Abstract
The control of food-borne outbreaks caused by Listeria monocytogenes in humans relies on the timely identification of food or environmental sources and the differentiation of outbreak-related isolates from unrelated ones. This study illustrates the utility of whole-genome sequencing for examining the link between clinical and environmental isolates of L. monocytogenes associated with an outbreak of hospital-acquired listeriosis in Sydney, Australia. Comparative genomic analysis confirmed an epidemiological link between the three clinical and two environmental isolates. Single nucleotide polymorphism (SNP) analysis showed that only two SNPs separated the three human outbreak isolates, which differed by 19 to 20 SNPs from the environmental isolates and 71 to >10,000 SNPs from sporadic L. monocytogenes isolates. The chromosomes of all human outbreak isolates and the two suspected environmental isolates were syntenic. In contrast to the genomes of background sporadic isolates, all epidemiologically linked isolates contained two novel prophages and a previously unreported clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) locus subtype sequence. The mobile genetic element (MGE) profile of these isolates was distinct from that of the other serotype 1/2b reference strains and sporadic isolates. The identification of SNPs and clonally distinctive MGEs strengthened evidence to distinguish outbreak-related isolates of L. monocytogenes from cocirculating endemic strains.
INTRODUCTION
Listeria monocytogenes is an important food-borne pathogen causing life-threatening infections in at-risk populations. Pregnant women, the elderly, and immunocompromised individuals are at high risk for listeriosis, which can present as an invasive systemic infection or gastroenteritis. L. monocytogenes has become one of the major food safety concerns worldwide, due to its ubiquitous presence in the environment. The bacterium can be introduced into food-processing facilities and food products, and it persists for long periods of time, owing to its ability to survive at a wide range of temperatures, including underrefrigeration and in low-pH (∼5.2 to 5.5) environments (1–3). Outbreaks of listeriosis have been associated with the consumption of contaminated ready-to-eat foods, dairy products, seafood, and fresh produce (4–8). Hospital-acquired listeriosis outbreaks were frequently reported worldwide. Eight outbreak cases reported between 1998 and 2010, mostly from major U.S. cities and European countries; all had strong evidence for food-borne acquisition in hospital-implicated foods, and cold sandwiches have been a common food source (9–11); however, in many cases, it was difficult to determine the specific origins and reservoirs of L. monocytogenes (12–14).
Investigations of listeriosis outbreaks often present a challenge to public health because of the relatively long incubation period of the disease. It can vary from 14 days to 6 weeks and can affect the quality of food histories (15). Therefore, the molecular subtyping of clinical and food or environmental L. monocytogenes isolates has played an increasingly important role in establishing epidemiological links and supporting public health investigations. A multiplex PCR molecular serotyping scheme was developed to characterize L. monocytogenes isolates (16); however, this technique is of limited use in outbreaks, due to its poor discriminatory power. Of the 14 serotypes that have been described for L. monocytogenes (17), four major serotypes (1/2a, 1/2c, 1/2b, and 4b) have been involved in 98% of the documented human cases (18, 19). Other subtyping methods have been applied to further characterize L. monocytogenes isolates associated with outbreaks. A pulsed-field gel electrophoresis (PFGE) typing scheme has been adopted by PulseNet as an internationally standardized method (20). PFGE is a relatively inexpensive approach with excellent discriminatory power, but it is labor-intensive, time-consuming, and technically demanding. Sequence-based typing techniques have been developed to enable interlaboratory harmonization and high-throughput processing. A multilocus sequence typing (MLST) assay based on seven housekeeping genes demonstrated consistency with the PFGE typing method (21). A Web-based MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Lmono.html) containing the reference allele sequences was established to assist with the rapid identification of sequence types. MLST is a valuable tool for global epidemiology studies and for studies on the molecular evolution of pathogens. However, it provides insufficient discriminatory power for outbreak investigations. Multilocus variable-number tandem-repeat analysis (MLVA), a method that determines the number of tandem-repeat sequences at different loci in the bacterial genome, has been developed for several food-borne bacteria, including L. monocytogenes. Several MLVA typing schemes, each targeting different numbers of repeat regions ranging from eight to 11 loci, have been applied successfully in outbreak investigations (22–26). As a typing method, MLVA has higher discriminatory power than that of MLST, with its main limitation being that it is not 100% reproducible between laboratories unless the allele amplicons are sequenced, and the beginnings and ends of the amplicons have to be agreed on by different users. This method therefore needs to be better harmonized using a set of strains with known allele sizes at each locus for intralaboratory comparison.
The introduction of next-generation sequencing bench-top analyzers has enabled rapid whole-genome sequencing (WGS) and comparative analysis of L. monocytogenes genomes in a clinically relevant time frame (27, 28). The costs of bacterial WGS continue to decline each year. Currently, the cost of WGS is close to the price of MLST and MLVA carried out by traditional Sanger sequencing (29, 30). WGS is increasingly being used for the detection and characterization of viral and bacterial pathogens during community- and hospital-acquired outbreaks (31–35). It promises the ultimate level of high-resolution molecular typing, with simultaneous characterization of antibiotic resistance determinants and virulence factors (36). The main criterion for determining the relatedness between isolates has been the number of single nucleotide polymorphisms (SNPs) between microbial genomes (30, 37–39). However, the cutoff SNP number to be used varies significantly for different bacterial species or clonal lineages and has been difficult to validate for pathogens with fairly stable genomes, such as L. monocytogenes (35, 40).
WGS offers the opportunity to compare the genomic contents of strains recovered from cases associated with outbreaks and to assist with testing epidemiological hypotheses. Recently, Kuenne et al. (41) performed a genomic comparison of 16 L. monocytogenes strains, including representatives of each of the major serotypes. Analysis of the core and accessory genomes revealed that the core genomes of all isolates were highly syntenic and that strain-specific genes were not spread throughout the chromosome but clustered into only nine defined hot spot regions (41). The other major source of genomic diversity resulted from the varied carriage of mobile genetic elements (MGEs). Thirteen different MGEs, including prophages, genomic islands, transposons, and insertion sequences, were identified at 13 specific chromosomal integration sites. Each of the 16 strains was found to contain between one and five MGEs (41). A subset of strains also contained different subtypes of clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas) genes. These genes are involved in the encoding of diverse proteins to constitute genomic defense barriers, which protect against the invasion of foreign prophages and conjugative plasmids (42).
In this study, we applied WGS to substantiate the epidemiological links between human cases of listeriosis and environmental samples from a company supplying food to three different hospitals involved in a hospital-acquired listeriosis outbreak. Our findings demonstrate that comparative genomic analyses should not focus solely on SNP-based comparisons. The identification of uncommon mobile genetic elements and distinctive genetic loci can improve the resolution of analysis and provide important evidence on the potential relationships between clinical and environmental outbreak isolates.
MATERIALS AND METHODS
Hospital-acquired outbreak.
The detailed epidemiological investigation of this hospital-acquired outbreak was reported in a previous publication (43). Briefly, in April 2013, the Public Health Unit in Sydney, Australia, was notified of three inpatients from three tertiary hospitals within two local health districts in Sydney, which were blood culture positive for L. monocytogenes within an 8-day period. The first reported case was an elderly female patient admitted to hospital A. She had a background of heart failure and chronic obstructive pulmonary disease. A positive blood culture for L. monocytogenes was obtained on 2 April 2013. Case 2 was a middle-age female inpatient of hospital B with myelofibrosis. She had a positive blood culture for L. monocytogenes 4 days after case 1 (6 April 2013). Case 3 was an elderly male with a background of end-stage hepatocellular carcinoma. He had a history of two recent admissions to hospital C and was readmitted on 8 April 2013 following a fall. The blood culture collected on this day was also positive for L. monocytogenes.
Following notifications from different diagnostic laboratories, a public health investigation was initiated in all three hospitals. Common food items consumed by all three cases during the overlapping periods of their admission (20 to 26 March 2013) in each hospital were identified by reviewing the food menus of the hospitals. A chocolate profiterole was identified as the common food consumed by all three cases on the same day (24 March 2013) of their admission. A large food company that supplied all of the hospitals, company X, was identified, and leftover food samples of interest, including chocolate profiteroles, were tested by an approved food test laboratory for the presence of L. monocytogenes. Although the results were all negative, monthly testing records from company X indicated that environmental swabs and a sample from a chocolate profiterole had been positive for Listeria innocua and Listeria species during routine quality screening 3 weeks prior to this outbreak. Subsequent inspection was conducted at the company X premises, and multiple environmental samples from a cool-room bench, floor, and boots were taken from the premises. Listeria species was detected in seven swabs, and L. monocytogenes was cultured from two boot swabs collected in the cool-room food production area.
Bacterial isolates.
Five isolates obtained during the outbreak (three human isolates, L. monocytogenes Lm4664, Lm2128, and Lm4447, and two environmental isolates from the cool room of company X, L. monocytogenes Lm4422 and Lm4424) were included in the study. Four other isolates of L. monocytogenes that were not related to the outbreak were added for comparison. These included three epidemiologically unrelated sporadic human isolates, of which one (L. monocytogenes Lm4370) was recovered from a blood culture during the same time period as the outbreak, another (L. monocytogenes Lm4082) was obtained nearly a year prior to the outbreak, and the third one (L. monocytogenes Lm1414) was obtained 5 months after the outbreak. One isolate (L. monocytogenes Lm3554) obtained from cheese as part of a different public health investigation 2 months prior to the outbreak was also included in the study (Table 1).
TABLE 1.
L. monocytogenes isolates included in the study and relevant subtyping results
| Isolate | Source | Specimen type | Molecular serotype | MLST | BTa | MLVA | PFGE typeb |
|---|---|---|---|---|---|---|---|
| Lm4664 | Case 1 | Blood culture | 1/2b, 3b, 7 | 3 | 223 | 04-17-16-05-03-11-14-00-16 | 4A:4:1 |
| Lm2128 | Case 2 | Blood culture | 1/2b, 3b, 7 | 3 | 223 | 04-17-16-05-03-11-14-00-16 | 4A:4:1 |
| Lm4447 | Case 3 | Blood culture | 1/2b, 3b, 7 | 3 | 223 | 04-17-16-05-03-11-14-00-16 | 4A:4:1 |
| Lm4422 | Cool room | Environmental swab | 1/2b, 3b, 7 | 3 | 223 | 04-17-16-05-03-11-14-00-16 | 4A:4:1 |
| Lm4424 | Cool room | Environmental swab | 1/2b, 3b, 7 | 3 | 223 | 04-17-16-05-03-11-14-00-16 | 4A:4:1 |
| Lm1414 | Sporadic | Blood culture | 1/2b, 3b, 7 | 3 | 158 | 04-17-16-05-03-12-14-00-16 | 4:4A:1 |
| Lm4370 | Sporadic | Blood culture | 1/2b, 3b, 7 | 3 | 222 | 04-17-15-05-03-10-14-00-16 | 4:4A:1 |
| Lm4082 | Sporadic | Blood culture | 4b, 4d, 4e | 1 | 255 | 03-16-12-05-03-05-15-00-18 | 86A:46A:37 |
| Lm3554 | Cheese | Food swab | 4b, 4d, 4e | 1 | 255 | 03-16-14-06-03-05-15-00-18 | 119A:44A:1 |
BT, binary typing (44).
Three restriction enzymes used in PFGE with an order of ApaI:SmaI:NotI.
Molecular subtyping.
All isolates were characterized at the NSW Enteric Reference Laboratory, Pathology West, Westmead Hospital in Sydney, and then subtyped by a binary typing assay (44). The assay is a PCR-based rapid method with a turnaround time of 2 to 3 h after the bacterial cultures are harvested. It detects the presence of eight gene loci selected from 44 genes that give the most variable binary results. These eight genes, mostly encoding hypothetical proteins, are distributed evenly throughout the genome of L. monocytogenes (44). MLVA was conducted to detect variable numbers of repeats at nine selected tandem-repeat regions. This method has a turnaround time of 24 to 48 h and has also been adopted in the four enteric reference laboratories in Australia (25). A modified PFGE protocol with ApaI, SmaI, and NotI as restriction enzymes (45) was performed by the Microbiological Diagnostic Unit (MDU), The University of Melbourne, Victoria, Australia. Molecular serotyping based on a multiplex PCR scheme (16) and a seven-housekeeping gene MLST scheme developed by the Institut Pasteur (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Lmono.html) were also performed by MDU.
DNA extraction.
A single colony of each isolate was cultured on blood agar plates at 37°C for 24 h. DNA extraction for binary typing and MLVA were performed by suspending 3 to 5 colonies in 150 μl of molecular-grade water and boiling for 5 min. The supernatant was collected and immediately used for PCR testing or stored at −20°C. For WGS, DNA was extracted using the DNeasy blood and tissue kit (Qiagen), according to the manufacturer's instructions. DNA quantities were estimated using the Qubit double-stranded DNA (dsDNA) BR assay kit and the Qubit fluorometer (Life Technologies, Germany), according to the manufacturer's instructions.
Genome sequencing.
WGS was performed on an Ion Torrent PGM (Life Technologies, Guilford, CT, USA). Genomic libraries were prepared using 1 μg of input genomic DNA and the Ion Xpress Plus genomic DNA (gDNA) fragment library kit. Size selection was done using E-Gel SizeSelect 2% agarose gels (Invitrogen). The concentrations and fragment size distributions of the libraries were measured by a 2100 Bioanalyzer on a high-sensitivity DNA chip (Agilent Technologies, Inc.). Diluted libraries were used as the template for emulsion PCR using the Ion OneTouch 200 template kit. Sequencing was performed using the Ion PGM 200 sequencing kit and either an Ion 314, Ion 316, or Ion 318 Chip (Life Technologies), according to the manufacturer's instructions.
Assembly and phylogenetic analysis.
De novo assembly was performed on sequencing reads using the Torrent suite assembler plugin version 3.4.2.0, which uses MIRA (version 3.9.9) to assemble contigs. The contigs were then scaffolded against the reference chromosome of L. monocytogenes strain SLCC2755 (GenBank accession no. FR733646) using Mauve version 2.3.1 (46). Contig files were submitted to snpTree 1.1, a Web server (http://cge.cbs.dtu.dk/services/snpTree/) that identifies single nucleotide polymorphisms (SNPs) present in WGS data (47). An SNP-based phylogenetic tree was created by comparing the sequencing data with 14 reference genomes using the default parameters. The resulting multialignment FASTA file was imported to MEGA6 (48) to generate a maximum parsimony tree. To illustrate the features of the identified SNPs, the FASTQ files were imported into CLC Genomics Workbench version 7.0 (CLC bio, Aarhus, Denmark), and reads were mapped to the reference genome of L. monocytogenes SLCC2755. Quality-based variant detection was performed using settings of a minimum neighborhood quality of 15 and minimum central quality of 20. Variant detection thresholds were set for a minimum coverage of 10 reads and minimum variant frequency of 75%. The de novo assembly of environmental isolate Lm4424 was also performed in CLC and used as a reference for read mapping and variant detection, as described above, to validate SNPs that were identified by the read mapping against the reference strain SLCC2755.
The L. monocytogenes reference sequences used in this study and their corresponding GenBank accession numbers were SLCC2755, FR733646 (read mapping reference, serotype 1/2b); SLCC2482, FR720325 (serotype 7); Clip80459, FM242711 (serotype 4b); F2365, AE017262 (serotype 4b); L99, FM211688.1 (serotype 4a); HCC23, CP001175 (serotype 4a); M7, CP002816 (serotype 4a); J0161, CP002001 (serotype 1/2a); 10403S, CP002002 (serotype 1/2a); Finland 1998, CP002004 (serotype 3a); 08-5578, CP001602 (serotype 1/2a); 08-5923, CP001604 (serotype 1/2a); FSL R2-561, CP002003 (serotype 1/2c); and EGD-e, AL591824 (serotype 1/2a).
Comparative genomics.
The mapping coverage of each of the isolates to the reference genome was visualized in Artemis (www.sanger.ac.uk/Software/Artemis/). Scaffolded contig files were uploaded to WebACT (www.webact.org/WebACT/home) to produce BLAST comparisons of sequenced isolate scaffolds to the reference strain. Genomic structural variation was visualized using ACT (http://www.sanger.ac.uk/Software/ACT/) (49) and the CLC Genomics Workbench microbial genome finishing module (CLC bio Aarhus, Denmark). The genomes were annotated using the RAST Web-based server (http://rast.nmpdr.org) (50).
Whole-genome sequence in silico MLST.
A conventional seven-loci MLST, based on the Institut Pasteur MLST scheme (http://www.pasteur.fr/mlst), was inferred from the sequenced reads. A FASTA format pseudomolecule containing the sequences of alleles from each of the seven loci provided by the MLST scheme was imported into CLC Genomics Workbench (CLC bio, Aarhus, Denmark) as a simulated MLST locus reference, and the sequenced reads from all isolates were mapped against this reference. The consensus sequence for each locus was extracted and submitted to the Listeria MLST website (http://www.pasteur.fr/mlst) for determination of the in silico MLST.
Nucleotide sequence accession numbers.
All sequencing FASTQ files were deposited to the Sequence Read Archive in the National Center for Biotechnology Information (NCBI) with accession numbers SRX992273, SRX992277, SRX993493, SRX993494, SRX993496, SRX993497, SRX993498, SRX993722, and SRX993724.
RESULTS
Molecular typing and genome sequencing.
The three isolates from the outbreak-related clinical cases and the two isolates from the environmental source shared the same molecular subtyping profiles and belonged to molecular serotype 1/2b, 3b, and 7, sequence type 3 (ST3), PFGE type 4A(ApaI):4(SmaI):1(NotI), binary type 223, and an MLVA profile of 04-17-16-05-03-11-14-00-16 (Table 1). Two sporadic isolates (Lm1414 and Lm4370) had the same molecular serotype and MLST as the outbreak-related isolates but had different PFGE, binary, and MLVA profiles. The other two sporadic and environmental isolates (Lm4082 and Lm3554) belonged to molecular serotype 4b and ST1 and produced distinct molecular profiles by all other typing methods (Table 1).
WGS generated various numbers of reads, between 375,305 and 1,987,282 for the nine L. monocytogenes isolates. These had reference genome depths of coverage ranging from 21.9- to 124.91-fold. De novo assembly resulted in contig numbers between 35 and 66 per isolate. The consensus lengths of the assembled sequences were between 2,934,544 and 3,066,775 bp for the nine isolates, with N50 values ranging from 15,377 to 478,854 bp (see Table S1 in the supplemental material). In silico MLST performed on the sequence read data from all isolates showed that all five outbreak-related clinical and environmental isolates and the two nonoutbreak molecular serotype 1/2b, 3b, and 7 isolates were ST3. The serotype 7 reference SLCC2482 is also an ST3 strain and belongs to the same clonal complex (CC), CC3, as the serotype 1/2b reference strain SLCC2755 (41). The two sequenced molecular serotype 4b isolates were identified as ST1.
Variant detection, based on read mapping of each of the isolates to the reference strain SLCC2755, identified between 274 and 288 SNPs for the three clinical and the two environmental outbreak isolates and 181 and 256 for the two sporadic molecular serotype 1/2b, 3b, and 7 isolates Lm4370 and Lm1414, respectively. The two molecular serotype 4b isolates, Lm3554 and Lm4082, had 10,526 and 11,487 SNPs, respectively. A further comparison of the SNPs between the outbreak isolates indicated that the isolate Lm4664 from case 3 was separated from the other two clinical isolates, Lm2128 from case 2 and Lm4447 from case 1, by two SNP differences (Table 2). There were no SNP differences between the two environmental isolates, while there were 19 to 20 SNPs observed between the environmental isolates and the clinical isolates associated with the outbreak. Of the 20 SNPs identified, 15 were nonsynonymous (Table 2; see also Fig. S1A in the supplemental material). The number of SNPs identified between nonoutbreak and outbreak isolates ranged from 67 to 70 for Lm4370 and Lm1414 to >10,000 for Lm3354 and Lm4028.
TABLE 2.
SNPs identified in human and environmental isolates of L. monocytogenes
| No. | Positiona | Consensus | SNP in isolate: |
Affected gene/gene reference | Amino acid change | Gene producta | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lm4422 | Lm4424 | Lm4664 | Lm2128 | Lm4447 | ||||||
| 1 | 210356 | A | T | T | T | actA | Actin assembly-inducing protein | |||
| 2 | 273330 | C | T | T | T | LMOSLCC2755_0254 | ABC transporter | |||
| 3 | 356520 | C | T | T | LMOSLCC2755_0327 | Val→Ile | Endonuclease/exonuclease/phosphatase family protein | |||
| 4 | 465953 | G | A | A | A | inlB | Gly→Arg | Internalin B | ||
| 5 | 801293 | G | A | A | A | LMOSLCC2755_0773 | Ala→Thr | Transcriptional regulator, GntR family | ||
| 6 | 868456 | T | A | A | A | LMOSLCC2755_0835 | Phe→Ile | ABC transporter | ||
| 7 | 916082 | A | —e | — | LMOSLCC2755_0875 | Lys→fsd | Phosphotransferase system | |||
| 8 | 1027362 | G | A | A | A | ogt-1 | Asp→Asn | Methylated-DNA-protein-cysteine methyltransferase | ||
| 9 | 1101468 | A | G | G | tagH | Asp→Gly | Teichoic acid ABC transporter | |||
| 10 | 1274142 | C | T | T | ||||||
| 11 | 1296522 | G | A | A | A | parE | Ser→Asn | DNA topoisomerase IV | ||
| 12 | 1413182 | T | C | C | C | LMOSLCC2755_1405 | Val→Ala | Phosphoesterase family protein | ||
| 13 | 1783102 | T | C | C | LMOSLCC2755_1740 | HNH endonuclease | ||||
| 14 | 1915979 | G | T/Gb | T | T | smc | Ser→Tyr | Chromosome condensation and segregation protein | ||
| 15 | 1941390 | C | T | T | T | gmk | Arg→Lys | Guanylate kinase | ||
| 16 | 1996953 | A | T/Ac | xpt | Ile→Asn | Xanthine phosphoribosyltransferase | ||||
| 17 | 2374784 | C | A | A | A | |||||
| 18 | 2438927 | C | A | A | A | LMOSLCC2755_2364 | Gly→Val | Cof-like hydrolase | ||
| 19 | 2566331 | C | T | T | T | LMOSLCC2755_2484 | Gly→Glu | Tetratricopeptide repeat domain protein | ||
| 20 | 2931810 | C | T | T | T | LMOSLCC2755_2844 | Asp→Asn | Hypothetical protein | ||
Refers to genomic nucleotide positions and gene products from the mapping reference sequence of L. monocytogenes SLCC2755 (GenBank accession no. FR733646).
T/G, mixed base calls were present at this position, with T at 86% and A at 14%.
T/A, mixed base calls were present at this position, with T at 81% and G at 19%.
fs, frameshift due to deletion.
—, deletion.
Phylogenomic relationships of the isolates.
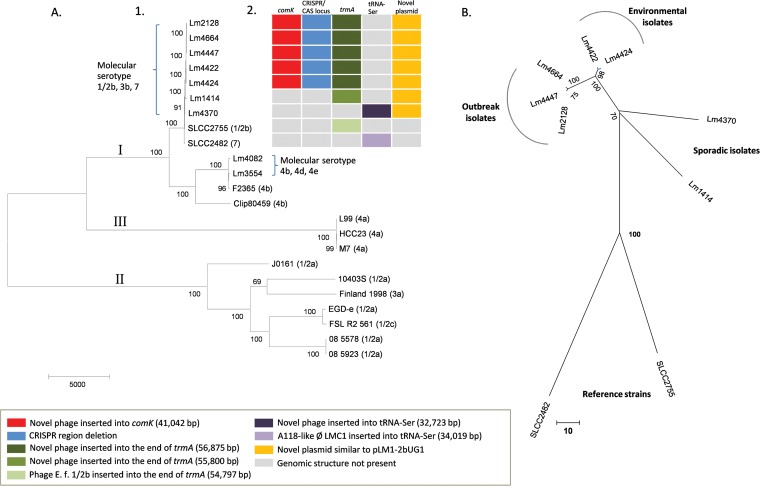
The phylogenetic analysis performed using the snpTree Web server included two reference sequences of SLCC2755 and SLCC2482, nine studied isolates, and 12 full-genome sequences belonging to all three main lineages of L. monocytogenes (51, 52), which were provided in the snpTree database. All positions containing gaps and missing data were eliminated. There was a total of 92,518 concatenated SNP positions in the final data set. As expected from previous studies (51, 52), the seven molecular serotype 1/2b, 3b, and 7, and two molecular serotype 4b isolates were all clustered within lineage I (Fig. 1A). Deep branching, reflecting the large genomic distances between the three main lineages, makes it difficult to visualize the relationships between closely related isolates. Therefore, a further SNP-based comparison was performed on the seven sequenced molecular serotype 1/2b, 3b, and 7 isolates and the reference strains SLCC2755 (serotype 1/2b) and SLCC2482 (serotype 7). This tree indicates that the three clinical isolates (Lm2128, Lm4447, and Lm4664) and two environmental isolates (Lm4422 and Lm4424) belong to a cluster that is distinct from the other nonclustered molecular serotype 1/2b, 3b, and 7 sporadic isolates and the reference strains (Fig. 1B).
FIG 1.
SNP-based phylogenetic trees of L. monocytogenes isolates and genomic diversity among outbreak isolates and related molecular serotype 1/2b, 3b, 7 strains. (A) 1, maximum parsimony analysis of outbreak and sporadic isolates compared to reference strains. SNP differences were determined using snpTree (47) and subsequent phylogenetic analyses performed using MEGA6 (48). Reference strains (serotypes indicated in brackets) clustered into three distinct lineages, I, II, and III (51, 52). Outbreak and sporadic isolates all belonged to lineage I. 2, presentation of unique prophages inserted into comK and the end of trmA and a CRISPR/Cas-related deletion that distinguishes the outbreak-associated isolates from closely related reference strains and sporadic temporally related molecular serotype 1/2b, 3b, and 7 isolates. (B) Genetic distances between molecular serotype 1/2b, 3b, and 7 isolates. Sequencing reads from all molecular serotype 1/2b, 3b, and 7 isolates were mapped to the reference L. monocytogenes strains SLCC2755 and SLCC2482 using CLC Genomics Workbench. Concatenated SNP profiles were then used to perform a maximum parsimony analysis using MEGA6. This analysis excluded the variable bacteriophage region spanning nucleotide positions 1752576 to 1807472 of the reference genome. The branch lengths indicate the number of SNP differences, shown by the scale bars. Bootstrap support values (1,000 replicates) are shown.
Characterization of MGEs and regions of chromosomal variation.
The contig scaffolds of all sequenced molecular serotype 1/2b, 3b, and 7 isolates were compared to reference strains SLCC2755 (serotype 1/2b) and SLCC2482 (serotype 7) using WebACT and ACT (49) to identify regions of broken synteny in the chromosomal backbone (see Fig. S2 in the supplemental material). The chromosomes of the three clinical and two environmental outbreak isolates were completely syntenic and clearly distinct from the two background molecular serotype 1/2b, 3b, and 7 isolates and the reference strains (see Fig. S1 in the supplemental material). The five outbreak-associated strains were the only molecular serotype 1/2b, 3b, and 7 isolates sequenced to date with a prophage inserted in the comK gene (Fig. 1A2, comK). This is the known site of integration for bacteriophage A118 in L. monocytogenes serotype 1/2a strain EGD-e (53) and bacteriophage A006:φLMC3 in some sequenced serotype 1/2a, 3c, and 1/2c isolates (41). The outbreak-associated isolates all contained an insertion of ∼41,042 bp in the comK gene, which shared regions of similarity with bacteriophage A118. This sequence was annotated using the RAST server and was identified as a novel bacteriophage containing 67 coding sequences (CDSs). The complete structure of this novel bacteriophage p008 from isolate Lm2128 is illustrated in Fig. S3 in the supplemental material, and the genome features, including the CDS locations, sizes, and functions and nucleotide and amino acid sequences, are detailed in Table S2 in the supplemental material.
Interestingly, all five outbreak-associated isolates also shared another prophage (Fig. 1A2, trmA). This phage was similar to the prophage annotated as E.f. (1/2b) that inserted into the end of trmA in SLCC2755 and integrated at the same attachment site at the 3′ end of open reading frame (ORF) LMOSLCC2755_1765 (41), which encodes a TrmA/RumA/YfjO family RNA methyltransferase. The prophages from the outbreak-associated isolates (56,875 bp in size) and SLCC2755 (54,797 bp in size) were very similar, apart from the region immediately adjacent to the left chromosome/prophage (oriC-distal) junction, where the large ORF LMOSLCC_1715 was absent and replaced by four ORFs not previously found in L. monocytogenes genome sequences. Sporadic molecular serotype 1/2b, 3b, and 7 isolate Lm1414 also had a prophage integrated into this attachment site. The left junction region of this prophage was similar to that of the outbreak-associated isolates (55,800 bp in size), but the remainder of the phage sequence shared low degrees of similarity with the prophages from SLCC2755 or the outbreak isolates (see Fig. S2 in the supplemental material). All of the serotype 1/2b genome reference sequence SLCC2755 and serotype 7 reference SLCC2482, and molecular serotype 1/2b, 3b, and 7 nonoutbreak isolates, had distinctive patterns of prophage carriage, which clearly distinguished them from the outbreak-associated isolates (see Fig. S1 in the supplemental material).
The third novel chromosomal feature that distinguished the outbreak isolates from other sequenced strains was the sequence of the CRISPR/Cas locus (Fig. 1A2), which had a deletion of 451 bp compared to the sequences in SLCC2755, SLCC2482, and two nonoutbreak isolates, Lm1414 and Lm4370 (see Fig. S1B in the supplemental material). This CRISPR subtype sequence is unique to the outbreak-associated isolates.
A plasmid structure similar to that of the plasmid pLM1-2bUG1 (54) obtained from the reference strain SLCC2755 was identified in all outbreak-related isolates, together with two nonoutbreak isolates, Lm1414 and Lm4370 (Fig. 1A2, novel plasmid). From this plasmid structure, a deletion of 6.7 kb was found between CDSs p0011 to p0019, which was adjacent to the part of transposing insertion IS1216 relating to a tetR gene. There were no SNP differences in this plasmid structure between the three human outbreak isolates and the two environmental isolates, but all these isolates had one SNP difference from the two nonoutbreak isolates. The gene structure and summary of the plasmid pl007 genome assembled by de novo assembly from the outbreak human isolate Lm4664 are shown in Fig. S4 and Table S3 in the supplemental material). From the nonoutbreak isolate Lm4370, a novel phage sequence similar to A118-like phage LMC1 that inserted into tRNA-Ser in the SLCC2482 was identified, which was not presented in all other isolates and the SLCC2755 strain (Fig. 1A2, tRNA-Ser).
DISCUSSION
Our comparison of the genomes of L. monocytogenes has demonstrated the added value of examining chromosomal structures when resolving epidemiological questions related to the investigation of listeriosis outbreaks. These observations opened new lines of evidence to test epidemiological hypotheses, supplementing accepted SNP-based analyses that are used for estimating the distance between strains of public health interest. Our findings indicated that the combination of SNP-based analysis and the comparison of mobile genomic elements offered higher discriminatory power than that with conventional serotyping, binary typing, or MLST of L. monocytogenes. These methods can detect genetic differences between isolates based on only a few genes or gene loci of the genome. Although PFGE addresses a large portion of an investigated genome (>90%), its resolution power depends only on the restriction enzyme sites, and it also lacks the resolution power necessary to distinguish bands of nearly identical size. A comparison of the variation across the wider genome offers the highest resolution power by providing a cost-effective way to examine genome-wide variations, which has been showcased by the fact that the cost and the time frame of WGS are close to those of MLST and MLVA (29, 30). The stable nature of the L. monocytogenes genome and limited resolution power of historical molecular subtyping methods make genome sequencing very appealing, especially in situations in which the testing of implicated food returns negative results but bacterial isolates are recovered from environmental sources in food-processing facilities (43).
The molecular subtyping of L. monocytogenes isolates from three human cases from three different hospitals found that all isolates shared identical molecular serotypes and binary, MLVA, and PFGE types. This established a possible link between the clinical isolates from the three patients. A subsequent public health investigation implicated a food source, but at the time of public health follow-up (two weeks after the first clinical case was reported), no leftover food was available for testing, and no isolates of L. monocytogenes were recovered from food samples of the same batches ordered by the patients in the hospitals, which were supplied by the food companies between 24 March (the overlapping admission time of the three patients) and 2 April 2013 (the date of the first reported clinical case). However, environmental isolates with the same typing profile linked the outbreak cases to company X, which supplied food to all three hospitals. The WGS-enabled discovery that clinical and environmental outbreak isolates possessed genome synteny and shared novel genomic features indicated a link between the food-processing facility and the human cases associated with this hospital-acquired outbreak of listeriosis (43).
Phylogenetic analyses based on SNP differences clustered the five outbreak isolates together and separated them from the two nonoutbreak isolates and the reference strains. All studied isolates were grouped into the lineage I cluster when they were compared with the reference strain sequences provided by the snpTree Web server. These reference sequences were selected from three evolutionary lineages of L. monocytogenes that were classified according to MLST results combined with other typing methods (55, 56). The lineages reflect the serotype distribution, and lineage I consists of serotypes l/2b, 3b, 4d, 4e, and most 4b isolates, which covers the majority of human clinical isolates (57). The isolates obtained in this study were molecular serotype 1/2b, 3b, 7, or 4b. These serotype groupings were supported by SNP-based phylogenetic analysis through clustering of all isolates in the lineage I group.
The SNP analysis indicated that the three clinical isolates differed from each other by ≤2 SNPs, suggesting that they had originated from the same source. Further inspection of read-mapping files showed that there were mixed base calls at both of the two SNP positions; in each case, one of the variants was the same as the other two clinical isolates. This observation may represent clones of L. monocytogenes during different stages of intrahost microevolution or a mix of closely related subpopulations present in the contaminated source. The presence of such transition-stage variants in Lm4664 (data not shown) was observed in this study. Two environmental isolates recovered from the implicated factory were closely related to these clinical isolates, with just 20 SNP differences between them. During infection in humans, the bacterial pathogens are in a relatively stable environment with similar temperature, pH, and nutrients. Outside the human host, these pathogens are under increased survival pressure due to the less favorable conditions. Once a pathogen is successfully adapted in the human host, less survival pressure is present between hosts due to the similar host conditions. This may explain why the SNPs identified in human isolates are less divergent than those found in environmental isolates in an outbreak. This was consistent with a previous listeriosis outbreak in Austria and Germany, during which clinical L. monocytogenes isolates from an outbreak cluster during a period of 2 years presented allelic differences at ≤6 out of 2,298 genes, and food isolates from implicated soft cheese and meat had allelic differences at 8 to 19 genes (40). However, the threshold estimate of SNPs in L. monocytogenes genomes that determines the epidemiological links between isolates remains uncertain (38, 40, 58, 59), and other lines of evidence to rule in or rule out the relationship between genomes might be of value.
We therefore supplemented this SNP-based comparison with the structural analysis of genomic content for the isolates associated with the outbreak and the background isolates that cocirculated in Sydney at the time of the outbreak. Two novel bacteriophages were identified in all three clinical isolates and the potential environmental source isolates. All five isolates also contained a novel CRISPR/Cas locus 2 (41) subtype sequence. Across the whole mapped genome, all human outbreak and environmental isolates were completely syntenic. This separated the outbreak associated isolates from all other comparison isolates, which had unique mobile genetic elements, CRISPR subtype sequences, and patterns of insertions and deletions.
In conclusion, the comparison of mobile genetic elements and deletions in L. monocytogenes genomes supplemented SNP-based analysis in the establishment of a probable epidemiological link between a hospital-acquired L. monocytogenes outbreak and the common source of food supplied to the affected inpatients. All isolates from the clinical cases and environmental samples obtained from the outbreak clustered together by SNP profiles and phylogenomic tree analyses. The unique genomic features present in all isolates further suggested that they represent the same clone; therefore, it is highly likely that the environmental and clinical isolates are related. Combining SNP analysis with the identification of clonally distinctive genomic features, including novel MGEs, helped to distinguish outbreak-related isolates from unrelated background isolates and improved the resolution of public health laboratory surveillance.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Microbiological Diagnostic Unit (MDU), The University of Melbourne, Victoria, Australia, for providing results of molecular serotyping, MLST, and PFGE typing. We also thank Z. Najjar, L. Gupta, F. Spechler, S. Kennewell, G. Kennedy, N. Bansal, and G. Arnold for their expert advice about the epidemiological context of the outbreak.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00202-15.
REFERENCES
- 1.Beresford MR, Andrew PW, Shama G. 2001. Listeria monocytogenes adheres to many materials found in food-processing environments. J Appl Microbiol 90:1000–1005. doi: 10.1046/j.1365-2672.2001.01330.x. [DOI] [PubMed] [Google Scholar]
- 2.Walker SJ, Archer P, Banks JG. 1990. Growth of Listeria monocytogenes at refrigeration temperatures. J Appl Bacteriol 68:157–162. doi: 10.1111/j.1365-2672.1990.tb02561.x. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi M, Chikindas ML. 2007. Listeria: a foodborne pathogen that knows how to survive. Int J Food Microbiol 113:1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Lianou A, Sofos JN. 2007. A review of the incidence and transmission of Listeria monocytogenes in ready-to-eat products in retail and food service environments. J Food Prot 70:2172–2198. [DOI] [PubMed] [Google Scholar]
- 5.Evans MR, Swaminathan B, Graves LM, Altermann E, Klaenhammer TR, Fink RC, Kernodle S, Kathariou S. 2004. Genetic markers unique to Listeria monocytogenes serotype 4b differentiate epidemic clone II (hot dog outbreak strains) from other lineages. Appl Environ Microbiol 70:2383–2390. doi: 10.1128/AEM.70.4.2383-2390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLauchlin J, Hall SM, Velani SK, Gilbert RJ. 1991. Human listeriosis and pate: a possible association. BMJ 303:773–775. doi: 10.1136/bmj.303.6805.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O'Connor KA, Cosgrove S, Gossack JP, Parachini SS, Jain NS, Ettestad P, Ibraheem M, Cantu V, Joshi M, DuVernoy T, Fogg NW Jr, Gorny JR, Mogen KM, Spires C, Teitell P, Joseph LA, Tarr CL, Imanishi M, Neil KP, Tauxe RV, Mahon BE. 2013. Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med 369:944–953. doi: 10.1056/NEJMoa1215837. [DOI] [PubMed] [Google Scholar]
- 8.Bula CJ, Bille J, Glauser MP. 1995. An epidemic of food-borne listeriosis in western Switzerland: description of 57 cases involving adults. Clin Infect Dis 20:66–72. doi: 10.1093/clinids/20.1.66. [DOI] [PubMed] [Google Scholar]
- 9.Dawson SJ, Evans MR, Willby D, Bardwell J, Chamberlain N, Lewis DA. 2006. Listeria outbreak associated with sandwich consumption from a hospital retail shop, United Kingdom. Euro Surveill 11:89–91. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=632. [PubMed] [Google Scholar]
- 10.Silk BJ, McCoy MH, Iwamoto M, Griffin PM. 2014. Foodborne listeriosis acquired in hospitals. Clin Infect Dis 59:532–540. doi: 10.1093/cid/ciu365. [DOI] [PubMed] [Google Scholar]
- 11.Shetty A, McLauchlin J, Grant K, O'Brien D, Howard T, Davies EM. 2009. Outbreak of Listeria monocytogenes in an oncology unit associated with sandwiches consumed in hospital. J Hosp Infect 72:332–336. doi: 10.1016/j.jhin.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Elsner HA, Tenschert W, Fischer L, Kaulfers PM. 1997. Nosocomial infections by Listeria monocytogenes: analysis of a cluster of septicemias in immunocompromised patients. Infection 25:135–139. doi: 10.1007/BF02113599. [DOI] [PubMed] [Google Scholar]
- 13.Green HT, Macaulay MB. 1978. Hospital outbreak of Listeria monocytogenes septicaemia: a problem of cross infection? Lancet ii:1039–1040. [DOI] [PubMed] [Google Scholar]
- 14.Martins IS, Faria FC, Miguel MA, Dias MP, Cardoso FL, Magalhães AC, Mascarenhas LA, Nouer SA, Barbosa AV, Vallim DC, Hofer E, Rabello RF, Riley LW, Moreira BM. 2010. A cluster of Listeria monocytogenes infections in hospitalized adults. Am J Infect Control 38:e31–e36. doi: 10.1016/j.ajic.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulet V, King LA, Vaillant V, de Valk H. 2013. What is the incubation period for listeriosis? BMC Infect Dis 13:11. doi: 10.1186/1471-2334-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graves LM, Swaminathan B, Hunter S. 1999. Subtyping Listeria monocytogenes, p 279–298. In Ryser ET, Marth EH (ed), Listeria, listeriosis, and food safety. Marcel Dekker, Inc., New York, NY. [Google Scholar]
- 18.Wiedmann M. 2002. Molecular subtyping methods for Listeria monocytogenes. J AOAC Int 85:524–531. [PubMed] [Google Scholar]
- 19.Swaminathan B, Gerner-Smidt P. 2007. The epidemiology of human listeriosis. Microbes Infect 9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Graves LM, Swaminathan B. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int J Food Microbiol 65:55–62. doi: 10.1016/S0168-1605(00)00501-8. [DOI] [PubMed] [Google Scholar]
- 21.Salcedo C, Arreaza L, Alcala B, de la Fuente L, Vazquez JA. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J Clin Microbiol 41:757–762. doi: 10.1128/JCM.41.2.757-762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sperry KE, Kathariou S, Edwards JS, Wolf LA. 2008. Multiple-locus variable-number tandem-repeat analysis as a tool for subtyping Listeria monocytogenes strains. J Clin Microbiol 46:1435–1450. doi: 10.1128/JCM.02207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindstedt BA, Tham W, Danielsson-Tham ML, Vardund T, Helmersson S, Kapperud G. 2008. Multiple-locus variable-number tandem-repeats analysis of Listeria monocytogenes using multicolour capillary electrophoresis and comparison with pulsed-field gel electrophoresis typing. J Microbiol Methods 72:141–148. doi: 10.1016/j.mimet.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Murphy M, Corcoran D, Buckley JF, O'Mahony M, Whyte P, Fanning S. 2007. Development and application of multiple-locus variable number of tandem repeat analysis (MLVA) to subtype a collection of Listeria monocytogenes. Int J Food Microbiol 115:187–194. doi: 10.1016/j.ijfoodmicro.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Huang B, Eglezos S, Graham T, Blair B, Bates J. 2013. Identification of an optimized panel of variable number tandem-repeat (VNTR) loci for Listeria monocytogenes typing. Diagn Microbiol Infect Dis 75:203–206. doi: 10.1016/j.diagmicrobio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Chenal-Francisque V, Diancourt L, Cantinelli T, Passet V, Tran-Hykes C, Bracq-Dieye H, Leclercq A, Pourcel C, Lecuit M, Brisse S. 2013. Optimized multilocus variable-number tandem-repeat analysis assay and its complementarity with pulsed-field gel electrophoresis and multilocus sequence typing for Listeria monocytogenes clone identification and surveillance. J Clin Microbiol 51:1868–1880. doi: 10.1128/JCM.00606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medini D, Serruto D, Parkhill J, Relman DA, Donati C, Moxon R, Falkow S, Rappuoli R. 2008. Microbiology in the post-genomic era. Nat Rev Microbiol 6:419–430. [DOI] [PubMed] [Google Scholar]
- 28.Shendure J, Ji H. 2008. Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 29.Vernet G, Saha S, Satzke C, Burgess DH, Alderson M, Maisonneuve JF, Beall BW, Steinhoff MC, Klugman KP. 2011. Laboratory-based diagnosis of pneumococcal pneumonia: state of the art and unmet needs. Clin Microbiol Infect 17: Suppl 3:1–13. [DOI] [PubMed] [Google Scholar]
- 30.Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundu S, Lockwood J, Depledge DP, Chaudhry Y, Aston A, Rao K, Hartley JC, Goodfellow I, Breuer J. 2013. Next-generation whole genome sequencing identifies the direction of norovirus transmission in linked patients. Clin Infect Dis 57:407–414. doi: 10.1093/cid/cit287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavezzo E, Toppo S, Franchin E, Di Camillo B, Finotello F, Falda M, Manganelli R, Palu G, Barzon L. 2013. Genomic comparative analysis and gene function prediction in infectious diseases: application to the investigation of a meningitis outbreak. BMC Infect Dis 13:554. doi: 10.1186/1471-2334-13-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherry NL, Porter JL, Seemann T, Watkins A, Stinear TP, Howden BP. 2013. Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J Clin Microbiol 51:1396–1401. doi: 10.1128/JCM.03332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel U, Szczepanowski R, Claus H, Junemann S, Prior K, Harmsen D. 2012. Ion Torrent personal genome machine sequencing for genomic typing of Neisseria meningitidis for rapid determination of multiple layers of typing information. J Clin Microbiol 50:1889–1894. doi: 10.1128/JCM.00038-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pallen MJ, Loman NJ, Penn CW. 2010. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol 13:625–631. doi: 10.1016/j.mib.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Bakker HC, Switt AI, Cummings CA, Hoelzer K, Degoricija L, Rodriguez-Rivera LD, Wright EM, Fang R, Davis M, Root T, Schoonmaker-Bopp D, Musser KA, Villamil E, Waechter H, Kornstein L, Furtado MR, Wiedmann M. 2011. A whole-genome single nucleotide polymorphism-based approach to trace and identify outbreaks linked to a common Salmonella enterica subsp. enterica serovar Montevideo pulsed-field gel electrophoresis type. Appl Environ Microbiol 77:8648–8655. doi: 10.1128/AEM.06538-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gieraltowski L, Julian E, Pringle J, Macdonald K, Quilliam D, Marsden-Haug N, Saathoff-Huber L, Von Stein D, Kissler B, Parish M, Elder D, Howard-King V, Besser J, Sodha S, Loharikar A, Dalton S, Williams I, Barton Behravesh C. 2013. Nationwide outbreak of Salmonella Montevideo infections associated with contaminated imported black and red pepper: warehouse membership cards provide critical clues to identify the source. Epidemiol Infect 141:1244–1252. doi: 10.1017/S0950268812001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, Kleta S, Prager R, Preussel K, Aichinger E, Mellmann A. 2014. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect 20:431–436. doi: 10.1111/1469-0691.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, Barbuddhe S, Hain T, Chakraborty T. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47. doi: 10.1186/1471-2164-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sesto N, Touchon M, Andrade JM, Kondo J, Rocha EP, Arraiano CM, Archambaud C, Westhof E, Romby P, Cossart P. 2014. A PNPase dependent CRISPR system in Listeria. PLoS Genet 10:e1004065. doi: 10.1371/journal.pgen.1004065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Najjar Z, Gupta L, Sintchenko V, Shadbolt C, Wang Q, Bansal N. 2015. Listeriosis cluster in Sydney linked to hospital food. Med J Aust 202:448–449. doi: 10.5694/mja14.00913. [DOI] [PubMed] [Google Scholar]
- 44.Huang B, Schleehauf JK, Eglezos S, Bates J. 2007. Binary typing of Listeria monocytogenes isolates from patients and food through multiplex PCR and reverse line hybridisation, abstr PP18.12, p 136–137. Ann Sci Meet Aust Soc Microbiol. Australian Society for Microbiology, SA, Australia. [Google Scholar]
- 45.Carriere C, Allardet-Servent A, Bourg G, Audurier A, Ramuz M. 1991. DNA polymorphism in strains of Listeria monocytogenes. J Clin Microbiol 29:1351–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leekitcharoenphon P, Kaas RS, Thomsen MC, Friis C, Rasmussen S, Aarestrup FM. 2012. snpTree–a Web-server to identify and construct SNP trees from whole genome sequence data. BMC Genomics 13(Suppl 7):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream MA. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Ward TJ, Ducey TF, Usgaard T, Dunn KA, Bielawski JP. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl Environ Microbiol 74:7629–7642. doi: 10.1128/AEM.01127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loessner MJ, Inman RB, Lauer P, Calendar R. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol Microbiol 35:324–340. doi: 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- 54.Kuenne C, Voget S, Pischimarov J, Oehm S, Goesmann A, Daniel R, Hain T, Chakraborty T. 2010. Comparative analysis of plasmids in the genus Listeria. PLoS One 5:e12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun 65:2707–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeffers GT, Bruce JL, McDonough PL, Scarlett J, Boor KJ, Wiedmann M. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095–1104. [DOI] [PubMed] [Google Scholar]
- 58.Hawkey J, Edwards DJ, Dimovski K, Hiley L, Billman-Jacobe H, Hogg G, Holt KE. 2013. Evidence of microevolution of Salmonella Typhimurium during a series of egg-associated outbreaks linked to a single chicken farm. BMC Genomics 14:800. doi: 10.1186/1471-2164-14-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Octavia S, Wang Q, Tanaka MM, Kaur S, Sintchenko V, Lan R. 2015. Delineating community outbreaks of Salmonella enterica serovar Typhimurium by use of whole-genome sequencing: insights into genomic variability within an outbreak. J Clin Microbiol 53:1063–1071. doi: 10.1128/JCM.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.