Abstract
We describe the selection of reduced chlorhexidine susceptibility during chlorhexidine use in a patient with two episodes of cutaneous USA300 methicillin-resistant Staphylococcus aureus abscess. The second clinical isolate harbors a novel plasmid that encodes the QacA efflux pump. Greater use of chlorhexidine for disease prevention warrants surveillance for resistance.
CASE REPORT
An 18-year-old man undergoing infantry basic training at Fort Benning, GA, presented to the outpatient clinic in July complaining of a painful skin lesion on his left knee. On physical examination, the patient was afebrile (37°C) and examination was remarkable only for a prepatellar nodule that was erythematous, warm, indurated, tender, and fluctuant. The patient had no other skin lesions and no lymphadenopathy. The remainder of his examination was normal, and the patient did not report a history of cutaneous abscesses. The patient was diagnosed with a cutaneous abscess without joint involvement and underwent incision and drainage. The purulent material underwent standard wound culture and yielded methicillin-resistant Staphylococcus aureus (MRSA) with the BD Phoenix automated microbiology system (Becton, Dickinson, Sparks, MD). The isolate was resistant to oxacillin and erythromycin and had inducible clindamycin resistance, as determined by double-disk diffusion (1), but was susceptible to trimethoprim-sulfamethoxazole (TMP-SMX), doxycycline, levofloxacin, linezolid, daptomycin, and vancomycin (Table 1). With no known medication allergies, the patient was treated with a 10-day course of twice-daily double-strength TMP-SMX and underwent serial follow-up examinations and wound care with complete resolution of his abscess within 14 days.
TABLE 1.
Molecular characteristics and antimicrobial susceptibilities of clinical MRSA isolates
| Characteristic | C01 | C02 |
|---|---|---|
| MLST result | ST8 | ST8 |
| PFGE type | USA300 | USA300 |
| Susceptibility to: | ||
| Oxacillin | Resistant | Resistant |
| Erythromycin | Resistant | Resistant |
| Clindamycin | Inducible resistance | Inducible resistance |
| TMP-SMX | Susceptible | Susceptible |
| Doxycycline | Susceptible | Susceptible |
| Daptomycin | Susceptible | Susceptible |
| Vancomycin | Susceptible | Susceptible |
| Levofloxacin | Susceptible | Resistant |
| qacA | Negative | Positive |
| mecA | Positive | Positive |
| SCCmec type | IV | IV |
| PVL | Positive | Positive |
| mupA | Negative | Negative |
| norA | Positive | Positive |
| Chlorhexidine MIC (μg/ml) | 0.3 | 0.8 |
Nine weeks after his initial presentation, the patient returned to the clinic. This time, he complained of a similar painful lesion on his left foot. On physical examination, the patient was again afebrile (37°C) and examination was remarkable only for an inflamed and fluctuant nodule on the dorsum of his left foot. The patient was again diagnosed with a cutaneous abscess and underwent incision and drainage, with standard wound cultures yielding MRSA. This second clinical isolate had the same antibiotic susceptibility pattern as the first MRSA isolate; however, it was resistant to levofloxacin (Table 1). The patient was treated in a similar fashion as for the first episode and recovered without additional recurrences.
The patient was a soldier participating in a prospective cluster-randomized trial aimed at preventing skin and soft tissue infections (SSTIs), which are common in this population (2). The patient was in a study group that received chlorhexidine for weekly showering (4% chlorhexidine gluconate, Hibiclens; Mölnlycke Health Care, Norcross, GA). As part of the protocol, the patient completed a questionnaire at the time of his second episode that queried his chlorhexidine use; he reported using the agent every other week throughout his training. The patient would have used chlorhexidine once or twice before his first episode and four or five times prior to the second episode.
The two MRSA isolates underwent molecular analysis, including typing by pulsed-field gel electrophoresis (PFGE) (3), multilocus sequence typing (MLST) (4), and PCR assays for toxin (5) and resistance genes (6, 7). PFGE findings were resolved and analyzed with BioNumerics (Applied Math, Austin, TX). Both MRSA isolates were sequence type 8 (ST8), USA300, staphylococcal cassette chromosome mec type IV, positive for Panton-Valentine leukocidin (PVL, encoded by lukS-PV), and negative for high-level mupirocin resistance (mupA). The first clinical isolate (C01) was negative for the chlorhexidine resistance genes (qacA/B), but the second clinical isolate (C02) was positive for qacA/B. As part of the research protocol (2), sampling of the anterior nares at the second episode revealed that the patient was colonized with a separate, unrelated, methicillin-susceptible S. aureus strain (ST30).
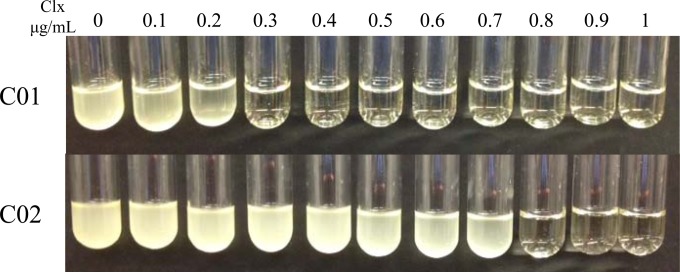
Both clinical isolates underwent chlorhexidine susceptibility testing. Briefly, about 4 × 104 CFU from an overnight culture were inoculated into 1 ml of cation-adjusted Mueller-Hinton II broth (BD BBL) containing purified chlorhexidine (Sigma) at concentrations ranging from 0 to 1 μg/ml. The cultures were grown overnight with shaking (220 rpm). Upon visual analysis (Fig. 1), the MIC of chlorhexidine for the C01 isolate was 0.3 μg/ml, while that for the C02 isolate was 0.8 μg/ml, an approximate 2.7-fold increase over that for C01.
FIG 1.
MICs of chlorhexidine (Clx) for S. aureus abscess isolates C01 and C02. The chlorhexidine concentrations tested ranged from 0 to 1 μg/ml. All cultures were inoculated with approximately 4 × 104 CFU and grown overnight at 37°C with shaking at 220 rpm. The MIC for the C01 strain was determined to be 0.3 μg/ml, while that for the C02 isolate was 0.8 μg/ml.
Previous reports have found that mutations in the promoter of the norA efflux pump can lead to increases in norA transcription, which result in increased resistance to antiseptic agents such as chlorhexidine (8, 9). The promoter region of norA was PCR amplified and sequenced with the 5′-GTCTTGGTCATCTGCAAAGGTTG-3′ and 5′-GACTGGTATTACTAAACCGATACC-3′ primers. Additionally, the 5′-GGTGGTATGAGTGCTGGTATGG-3′ and 5′-GCATACGATGTGAAACTTCTGCC-3′ primers were used to assess norA transcription via reverse transcription (RT)-PCR. Total RNA was extracted from the C01 and C02 isolates with the Qiagen EasyRNA kit. Prior to RNA extraction, S. aureus was lysed by incubation for 1 h in lysis buffer containing Tris-EDTA buffer, lysostaphin (20 μg/ml), and proteinase K (200 μg/ml). cDNA was synthesized from total RNA with the QuantiTect RT kit (Qiagen). All PCR and sequencing reactions were performed as previously described (10). RT-PCR was performed with 10-μl reaction mixtures containing 1× SYBR green (Qiagen) and 1.5 μM each primer. Reaction mixtures were incubated for 5 min at 95°C, followed by 35 cycles of 95°C for 10 s and then 50°C for 10 s. Fluorescence readings were acquired at the end of each cycle with the Qiagen Rotor-Gene Q RT-PCR machine. Sequencing data analysis revealed that the C01 and C02 norA promoters were identical in nucleotide composition. Not surprisingly, norA expression in the C02 isolate was indistinguishable from that in the C01 isolate.
Plasmid extraction with the Qiagen Plasmid Purification kit and subsequent PCR amplification (5′-GCTGCATTTATGACAATGTTTG-3′ and 5′-AATCCCACCTACTAAAGCAG-3′) (11) and visualization on a 1% agarose gel revealed that the qacA gene was detectable only in the C02 isolate. Given the high level of nucleotide sequence similarity between the qacA and qacB genes, we determined the qacA DNA sequence from the C02 plasmid and then compared it to the canonical qacA (accession no. GU565967.1) and qacB (accession no. AF053772.1) sequences. Of the seven amino acid differences described by Paulsen et al. that distinguish qacA from qacB, six of the qacA residues were observed in the qacA gene from C02; this includes a key aspartic acid residue at amino acid position 323 (12). The one amino acid residue that differed from the qacA consensus occurred at the first position, where an alternative lysine start codon was found. In total, the data suggest that C02 harbors the qacA gene. Finally, to ensure that the C02 isolate was the only strain that expressed the qacA efflux pump, we utilized cDNA as a template for qacA PCR amplification with the same qacA detection/sequencing primers as mentioned before. Amplicon detection on a 1% agarose gel confirmed that the C02 isolate actively expressed the qacA gene while the C01 isolate did not.
Previous reports have found that qacA is typically carried on the pSK1 family of plasmids (13, 14). We therefore sequenced approximately 550 bp upstream of the qacA gene with the 5′-CTCCAATCCTTATAGACCGTGC-3′ primer and found a high level of nucleotide sequence similarity to the pSK1 DNA sequence (GenBank accession no. NC_014369) (>99% nucleotide sequence similarity). To determine if the plasmid was indeed pSK1, we next used the pSK1 plasmid sequence to design a series of 10 PCR primer pairs that span the entire plasmid (Table 2). We found that only the primer pair that encompassed the qacA gene (primer pair 9) yielded a PCR amplicon of the correct size in the C02 isolate. This led us to conclude that the qacA gene found in the C02 isolate may be carried on a pSK1-like plasmid but not on pSK1 itself.
TABLE 2.
pSK1-like plasmid PCR primer panel
| Primer pair | Primer (5′-3′) |
|
|---|---|---|
| Forward | Reverse | |
| 1 | GGAGCACTAGTAGCAACTTTCATC | CCAGAGCCGATGCTACGC |
| 2 | GCCTTAAAATTCCAGGCGC | GCTGAAAGTTATAGAGCGGC |
| 3 | GAAGCACTTGCATACGATAGTG | GCTCACGCTATACCGACATTC |
| 4 | CTAACGTGCGATCAGATGCTTG | GCACCCTCAGAAGCCATTC |
| 5 | CGCAGTTGGAGCAAGTGAG | CTTTATCTTCGACTCTATCACGAAC |
| 6 | CATCATAGCACCAGTCATCAG | GTGTGCGATCATCGCGTCTATTC |
| 7 | CAATTACCTTGGCACTTACCAAATG | GGTTGGAAGAACGCACATATG |
| 8 | CTTAGATAGTAGCCAACGGCTAC | CATCGTATCGATCTTGTTGTCC |
| 9 | CGATCGCACGGTCTATAAGG | GCTTTGAATCTCTTCGCTTTTCAG |
| 10 | CGAAGACGCCTTTCAATATACCG | CCTAGAGCTTGCCATGTATATG |
To assess clonality between the two isolates, total DNA from both isolates was prepared via phenol-chloroform extraction and subjected to Pacific Biosciences RS II SMRT whole-genome sequencing (University of Maryland, School of Medicine, Institute for Genome Sciences). A single closed circular chromosome and plasmid were obtained for each isolate. A comprehensive list of the chromosome and plasmid characteristics is included in Table 3. Genome analysis revealed that the C01 isolate contained 2,770 chromosomal open reading frames (ORFs) and was approximately 2.92 Mbp in length. Conversely, the C02 isolate had a reduced chromosome of 2.86 Mbp encoding 2,704 ORFs. In addition to whole gene changes, we observed >140 single nucleotide polymorphisms (SNPs) between the two chromosomes, which suggests that the two isolates are genetically distinct. Of note, two nonsynonymous SNPs in the C02 gyrA and grlA genes (C251T and T239A, respectively) were detected. These SNPs result in an S84L amino acid mutation in GyrA and an F80Y mutation in GrlA, which have previously been shown to contribute to quinolone resistance (15–17) and therefore likely explain the levofloxacin resistance of the C02 isolate.
TABLE 3.
Genome and plasmid statistics
| Cell component | Size (bp) | % GC | No. of ORFs | Avg gene length (bp) | % Coding |
|---|---|---|---|---|---|
| Chromosome | |||||
| C01 | 2,918,599 | 32.8 | 2,770 | 879 | 84.5 |
| C02 | 2,864,998 | 32.8 | 2,704 | 882 | 84.4 |
| Plasmid | |||||
| pC01 | 27,044 | 30.6 | 32 | 592 | 70.1 |
| pC02 | 61,537 | 29.5 | 71 | 677 | 78.2 |
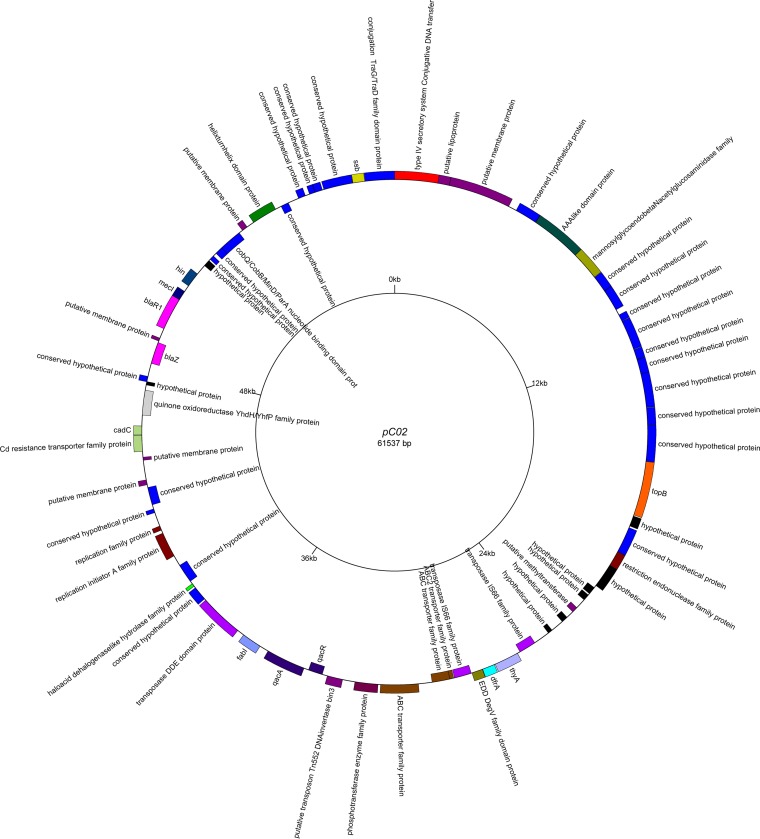
The pC01 and pC02 plasmid sequences were analyzed with the Basic Local Alignment Search Tool (BLAST; NCBI). While the pC01 sequence was nearly perfectly identical to known S. aureus plasmids such as SAP046A (GenBank accession no. NC_013294.1), the pC02 plasmid was significantly larger and less than 40% of the plasmid contained regions similar to those of other sequences in the NCBI database. Annotation of the pC02 plasmid revealed the presence of numerous proteins involved in a range of cellular processes, including DNA replication (Ssb, TopB), transcriptional regulation (QacR), and substrate translocation (CadC). The fully annotated pC02 map is depicted in Fig. 2 (18). The qacA-containing pC02 plasmid appears to be novel and may represent an additional class of antimicrobial resistance plasmids in S. aureus.
FIG 2.
Annotated map of the pC02 plasmid. Sequencing and annotation were performed at the University of Maryland Institute for Genome Sciences and visualized with GenomeVX.
Chlorhexidine is a broad-spectrum topical biguanide cationic antiseptic agent with activity against S. aureus (19, 20). Although it has been used for decades in various roles, ranging from hand washing to preoperative skin preparation, chlorhexidine has been increasingly employed for the prevention of both nosocomial (21–24) and community-associated infections (2, 25–27). Evidence from large randomized-control trials points to the importance of chlorhexidine in the prevention of the spread of drug-resistant organisms and hospital-acquired infections (21, 24, 28). Indeed, chlorhexidine has also been an integral component of strategies aimed at the prevention of recurrent MRSA SSTIs in individuals and households (26, 29).
Despite its widespread use, the prevalence of chlorhexidine resistance in the United States is low (approximately 1%) (25, 30, 31); this is in contrast to observations in other countries (11, 32). When used in large trials in both community and hospital settings, chlorhexidine resistance has been only rarely reported (21, 24, 27, 31). Nevertheless, with the widespread and increasing use of this agent, experience has shown that concern about the potential emergence of chlorhexidine resistance is appropriate (32). Additional studies that investigate the frequency of chlorhexidine use and selection of chlorhexidine-resistant strains must be conducted to ensure proper chlorhexidine stewardship.
The plasmid-borne qacA gene, in particular, encodes an efflux pump that has been shown in numerous reports to confer resistance to numerous hydrophobic compounds, including cationic biocides such as chlorhexidine (20, 33, 34). While there are no established breakpoints for chlorhexidine resistance, the presence of these genes has been associated both with increased MICs and with untoward clinical outcomes (35–38). Interestingly, multiple reports have identified chlorhexidine-resistant S. aureus isolates with MICs of ≥4 μg/ml (30, 31, 38). The isolate described in this report, however, showed a reduced MIC (≤0.8 μg/ml). While the reason for the lower MIC is not clear, this may be due to reduced translation efficiency due to the alternate start codon found in the C02 qacA gene (39). The presence of qacA/B and an increased MIC are sometimes poorly correlated (30); however, in our isolates, an increase in the MIC for the qacA-positive C02 strain was clearly observed. We cannot determine the overall clinical impact of this reduced chlorhexidine susceptibility in our patient other than to note that he developed a second USA300 MRSA abscess.
Chlorhexidine has residual antibacterial activity, which may be beneficial in reducing the bacterial burden or preventing the spread of resistant organisms (40); however, this residual activity may also contribute to an environment that ultimately fosters resistance (11). Since our patient utilized chlorhexidine every other week, this may have played a part in the selection of reduced chlorhexidine susceptibility in the patient. Evidence suggests that the qacA gene may be able to be horizontally transferred across various staphylococcal species (33, 41). This typically is plasmid mediated since numerous reports have shown that the qacA gene is often carried on a plasmid from the pSK1 family of vectors (34, 42). Although we do not know its original source, qacA in our identified clinical MRSA isolate was carried on a large, uncharacterized plasmid that shows limited similarity to pSK1. This finding suggests that transmission of qacA is not limited to the pSK1-like vectors. Furthermore, the identification of this novel qacA-containing plasmid combined with the now ubiquitous use of chlorhexidine, highlights the need for increased surveillance programs that would seek to understand the evolution of qacA transmission across MRSA isolates and potentially across other staphylococcal species.
In summary, to our knowledge, this is the first report of selection for increased chlorhexidine MICs while using chlorhexidine in a community-based patient with recurrent USA300 MRSA SSTIs. In light of recent clinical trials that show the benefit of chlorhexidine in the prevention of drug-resistant infections, the medical community should anticipate greater use of this agent and consequently increased resistance. Further study and surveillance for the emergence of chlorhexidine resistance should be considered in health care and community settings that use chlorhexidine for disease prevention.
Nucleotide sequence accession numbers.
The C01 and C02 genomes and plasmids were submitted to NCBI and given accession no. CP012118, CP012119, CP012120, and CP012121.
ACKNOWLEDGMENTS
This work (IDCRP-055) was supported by the Infectious Disease Clinical Research Program, a Department of Defense (DoD) program executed through the Uniformed Services University (USU) of the Health Sciences. This project has been funded in whole, or in part, with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), under interagency agreement Y1-AI-5072. Additional funding was provided by Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases, Division of Healthcare Quality Promotion interagency agreement 09FED914272 (M.W.E.); DoD Global Emerging Infections Surveillance program C0366-11-HS (M.W.E.); and USU DoD program project HT9404-12-1-0019 (D.S.M.). R.C.J. is supported by a fellowship from the Henry M. Jackson Foundation.
The views expressed in this paper are those of the authors and do not necessarily represent the views of the USU of the Health Sciences, the DoD, or other federal agencies.
We thank Kimberly Bishop-Lilly for her expertise and valuable discussions.
REFERENCES
- 1.Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH. 2003. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol 41:4740–4744. doi: 10.1128/JCM.41.10.4740-4744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis MW, Schlett CD, Millar EV, Wilkins KJ, Crawford KB, Morrison-Rodriguez SM, Pacha LA, Gorwitz RJ, Lanier JB, Tribble DR. 2014. Hygiene strategies to prevent methicillin-resistant Staphylococcus aureus skin and soft-tissue infections: a cluster-randomized controlled trial among high-risk military trainees. Clin Infect Dis 58:1540–1548. doi: 10.1093/cid/ciu166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fosheim GE, Nicholson AC, Albrecht VS, Limbago BM. 2011. Multiplex real-time PCR assay for detection of methicillin-resistant Staphylococcus aureus and associated toxin genes. J Clin Microbiol 49:3071–3073. doi: 10.1128/JCM.00795-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGann P, Milillo M, Kwak YI, Quintero R, Waterman PE, Lesho E. 2013. Rapid and simultaneous detection of the chlorhexidine and mupirocin resistance genes qacA/B and mupA in clinical isolates of methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis 77:270–272. doi: 10.1016/j.diagmicrobio.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Mediavilla JR, Oliveira DC, Willey BM, de Lencastre H, Kreiswirth BN. 2009. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec typing. J Clin Microbiol 47:3692–3706. doi: 10.1128/JCM.00766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noguchi N, Okada H, Narui K, Sasatsu M. 2004. Comparison of the nucleotide sequence and expression of norA genes and microbial susceptibility in 21 strains of Staphylococcus aureus. Microb Drug Resist 10:197–203. doi: 10.1089/mdr.2004.10.197. [DOI] [PubMed] [Google Scholar]
- 9.Fournier B, Truong-Bolduc QC, Zhang X, Hooper DC. 2001. A mutation in the 5′ untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J Bacteriol 183:2367–2371. doi: 10.1128/JB.183.7.2367-2371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis MW, Johnson RC, Crawford K, Lanier JB, Merrell DS. 2014. Molecular characterization of a catalase-negative methicillin-susceptible Staphylococcus aureus subsp. aureus strain collected from a patient with cutaneous abscess. J Clin Microbiol 52:344–346. doi: 10.1128/JCM.02455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vali L, Davies SE, Lai LL, Dave J, Amyes SG. 2008. Frequency of biocide resistance genes, antibiotic resistance and the effect of chlorhexidine exposure on clinical methicillin-resistant Staphylococcus aureus isolates. J Antimicrob Chemother 61:524–532. doi: 10.1093/jac/dkm520. [DOI] [PubMed] [Google Scholar]
- 12.Paulsen IT, Brown MH, Littlejohn TG, Mitchell BA, Skurray RA. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci U S A 93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguchi N, Hase M, Kitta M, Sasatsu M, Deguchi K, Kono M. 1999. Antiseptic susceptibility and distribution of antiseptic-resistance genes in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 172:247–253. doi: 10.1111/j.1574-6968.1999.tb13475.x. [DOI] [PubMed] [Google Scholar]
- 14.Mayer S, Boos M, Beyer A, Fluit AC, Schmitz FJ. 2001. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J Antimicrob Chemother 47:896–897. doi: 10.1093/jac/47.6.896. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M, Wang T, Onodera Y, Uchida Y, Sato K. 2000. Mechanism of quinolone resistance in Staphylococcus aureus. J Infect Chemother 6:131–139. doi: 10.1007/s101560070010. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz FJ, Hofmann B, Hansen B, Scheuring S, Luckefahr M, Klootwijk M, Verhoef J, Fluit A, Heinz HP, Kohrer K, Jones ME. 1998. Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12-8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J Antimicrob Chemother 41:481–484. doi: 10.1093/jac/41.4.481. [DOI] [PubMed] [Google Scholar]
- 17.Wang T, Tanaka M, Sato K. 1998. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob Agents Chemother 42:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conant GC, Wolfe KH. 2008. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24:861–862. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- 19.Milstone AM, Passaretti CL, Perl TM. 2008. Chlorhexidine: expanding the armamentarium for infection control and prevention. Clin Infect Dis 46:274–281. doi: 10.1086/524736. [DOI] [PubMed] [Google Scholar]
- 20.McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 12:147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, Weinstein RA, Sepkowitz KA, Jernigan JA, Sanogo K, Wong ES. 2013. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med 368:533–542. doi: 10.1056/NEJMoa1113849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans HL, Dellit TH, Chan J, Nathens AB, Maier RV, Cuschieri J. 2010. Effect of chlorhexidine whole-body bathing on hospital-acquired infections among trauma patients. Arch Surg 145:240–246. doi: 10.1001/archsurg.2010.5. [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Li S, Li L, Wu X, Zhang W. 2013. Effects of daily bathing with chlorhexidine and acquired infection of methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus: a meta-analysis. J Thorac Dis 5:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, Hayden MK, Jernigan JA, Weinstein RA, Fraser VJ, Haffenreffer K, Cui E, Kaganov RE, Lolans K, Perlin JB, Platt R, CDC Prevention Epicenters Program, AHRQ DECIDE Network and Healthcare-Associated Infections Program. 2013. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med 368:2255–2265. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fritz SA, Hogan PG, Camins BC, Ainsworth AJ, Patrick C, Martin MS, Krauss MJ, Rodriguez M, Burnham CA. 2013. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob Agents Chemother 57:559–568. doi: 10.1128/AAC.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller LG, Tan J, Eells SJ, Benitez E, Radner AB. 2012. Prospective investigation of nasal mupirocin, hexachlorophene body wash, and systemic antibiotics for prevention of recurrent community-associated methicillin-resistant Staphylococcus aureus infections. Antimicrob Agents Chemother 56:1084–1086. doi: 10.1128/AAC.01608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitman TJ, Schlett CD, Grandits GA, Millar EV, Mende K, Hospenthal DR, Murray PR, Tribble DR. 2012. Chlorhexidine gluconate reduces transmission of methicillin-resistant Staphylococcus aureus USA300 among Marine recruits. Infect Control Hosp Epidemiol 33:809–816. doi: 10.1086/666631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milstone AM, Elward A, Song X, Zerr DM, Orscheln R, Speck K, Obeng D, Reich NG, Coffin SE, Perl TM, Pediatric SCRUB Trial Study Group. 2013. Daily chlorhexidine bathing to reduce bacteraemia in critically ill children: a multicentre, cluster-randomised, crossover trial. Lancet 381:1099–1106. doi: 10.1016/S0140-6736(12)61687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fritz SA, Hogan PG, Hayek G, Eisenstein KA, Rodriguez M, Epplin EK, Garbutt J, Fraser VJ. 2012. Household versus individual approaches to eradication of community-associated Staphylococcus aureus in children: a randomized trial. Clin Infect Dis 54:743–751. doi: 10.1093/cid/cir919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDanel JS, Murphy CR, Diekema DJ, Quan V, Kim DS, Peterson EM, Evans KD, Tan GL, Hayden MK, Huang SS. 2013. Chlorhexidine and mupirocin susceptibilities of methicillin-resistant Staphylococcus aureus from colonized nursing home residents. Antimicrob Agents Chemother 57:552–558. doi: 10.1128/AAC.01623-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlett CD, Millar EV, Crawford KB, Cui T, Lanier JB, Tribble DR, Ellis MW. 2014. Prevalence of chlorhexidine-resistant methicillin-resistant Staphylococcus aureus following prolonged exposure. Antimicrob Agents Chemother 58:4404–4410. doi: 10.1128/AAC.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang JT, Sheng WH, Wang JL, Chen D, Chen ML, Chen YC, Chang SC. 2008. Longitudinal analysis of chlorhexidine susceptibilities of nosocomial methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J Antimicrob Chemother 62:514–517. doi: 10.1093/jac/dkn208. [DOI] [PubMed] [Google Scholar]
- 33.Horner C, Mawer D, Wilcox M. 2012. Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J Antimicrob Chemother 67:2547–2559. doi: 10.1093/jac/dks284. [DOI] [PubMed] [Google Scholar]
- 34.Leelaporn A, Paulsen IT, Tennent JM, Littlejohn TG, Skurray RA. 1994. Multidrug resistance to antiseptics and disinfectants in coagulase-negative staphylococci. J Med Microbiol 40:214–220. doi: 10.1099/00222615-40-3-214. [DOI] [PubMed] [Google Scholar]
- 35.Batra R, Cooper BS, Whiteley C, Patel AK, Wyncoll D, Edgeworth JD. 2010. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin Infect Dis 50:210–217. doi: 10.1086/648717. [DOI] [PubMed] [Google Scholar]
- 36.Lee AS, Macedo-Vinas M, Francois P, Renzi G, Schrenzel J, Vernaz N, Pittet D, Harbarth S. 2011. Impact of combined low-level mupirocin and genotypic chlorhexidine resistance on persistent methicillin-resistant Staphylococcus aureus carriage after decolonization therapy: a case-control study. Clin Infect Dis 52:1422–1430. doi: 10.1093/cid/cir233. [DOI] [PubMed] [Google Scholar]
- 37.Otter JA, Patel A, Cliff PR, Halligan EP, Tosas O, Edgeworth JD. 2013. Selection for qacA carriage in CC22, but not CC30, methicillin-resistant Staphylococcus aureus bloodstream infection isolates during a successful institutional infection control programme. J Antimicrob Chemother 68:992–999. doi: 10.1093/jac/dks500. [DOI] [PubMed] [Google Scholar]
- 38.Cookson BD, Bolton MC, Platt JH. 1991. Chlorhexidine resistance in methicillin-resistant Staphylococcus aureus or just an elevated MIC? An in vitro and in vivo assessment. Antimicrob Agents Chemother 35:1997–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donnell SM, Janssen GR. 2001. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J Bacteriol 183:1277–1283. doi: 10.1128/JB.183.4.1277-1283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson AF, Rosenberg A, Alatary SD. 1978. Comparative evaluation of surgical scrub preparations. Surg Gynecol Obstet 146:63–65. [PubMed] [Google Scholar]
- 41.Noguchi N, Nakaminami H, Nishijima S, Kurokawa I, So H, Sasatsu M. 2006. Antimicrobial agent of susceptibilities and antiseptic resistance gene distribution among methicillin-resistant Staphylococcus aureus isolates from patients with impetigo and staphylococcal scalded skin syndrome. J Clin Microbiol 44:2119–2125. doi: 10.1128/JCM.02690-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwong SM, Lim R, Lebard RJ, Skurray RA, Firth N. 2008. Analysis of the pSK1 replicon, a prototype from the staphylococcal multiresistance plasmid family. Microbiology 154:3084–3094. doi: 10.1099/mic.0.2008/017418-0. [DOI] [PubMed] [Google Scholar]