by dynamics along more than one reaction coordinate. This finding suggests a need for considerable caution when interpreting complex behavior in the force-dependent kinetics. Measurements that go beyond simple 1D projections may help clarify the picture and generally supply deeper insights into the folding landscape. For example, force probes may be attached at different locations on the molecule to generate varied projections and test competing explanations (70, 114). Combining force probes with other reporters, such as fluorophores for Förster resonance energy transfer attached to the biomolecule of interest, allows multiple internal coordinates to be monitored simultaneously (29, 59–61, 76) and holds promise for more detailed studies of multidimensional landscapes in the future.
These considerations lead to the more general question of how results from SMFS relate to results obtained from other assays. Direct comparisons of landscapes are not generally meaningful because of essential differences between the reaction coordinates and therefore the projections used. However, certain specific features can be compared. The free-energy change upon folding, as measured by SMFS, often agrees well with ensemble measurements that employ chemical or thermal denaturation, and in some cases, noticeable improvements in accuracy have been achieved (62).However, comparisons of energy barrier properties are less clear-cut. In some cases, the barrier height and even its position along the reaction coordinate are consistent with results obtained by traditional methods, as in studies of DNA hairpins (82, 110) and riboswitches (47), although recent work suggests that treatments of hairpin folding under tension may be somewhat simplistic (80). In other cases, even when the SMFS rates have agreed when extrapolated to zero force (20), molecular simulations and ϕ-analysis suggest that different barriers—and pathways—are probed by force and by chemical denaturants, as shown for the β-sandwich proteins titin (11, 108) and fibronectin (90). Some of these differences may be explained by a switch between different unfolding mechanisms in the low- and high-force regimes or by the influence of suboptimal pulling coordinates (13, 46). Such effects are likely to be most acute for brittle molecules, such as titin and fibronectin, which only unfold at high forces. That said, given the different effects at the molecular level between the various experimental modes of denaturation (mechanical, chemical, thermal, etc.), it is perhaps not surprising that discrepancies may occur. Indeed, the complementary nature of the information gleaned from various modes of denaturation suggests that simultaneously probing the effects of force and temperature, or force and chaotropic agents, could offer an even more powerful tool for characterizing complex landscapes (69, 74).
SUMMARY POINTS.
The profile of the folding landscape can be reconstructed from measurements of a single molecule placed under controlled tension.
Both equilibrium and nonequilibrium measurements can be used to reconstruct landscapes.
The properties of the measurement apparatus influence the reconstruction and should be removed by deconvolution.
Model-dependent approximations are effective at recovering critical properties of the landscape, such as the barrier height and position as well as the roughness of the landscape.
Landscape reconstructions allow otherwise elusive properties of folding, such as the configurational diffusion coefficient and the transition time for structural changes, to be determined.
Issues such as multiple pathways, reaction coordinate quality, and the effects of landscape multidimensionality are complicating factors in the interpretations of landscape reconstructions.
ACKNOWLEDGMENTS
We thank J. Brujić and A. La Porta for assistance with figure preparation, and we thank O. Dudko for creating Figure 6d.
Glossary
- Folding landscape
the multidimensional hypersurface describing the free energy of a biomolecule as a function of its conformation
- Reaction coordinate
a one-dimensional coordinate, which is used to describe the progress of a particular reaction along the landscape profile
- Landscape profile
the projection of the energy hypersurface onto a low-dimensional space, often 1D
- Single-molecule force spectroscopy (SMFS)
a class of assays probing the response of a single molecule to an applied load
- Atomic force microscope/microscopy (AFM)
an apparatus (or its use) for applying and measuring forces with a sharp tip attached to a compliant cantilever
- Optical tweezers
an apparatus for applying and measuring forces on small dielectric objects using optical gradient forces
- Feedback loop closure time
the time required to measure the signal of interest, respond to changes, and restore the desired set point
- Point-spread function (PSF)
the distribution of values expected from an ideal, single-valued signal owing to experimental effects such as noise
- Compliance
the extent to which an object deforms (extends) in response to an applied force
- Fluctuation theorems
a class of theorems relating the nonequilibrium properties of a system to equilibrium characteristics via statistical fluctuations
- Loading rate
the rate of change of the applied load
- Transition path
the microscopic path through conformational space taken as a molecule crosses the energy barrier for folding
- Compliant molecule
a molecule that undergoes substantial deformation before unfolding, exhibiting a large distance to the barrier, Δx‡
- Brittle molecule
a molecule that undergoes minimal deformation before unfolding, exhibiting a comparatively small distance to the barrier, Δx‡
- Förster resonance energy transfer
the distance-sensitive transfer of energy between nearby fluorophores, often used to monitor conformational changes
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Michael T. Woodside, Email: michael.woodside@ualberta.ca.
Steven M. Block, Email: sblock@stanford.edu.
LITERATURE CITED
- 1.Alemany A, Mossa A, Junier I, Ritort F. Experimental free-energy measurements of kinetic molecular states using fluctuation theorems. Nat. Phys. 2012;8:688–694. [Google Scholar]
- 2.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 3.Ansari A, Jones CM, Henry ER, Hofrichter J, Eaton WA. The role of solvent viscosity in the dynamics of protein conformational changes. Science. 1992;256:1796–1798. doi: 10.1126/science.1615323. [DOI] [PubMed] [Google Scholar]
- 4.Anthony PC, Perez CF, Garcia-Garcia C, Block SM. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc. Natl. Acad. Sci. USA. 2012;109:1485–1489. doi: 10.1073/pnas.1115045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball FG, Rice JA. Stochastic models for ion channels: introduction and bibliography. Math. Biosci. 1992;112:189–206. doi: 10.1016/0025-5564(92)90023-p. [DOI] [PubMed] [Google Scholar]
- 6.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 7.Berkovich R, Garcia-Manyes S, Klafter J, Urbakh M, Fernández JM. Hopping around an entropic barrier created by force. Biochem. Biophys. Res. Commun. 2010;403:133–137. doi: 10.1016/j.bbrc.2010.10.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkovich R, Garcia-Manyes S, Urbakh M, Klafter J, Fernández JM. Collapse dynamics of single proteins extended by force. Biophys. J. 2010;98:2692–2701. doi: 10.1016/j.bpj.2010.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkovich R, Hermans RI, Popa I, Stirnemann G, Garcia-Manyes S, et al. Rate limit of protein elastic response is tether dependent. Proc. Natl. Acad. Sci. USA. 2012;109:14416–14421. doi: 10.1073/pnas.1212167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Best RB. Atomistic molecular simulations of protein folding. Curr. Opin. Struct. Biol. 2012;22(1):52–61. doi: 10.1016/j.sbi.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Best RB, Fowler SB, Herrera JLT, Steward A, Paci E, Clarke J. Mechanical unfolding of a titin Ig domain: structure of transition state revealed by combining atomic force microscopy, protein engineering and molecular dynamics simulations. J. Mol. Biol. 2003;330:867–877. doi: 10.1016/s0022-2836(03)00618-1. [DOI] [PubMed] [Google Scholar]
- 12.Best RB, Hummer G. Coordinate-dependent diffusion in protein folding. Proc. Natl. Acad. Sci. USA. 2010;107:1088–1093. doi: 10.1073/pnas.0910390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Best RB, Paci E, Hummer G, Dudko OK. Pulling direction as a reaction coordinate for the mechanical unfolding of single molecules. J. Phys. Chem. B. 2008;112:5968–5976. doi: 10.1021/jp075955j. [DOI] [PubMed] [Google Scholar]
- 14.Blanco M, Walter NG. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 2010;472:153–178. doi: 10.1016/S0076-6879(10)72011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borgia A, Wensley BG, Soranno A, Nettels D, Borgia MB, et al. Localizing internal friction along the reaction coordinate of protein folding by combining ensemble and single-molecule fluorescence spectroscopy. Nat. Commun. 2012;3:1195. doi: 10.1038/ncomms2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockwell DJ, Paci E, Zinober RC, Beddard GS, Olmsted PD, et al. Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat. Struct. Biol. 2003;10:731–737. doi: 10.1038/nsb968. [DOI] [PubMed] [Google Scholar]
- 17.Brujić J, Hermans RI, Walther KA, Fernández JM. Single-molecule force spectroscopy reveals signatures of glassy dynamics in the energy landscape of ubiquitin. Nat. Phys. 2006;2:282–286. [Google Scholar]
- 18.Buchner J, Kiefhaber T, editors. Protein Folding Handbook. Weinheim: Wiley-VCH; 2005. [Google Scholar]
- 19.Bustamante C, Liphardt J, Ritort F. The nonequilibrium thermodynamics of small systems. Phys. Today. 2005;58:43–48. [Google Scholar]
- 20.Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter BC, Vershinin M, Gross SP. A comparison of step-detection methods: How well can you do? Biophys. J. 2008;94:306–319. doi: 10.1529/biophysj.107.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang J-C, de Messieres M, La Porta A. Effect of handle length and microsphere size on transition kinetics in single-molecule experiments. Phys. Rev. E. 2013;87:012721. doi: 10.1103/PhysRevE.87.012721. [DOI] [PubMed] [Google Scholar]
- 23.Chaudhury S, Makarov DE. A harmonic transition state approximation for the duration of reactive events in complex molecular rearrangements. J. Chem. Phys. 2010;133:034118. doi: 10.1063/1.3459058. [DOI] [PubMed] [Google Scholar]
- 24.Chen G, Wen JD, Tinoco I., Jr Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA. 2007;13:2175–2188. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Parrini C, Taddei N, Lapidus LJ. Conformational properties of unfolded HypF-N. J. Phys. Chem. B. 2009;113:16209–16213. doi: 10.1021/jp904189b. [DOI] [PubMed] [Google Scholar]
- 26.Chodera JD, Pande VS. Splitting probabilities as a test of reaction coordinate choice in single-molecule experiments. Phys. Rev. Lett. 2011;107:098102. doi: 10.1103/PhysRevLett.107.098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung HS, Louis JM, Eaton WA. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc. Natl. Acad. Sci. USA. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung HS, McHale K, Louis JM, Eaton WA. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335:981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comstock MJ, Ha T, Chemla YR. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat. Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Messieres M, Brawn-Cinani B, La Porta A. Measuring the folding landscape of a harmonically constrained biopolymer. Biophys. J. 2011;100:2736–2744. doi: 10.1016/j.bpj.2011.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Messieres M, Chang J-C, Brawn-Cinani B, La Porta A. Single-molecule study of G-quadruplex disruption using dynamic force spectroscopy. Phys. Rev. Lett. 2012;109:058101. doi: 10.1103/PhysRevLett.109.058101. [DOI] [PubMed] [Google Scholar]
- 32.Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu. Rev. Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du R, Pande VS, Grosberg AY, Tanaka T, Shakhnovich ES. On the transition coordinate for protein folding. J. Chem. Phys. 1998;108:334–350. [Google Scholar]
- 35.Dudko OK, Graham TGW, Best RB. Locating the barrier for folding of single molecules under an external force. Phys. Rev. Lett. 2011;107:208301. doi: 10.1103/PhysRevLett.107.208301. [DOI] [PubMed] [Google Scholar]
- 36.Dudko OK, Hummer G, Szabo A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys. Rev. Lett. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- 37.Dudko OK, Hummer G, Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. USA. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyer RB. Ultrafast and downhill protein folding. Curr. Opin. Struct. Biol. 2007;17:38–47. doi: 10.1016/j.sbi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Elms PJ, Chodera JD, Bustamante C, Marqusee S. The molten globule state is unusually deformable under mechanical force. Proc. Natl. Acad. Sci. USA. 2012;109:3796–3801. doi: 10.1073/pnas.1115519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl. Acad. Sci. USA. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forns N, de Lorenzo S, Manosas M, Hayashi K, Huguet JM, Ritort F. Improving signal/noise resolution in single-molecule experiments using molecular constructs with short handles. Biophys. J. 2011;100:1765–1774. doi: 10.1016/j.bpj.2011.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gebhardt JCM, Bornschloegla T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc. Natl. Acad. Sci. USA. 2010;107:2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gore J, Ritort F, Bustamante C. Bias and error in estimates of equilibrium free-energy differences from nonequilibrium measurements. Proc. Natl. Acad. Sci. USA. 2003;100:12564–12569. doi: 10.1073/pnas.1635159100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graham TGW, Best RB. Force-induced change in protein unfolding mechanism: discrete or continuous switch? J. Phys. Chem. B. 2011;115:1546–1561. doi: 10.1021/jp110738m. [DOI] [PubMed] [Google Scholar]
- 47.Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Passive all-optical force clamp for high-resolution laser trapping. Phys. Rev. Lett. 2005;95:208102. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc. Natl. Acad. Sci. USA. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta AN, Vincent A, Neupane K, Yu H, Wang F, Woodside MT. Experimental validation of free-energy-landscape reconstruction from non-equilibrium single-molecule force spectroscopy measurements. Nat. Phys. 2011;7:631–634. [Google Scholar]
- 52.Hagen SJ. Solvent viscosity and friction in protein folding dynamics. Curr. Protein Pept. Sci. 2010;11:385–395. doi: 10.2174/138920310791330596. [DOI] [PubMed] [Google Scholar]
- 53.Hagen SJ, Hofrichter J, Szabo A, Eaton WA. Diffusion-limited contact formation in unfolded cytochrome c: estimating the maximum rate of protein folding. Proc. Natl. Acad. Sci. USA. 1996;93:11615–11617. doi: 10.1073/pnas.93.21.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagen SJ, Qiu LL, Pabit SA. Diffusional limits to the speed of protein folding: fact or friction? J. Phys. Condens. Matter. 2005;17:S1503. [Google Scholar]
- 55.Hänggi P, Talkner P, Borkovec M. Reaction-rate theory: fifty years after Kramers. Rev. Mod. Phys. 1990;62:251–341. [Google Scholar]
- 56.Harris NC, Song Y, Kiang C-H. Experimental free energy surface reconstruction from single-molecule force spectroscopy using Jarzynski’s equality. Phys. Rev. Lett. 2007;99:068101. doi: 10.1103/PhysRevLett.99.068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hinczewski M, Gebhardt JCM, Rief M, Thirumalai D. From mechanical folding trajectories to intrinsic energy landscapes of biopolymers. Proc. Natl. Acad. Sci. USA. 2013;110:4500–4505. doi: 10.1073/pnas.1214051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoffmann A, Woodside MT. Signal-pair correlation analysis of single-molecule trajectories. Angew. Chem. Int. Ed. 2011;50:12643–12646. doi: 10.1002/anie.201104033. [DOI] [PubMed] [Google Scholar]
- 59.Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, et al. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hugel T, Holland NB, Cattani A, Moroder L, Seitz M, Gaub HE. Single-molecule optomechanical cycle. Science. 2002;296:1103–1106. doi: 10.1126/science.1069856. [DOI] [PubMed] [Google Scholar]
- 61.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, et al. Experimental test of connector rotation during DNA packaging into bacteriophage ϕ29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huguet JM, Bizarro CV, Forns N, Smith SB, Bustamante C, Ritort F. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proc. Natl. Acad. Sci. USA. 2010;107:15431–15436. doi: 10.1073/pnas.1001454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hummer G. From transition paths to transition states and rate coefficients. J. Chem. Phys. 2004;120:516–523. doi: 10.1063/1.1630572. [DOI] [PubMed] [Google Scholar]
- 64.Hummer G, Szabo A. Free energy reconstruction from nonequilibrium single-molecule pulling experiments. Proc. Natl. Acad. Sci. USA. 2001;98:3658–3661. doi: 10.1073/pnas.071034098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hummer G, Szabo A. Free energy profiles from single-molecule pulling experiments. Proc. Natl. Acad. Sci. USA. 2010;107:21441–21446. doi: 10.1073/pnas.1015661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyeon C, Morrison G, Pincus DL, Thirumalai D. Refolding dynamics of stretched biopolymers upon force quench. Proc. Natl. Acad. Sci. USA. 2009;106:20288–20293. doi: 10.1073/pnas.0905764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hyeon C, Morrison G, Thirumalai D. Force-dependent hopping rates of RNA hairpins can be estimated from accurate measurement of the folding landscapes. Proc. Natl. Acad. Sci. USA. 2008;105:9604–9609. doi: 10.1073/pnas.0802484105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hyeon C, Thirumalai D. Can energy landscape roughness of proteins and RNA be measured by using mechanical unfolding experiments? Proc. Natl. Acad. Sci. USA. 2003;100:10249–10253. doi: 10.1073/pnas.1833310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hyeon C, Thirumalai D. Multiple probes are required to explore and control the rugged energy landscape of RNA hairpins. J. Am. Chem. Soc. 2008;130:1538–1539. doi: 10.1021/ja0771641. [DOI] [PubMed] [Google Scholar]
- 70.Jagannathan B, Elms PJ, Bustamante C, Marqusee S. Direct observation of a force-induced switch in the anisotropic mechanical unfolding pathway of a protein. Proc. Natl. Acad. Sci. USA. 2012;109:17820–17825. doi: 10.1073/pnas.1201800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janovjak H, Knaus H, Muller DJ. Transmembrane helices have rough energy surfaces. J. Am. Chem. Soc. 2007;129:246–247. doi: 10.1021/ja065684a. [DOI] [PubMed] [Google Scholar]
- 72.Jansson PA. Deconvolution of Images and Spectra. 2nd ed. San Diego: Academic; 1997. [Google Scholar]
- 73.Jarzynski C. Nonequilibrium equality for free energy differences. Phys. Rev. Lett. 1997;78:2690–2693. [Google Scholar]
- 74.Klimov DK, Thirumalai D. Native topology determines force-induced unfolding pathways in globular proteins. Proc. Natl. Acad. Sci. USA. 2000;97:7254–7259. doi: 10.1073/pnas.97.13.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kramers HA. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica. 1940;7:284–304. [Google Scholar]
- 76.Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat. Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lannon H, Haghpanah JS, Montclare JK, Vanden-Eijnden E, Brujić J. Force-clamp experiments reveal the free-energy profile and diffusion coefficient of the collapse of protein molecules. Phys. Rev. Lett. 2013;110:128301. doi: 10.1103/PhysRevLett.110.128301. http://link.aps.org/abstract/PRL/v110/e128301. [DOI] [PubMed] [Google Scholar]
- 78.Li H, Wang H-C, Cao Y, Sharma D, Wang M. Configurational entropy modulates the mechanical stability of protein GB1. J. Mol. Biol. 2008;379:871–880. doi: 10.1016/j.jmb.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 79.Li PTX, Bustamante C, Tinoco I., Jr Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc. Natl. Acad. Sci. USA. 2007;104:7039–7044. doi: 10.1073/pnas.0702137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin J-C, Hyeon C, Thirumalai D. RNA under tension: folding landscapes, kinetic partitioning mechanism, and molecular tensegrity. J. Phys. Chem. Lett. 2012;3:3616–3625. doi: 10.1021/jz301537t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 82.Manosas M, Collin D, Ritort F. Force-dependent fragility in RNA hairpins. Phys. Rev. Lett. 2006;96:218301. doi: 10.1103/PhysRevLett.96.218301. [DOI] [PubMed] [Google Scholar]
- 83.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 84.Morrison G, Hyeon C, Hinczewski M, Thirumalai D. Compaction and tensile forces determine the accuracy of folding landscape parameters from single molecule pulling experiments. Phys. Rev. Lett. 2011;106:138102. doi: 10.1103/PhysRevLett.106.138102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nettels D, Gopich IV, Hoffmann A, Schuler B. Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc. Natl. Acad. Sci. USA. 2007;104:2655–2660. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neupane K, Ritchie DB, Yu H, Foster DAN, Wang F, Woodside MT. Transition path times for nucleic acid folding determined from energy-landscape analysis of single-molecule trajectories. Phys. Rev. Lett. 2012;109:068102. doi: 10.1103/PhysRevLett.109.068102. [DOI] [PubMed] [Google Scholar]
- 88.Neupane K, Yu H, Foster DAN, Wang F, Woodside MT. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011;39:7677–7687. doi: 10.1093/nar/gkr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nevo R, Brumfeld V, Kapon R, Hinterdorfer P, Reich Z. Direct measurement of protein energy landscape roughness. EMBO Rep. 2005;6:482–486. doi: 10.1038/sj.embor.7400403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ng SP, Rounsevell RWS, Steward A, Geierhaas CD, Williams PM, et al. Mechanical unfolding of TNfn3: the unfolding pathway of a fnIII domain probed by protein engineering, AFM and MD simulation. J. Mol. Biol. 2005;350:776–789. doi: 10.1016/j.jmb.2005.04.070. [DOI] [PubMed] [Google Scholar]
- 91.Oberhauser AF, Hansma PK, Carrion-Vazquez M, Fernández JM. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA. 2001;98:468–472. doi: 10.1073/pnas.021321798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q. Rev. Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 93.Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco I, Jr, Bustamante C. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Onuchic JN, Wolynes PG. Theory of protein folding. Curr. Opin. Struct. Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 95.Pereverzev YV, Prezhdo OV, Forero M, Sokurenko EV, Thomas WE. The two-pathway model for the catch-slip transition in biological adhesion. Biophys. J. 2005;89:1446–1454. doi: 10.1529/biophysj.105.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pfitzner E, Wachauf C, Kilchherr F, Pelz B, Shih WM, et al. Rigid DNA beams for high-resolution single-molecule mechanics. Angew. Chem. Int. Ed. 2013;52:7766–7771. doi: 10.1002/anie.201302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rief M, Gautel M, Oesterhelt F, Fernández JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 98.Ritchie DB, Foster DAN, Woodside MT. Programmed−1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc. Natl. Acad. Sci. USA. 2012;109:16167–16172. doi: 10.1073/pnas.1204114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schlierf M, Li H, Fernández JM. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl. Acad. Sci. USA. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 101.Stigler J, Ziegler F, Gieseke A, Gebhardt JCM, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 102.Suzuki Y, Dudko OK. Single-molecule rupture dynamics on multidimensional landscapes. Phys. Rev. Lett. 2010;104:048101. doi: 10.1103/PhysRevLett.104.048101. [DOI] [PubMed] [Google Scholar]
- 103.Taniguchi Y, Brockwell DJ, Kawakami M. The effect of temperature on mechanical resistance of the native and intermediate states of I27. Biophys. J. 2008;95:5296–5305. doi: 10.1529/biophysj.108.141275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 105.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 106.Watkins LP, Yang H. Detection of intensity change points in time-resolved single-molecule measurements. J. Phys. Chem. B. 2005;109:617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- 107.Wen J-D, Manosas M, Li PTX, Smith SB, Bustamante C, et al. Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophys. J. 2007;92:2996–3009. doi: 10.1529/biophysj.106.094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams PM, Fowler SB, Best RB, Toca-Herrera JL, Scott KA, et al. Hidden complexity in the mechanical properties of titin. Nature. 2003;422:446–449. doi: 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]
- 109.Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. USA. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Woodside MT, Garcia-Garcia C, Block SM. Folding and unfolding single RNA molecules under tension. Curr. Opin. Chem. Biol. 2008;12:640–646. doi: 10.1016/j.cbpa.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu H, Gupta AN, Liu X, Neupane K, Brigley AM, et al. Energy landscape analysis of native folding of the prion protein yields the diffusion constant, transition path time, and rates. Proc. Natl. Acad. Sci. USA. 2012;109:14452–14457. doi: 10.1073/pnas.1206190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu H, Liu X, Neupane K, Gupta AN, Brigley AM, et al. Direct observation of multiple misfolding pathways in a single prion protein molecule. Proc. Natl. Acad. Sci. USA. 2012;109:5283–5288. doi: 10.1073/pnas.1107736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu Z, Koirala D, Cui Y, Easterling LF, Zhao Y, Mao H. Click chemistry assisted single-molecule fingerprinting reveals a 3D biomolecular folding funnel. J. Am. Chem. Soc. 2012;134:12338–12341. doi: 10.1021/ja303218s. [DOI] [PubMed] [Google Scholar]
- 115.Yuan H, Orrit M. Reaction pathways from single-molecule trajectories. Chem Phys Chem. 2012;13:681–683. doi: 10.1002/cphc.201100983. [DOI] [PubMed] [Google Scholar]
- 116.Zhang Q, Brujić J, Vanden-Eijnden E. Reconstructing free energy profiles from nonequilibrium relaxation trajectories. J. Stat. Phys. 2011;144:344–366. [Google Scholar]
- 117.Zhang Y, Dudko OK. Complex biomolecular interactions: from mechanical fingerprints to the rate map to activation barriers. Proc. Natl. Acad. Sci. USA. 2013;110:16432–16437. doi: 10.1073/pnas.1309101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhurkov SN. Kinetic concept of the strength of solids. Int. J. Fract. Mech. 1965;1:311–322. [Google Scholar]
- 119.Žoldak G, Rief M. Force as a single molecule probe of multidimensional protein energy landscapes. Curr. Opin. Struct. Biol. 2013;23:48–57. doi: 10.1016/j.sbi.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 120.Zwanzig R. Diffusion in a rough potential. Proc. Natl. Acad. Sci. USA. 1988;85:2029–2030. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]