Abstract
Nocardia thailandica is a rare pathogen related to Nocardia asteroides, Nocardia neocaledoniensis, and Nocardia caishijiensis that, since its original description in 2004, has only been reported to cause wound and ocular infections in humans. We report a case of pulmonary nocardiosis caused by Nocardia thailandica in a 66-year-old solid organ transplant patient from Connecticut, which was identified at the molecular taxonomic level by secA1 analysis, 16S rRNA gene sequencing, and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). To our knowledge, this is the first reported case of N. thailandica in the United States and the first report of pulmonary infection by this pathogen in the literature.
CASE REPORT
We report a case of pulmonary nocardiosis caused by Nocardia thailandica in a 66-year-old patient from Connecticut with a history of ischemic cardiomyopathy and heart transplant in 2013.
The patient presented complaining of upper respiratory symptoms including sinus pressure, headaches, and decreased hearing in the left ear for a month prior to admission. There was no history of fevers, chills, exposure to sick contacts, or recent travel. Nine months earlier, he was admitted and underwent heart transplant surgery, for which he received and was maintained on immunosuppressive therapy with tacrolimus, mycophenolate, and prednisone, as well as prophylactic treatment with valganciclovir and trimethoprim-sulfamethoxazole (TMP-SMX). Four months after his cardiac transplant, the patient developed chronic cough. Chest X rays and computed tomography (CT) scanning performed at that time revealed bilateral patchy, ill-defined, dense nodular parenchymal opacities (Fig. 1A and B). Sputum Gram stains revealed few hyphae, and cultures performed from a bronchoalveolar lavage (BAL) fluid sample grew 1 CFU of Aspergillus fumigatus. A right upper lobe transbronchial biopsy specimen revealed mild chronic mucosal wall inflammation with no evidence of malignancy or granuloma, and the acid-fast bacillus (AFB) and Grocott-Gomori's methenamine silver (GMS) stains were negative. Routine bacterial, AFB, and mycology cultures from lavage samples were without growth, as were AFB cultures of the biopsy specimen. The patient was started on voriconazole and had subjective improvement after 2 weeks.
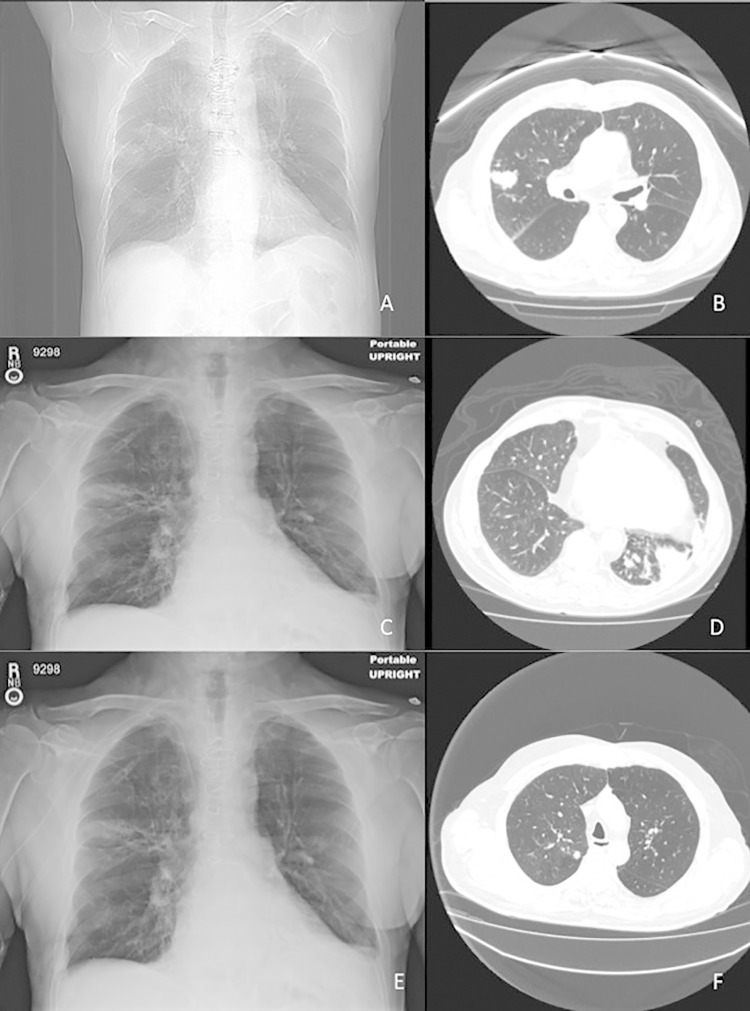
FIG 1.
(A, B) Bilateral patchy, ill-defined, dense nodular, parenchymal opacities in the lungs at 4 months after heart transplant. (C, D) Pulmonary embolism with large clot burden within the right pulmonary artery, multiple areas of consolidation with cavitation, evolving from larger denser areas and new left lower lobe consolidation at 8 months posttransplant. (E, F) Follow-up 6 weeks into therapy revealing nodular consolidative changes, some improved and others new and enlarged.
Four months later, and while still on voriconazole, he presented with acute shortness of breath. A chest CT scan showed pulmonary embolism with a large clot burden within the right pulmonary artery and multiple areas of consolidation with cavitation, evolving from larger, denser areas of consolidation seen in the prior study (Fig. 1C and D). In addition, a new left lower lobe consolidation was revealed. He was started on enoxaparin, and an inferior vena cava (IVC) filter was placed. A sputum Gram stain revealed Gram-negative bacilli later identified as Pseudomonas aeruginosa. Additionally, rhinovirus was detected in a BAL fluid specimen by multiplex PCR (Biofire FilmArray). A repeat right upper lobe transbronchial biopsy specimen confirmed the presence of focal chronic lymphocytic inflammation, again with no evidence of fungal elements or granulomas. AFB and mycology analyses of BAL fluid specimens were culture negative, and routine bacterial culture grew only <1+ normal flora. Voriconazole levels were performed, and the results were within the therapeutic range (voriconazole levels of 1.8 μg/ml). However, because of concern over voriconazole failure and/or superimposed bacterial infection, the patient was placed on ceftazidime (1 g every 8 h [q8h]), voriconazole (200 mg q8h), and caspofungin (50 mg q24h).
On a return visit a month later, radiographic studies showed progression of airspace disease despite improvement in his pulmonary symptoms. At this time, caspofungin was stopped and the patient continued on voriconazole. However, the patient returned again 2 weeks after with symptoms of viral rhinosinusitis and worsening pulmonary infiltrates on imaging, which prompted his admission.
He did not report fever, shortness of breath, or cough. His prior medical history was significant for chronic obstructive pulmonary disease (COPD), hypersensitivity lung disease (HLD), cerebral vascular accident, and upper gastrointestinal bleeding due to duodenal angioectasia. He was a former smoker and denied any allergies, exposure to pets or animals, or recent travel history. His physical examination was unremarkable except for swollen and hyperemic nasal turbinates and poor dentition. A complete blood count was notable for total leukocytes of 9,000/mm3, 82% neutrophils, 3.0% lymphocytes, decreased hemoglobin of 11 g/dl, and hematocrit of 33% but a normal platelet count of 463,000/mm3. Blood urea nitrogen, serum creatinine, and glucose were within reference ranges.
Shortly after admission, the patient underwent a percutaneous left lung biopsy and a biopsy specimen was obtained. An initial Gram stain revealed the presence of 1+ thin, branching, beaded Gram-positive rods (Fig. 2B). The routine microbiological cultures (blood agar, chocolate agar, and Sabouraud-dextrose agar at 35°C and 6% CO2) grew distinct, white/beige, hard, rough-surfaced colonies, pitting into the agar (Fig. 2A), highly suggestive of Nocardia species. Growth was observed on Lowenstein-Jensen medium after 3 days (at 35°C), revealing colonies with the same characteristics. Modified acid-fast stain confirmed the presence of partially acid-fast branching rods.
FIG 2.
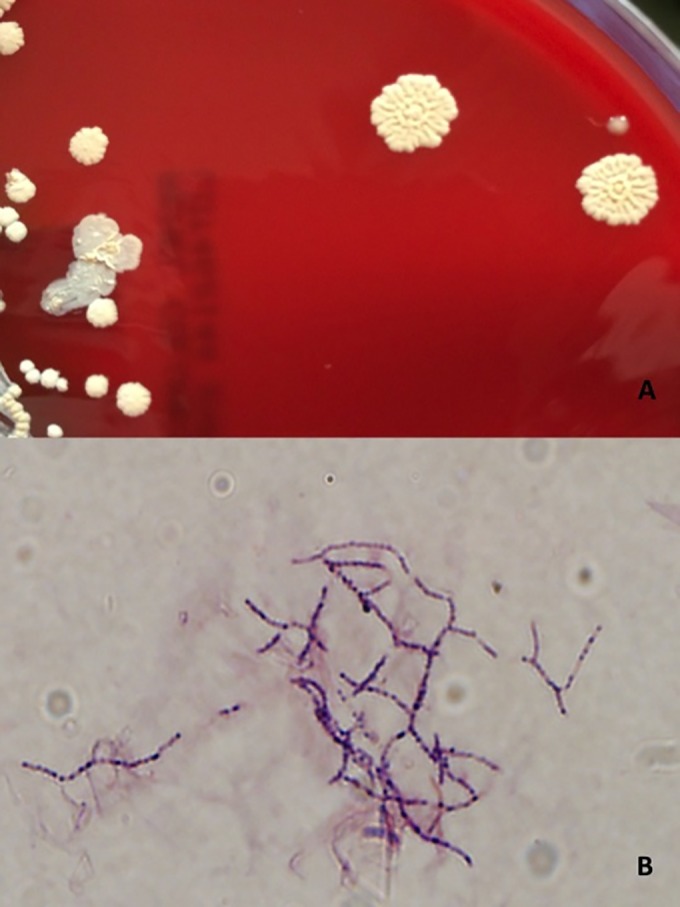
(A) Individual colonies were dome shaped, with a white/beige, hard, rough (coral-like) surface (blood agar, 35°C). (B) Characteristic thin, branching, beaded Gram-positive rods (Gram stain, ×100 magnification).
Direct colony method identification through matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was attempted but failed to render a reliable identification. Subsequently, the isolate was grown on blood agar and subjected to an extraction protocol that followed the inactivated-Mycobacteria/-Nocardia bead preparation method as suggested by the manufacturer (Bruker Daltonics, Inc., Billerica, MA). In brief, a 1-μl loopful of Nocardia colony biomass was collected in a 1.5-ml Eppendorf tube containing 300 μl of deionized water and 900 μl of absolute ethanol, which was then vortexed and incubated for 10 min at 100°C. The sample was then centrifuged at maximum speed for 2 min, followed by the addition of 500 μl of high-performance liquid chromatography (HPLC)-grade water, vortexing, and recentrifugation. Next, the supernatant was removed using a pipette, followed by the addition of 50 μl of deionized water, vortexing, and resuspension of the pellet. After a 60-min heat inactivation at 100°C, the tube was allowed to cool down, and 1,200 μl of precooled absolute ethanol was added. The specimen was centrifuged at maximum speed for 4 min, and ethanol was carefully removed by pipetting and allowing the pellet to air dry for 5 min. Later, 200 μl of 1-mm silica beads was added, with consequent resuspension of the pellet through vortexing with a combination of 30 μl of pure acetonitrile and 30 μl of 70% formic acid. After mixing by vortexing for 5 s and centrifuging at maximum speed for 2 min, 1 μl of the supernatant was placed on the MALDI target, allowed to dry, and then overlaid with 1 μl of matrix solution for subsequent analysis by MALDI-TOF MS using the MicroFlex LT mass spectrometer (Bruker Daltonics, Inc., Billerica, MA). Based on the spectral fingerprint of the isolate, this confirmed the identification (identification score of 1.651) as Nocardia asteroides, followed by this same species as the second (score of 1.507) and third (score of 1.303) best matches, using the mycobacterial Mycolib 1.03 and bacterial MALDI Biotyper database version 3.3.1.2 libraries (Bruker). However, this score was insufficient to identify the isolate to the species level (a score of 2.0 or better is recommended) or even definitively to the genus level (score of 1.7 or better).
Because the score was below the cutoff recommended by the manufacturer for confident identification to the genus level and the next-closest (but poor) matches were also to Nocardia species (Nocardia testacea, score of 1.179, and Nocardia thailandica, score of 1.102), we concluded that this isolate was most likely a Nocardia species either not or poorly represented in the database, and so we decided to further characterize this isolate at the molecular taxonomic level. Sequencing for identification was performed using the Fast MicroSeq 500 16S rRNA bacterial identification kit and 3130xl genetic analyzer (Life Technologies). It was identified as N. thailandica (100% sequence identity) using the BLAST search tool based on the GenBank database, followed by this same species as second and third best matches with 100% sequence identity.
Molecular taxonomic confirmation of the isolate was performed by SecA1 amino acid analysis based on secA1 gene sequencing. DNA was extracted from the clinical isolate using PrepMan ultra sample preparation reagent according to the manufacturer's protocol (Life Technologies, Carlsbad, CA). Following the procedure of Conville et al. (2), a region of the secA1 gene corresponding to bases 444 to 913 of the secA1 gene sequence of Nocardia farcinica IFM 10152 (3) was amplified, using the following primers with tails containing M13 binding sites: 5′ GTA AAA CGA CGG CCA GGA CAG YGA GTC GAT GGG YCG SGT GCA CCG 3′ and 5′ CAG GAA ACA GCT ATG ACG CGG ACG ATG TAG TCC TTG TC 3′. PCR was performed using 1 pmol of each primer, approximately 0.2 μg of extracted DNA, and the FailSafe enzyme kit (Epicentre, Madison, WI). The amplicon underwent cycle sequencing using primers M13-forward (5′ GTA AAA CGA CGG CCA G 3′) and M13 reverse (5′ CAG GAA ACA GCT ATG AC 3′) (ABI BigDye terminator version 3.1 cycle sequencing kit; Life Technologies, Carlsbad, CA) with the protocol according to the manufacturer's instructions. Sequencing analysis was performed on the ABI3500 genetic analyzer (Life Technologies, Carlsbad, CA). Sequence analysis and alignment of a 468-bp region of the secA1 gene was performed. Amino acid sequences were deduced from the secA1 gene sequences using the six-frame translation tool of San Diego Supercomputer Center (SDSC) Biology Workbench (La Jolla, CA).
An in-house validation was performed on approximately 20 isolates of each of the available Nocardia species in the UT Health Science Center Mycobacteria/Nocardia laboratory. The secA1 gene sequences of all the isolates in the validation were analyzed and compared to the Nocardia type strains using the cutoff values suggested by Conville et al. (2), and a database was created in-house for secA1 gene analysis. The amino acid sequences obtained were compared to the sequences in the in-house-validated secA1 database.
Alignment of the deduced amino acid sequence, comprised of 156 amino acid residues, showed 99.36% similarity (1 amino acid change) to the type strain of N. thailandica. There are no current CLSI guidelines for interpretation of secA1 sequences. Thus, the percent similarity and amino acid mismatch corresponding to ≥99% identity to the type strain were used, as in the method of Conville et al. (2).
16S rRNA sequencing.
Sequencing of the 1,500 bp of the 16S rRNA gene was then performed as previously described by Edwards et al. (1), using the ABI 3500 genetic analyzer. The following primers were used: pA-F, AGA GTT TGA TCC TGG CTC AG; pC-F, CTA CGG GAG GCA GCA GTG GG; pD-R, CAG CAG CCG CGG TAA TAC; pE-F, AAA CTC AAA GGA ATT GAC GG; and pH-R, AAG GAG GTG ATC CAG CCG CA. PCR was performed using primers pA-F and pH-R. Cycle sequencing was performed using primers pC-F, pD-R, and pE-F and the BigDye terminator version 3.1 cycle sequencing kit according to the manufacturer's instructions. Analysis of the full 16S rRNA gene using Ripseq software (Isentio A.S, Bergen, Norway) gave a 100% match (1422/1422 bp) to the 16S rRNA gene of N. thailandica type strain IFM 10145 (GenBank accession number AB126874).
The growth and morphology of the organism, along with the sequencing data, were consistent with the published description of N. thailandica, with the organism exhibiting tan/beige colony growth at 35°C and typical branched mycelium branching into bacteroid, rod-shaped, Gram-positive elements (4, 5). Susceptibility testing using the CLSI-recommended method of broth microdilution revealed that the organism was susceptible to amikacin, ceftriaxone, meropenem, tobramycin, imipenem, amoxicillin clavulanate, clarithromycin, cefepime, minocycline, linezolid, moxifloxacin, and trimethoprim-sulfamethoxazole (TMP-SMX) and resistant to ciprofloxacin (6).
The patient was treated initially with a combination of TMP-SMX (2 double-strength tablets twice a day [BID]) and meropenem (1 g q8h); however, after 1 week of therapy, he developed hyperkalemia, which required changing the TMP-SMX to minocycline (100 mg BID). Meropenem was continued for 1 month before discontinuation, and plans are to continue minocycline for 6 to 12 months. Follow-up imaging 6 weeks into therapy documented an overall good response to therapy despite minimal enlargement of the right and left upper lobe opacities, with significant interval reduction in the size of the left upper lobe and left lower lobe opacities (Fig. 1E and F).
N. thailandica was described by Kageyama et al. for the first time in a clinical wound specimen from a Thai patient in 2004 (4).
Microscopically, N. thailandica is characterized by staining Gram positive and partially acid fast (4). The organisms usually appear as branched mycelia, which fragment into bacteroid rod-shaped elements ranging in size from 0.4 by 0.8 to 0.6 by 1.6 μm (4, 5). The colonies are white and rough and grow rapidly (2 to 3 days) to later develop a gray, orange-to-tan appearance (4). The organism is aerobic, and growth occurs at 30 to 37°C but not at 45°C on Mueller-Hinton II (MHII) agar with 0.2% glucose (4), as well as on Lowenstein-Jensen medium. Isolates exhibit an aerial mycelium, which is long and visible in most media (4, 5). The organism utilizes glucose and mannose as carbon sources but not arabinose, galactose, inositol, maltose, rhamnose, sorbitol, xylose, or citrate, and it is urease positive (5).
Because of its strong similarity to other Nocardia species (4, 5), routine methods of identification may fail to recognize this agent; like many Nocardia species, it is relatively nonreactive biochemically, and many new species have been described that are indistinguishable using conventional methods (4, 5). Other methods, such as gene sequencing and mass spectrometry, are necessary for reliable identification of all the currently recognized species.
Nocardia thailandica is closely related to N. asteroides, N. neocaledoniensis, and N. caishijiensis based on DNA-DNA relatedness data (4) and can be distinguished from these based on its distinct phenotypic properties (4, 5). However, major phenotypic characteristics reveal a pattern identical to that of Nocardia asteroides, precluding its differentiation from the aforementioned related species (4, 5). In this case, MALDI-TOF analysis using a routine formic acid-acetonitrile method failed to accurately identify the isolate, rendering a nonreliable result, probably due to the nature of the Nocardia cell wall. However, tentative identification to the genus level was possible after preparing the samples using the inactivated-Mycobacteria bead preparation method (Bruker Daltonics, Inc., Billerica, MA) and the modified standard Mycobacteria and Nocardia protocol (Vitek MS; bioMérieux, France) which allowed breaking the cell wall of the organism to subsequently expose the targetable proteins by bead-beating mechanical disruption. Failure to identify this organism could also have resulted from the absence of representative reference spectra for certain species or protein variability among strains, as could have been the scenario for this case.
To date, this species has been isolated only from purulent secretion from an abscess of a Thai patient (4), as well as patients with choroiditis and keratitis (7) in Asia; however, no other environmental source has been described, nor has close-contact transmission been reported for this species. Nevertheless, like other Nocardia species, N. thailandica appears to be an environmental agent that may act as an opportunistic pathogen in immunocompromised hosts (5). Though infrequent, pulmonary nocardiosis is important to recognize and treat, as it can cause significant morbidity and mortality (5). The use of immunosuppressants, such as those used after hematopoietic stem cell transplants and solid organ transplantation (in this case), have been reported as major risk factors for nocardial infection (8). In addition, older age (9) and preexisting structural abnormalities of the lung (10) are also associated with a higher risk of mortality in pulmonary nocardiosis. Interestingly, pulmonary aspergillosis is also an independent and significant risk factor for overall mortality in pulmonary nocardiosis patients (9). Stereotypically, our patient had undergone a heart transplant 9 months prior, was on immunosuppressive treatment, had significant underlying compromise of his lung function due to COPD, and had recently been diagnosed and treated for pulmonary aspergillosis.
Nocardia infections cause a vast spectrum of potentially treatable disease that ranges from localized cutaneous infections to severe pulmonary and central nervous system involvement in immunocompromised hosts (5). Isolates that phenotypically resemble N. asteroides are usually divided into six different drug susceptibility patterns (5). However, while correct identification to species level allows the prediction of likely antimicrobial susceptibility patterns, infrequently reported species may pose a challenge, prompting susceptibility testing of all isolates (5, 11, 12). Our isolate proved susceptible to the majority of antimicrobials tested for Nocardia species, with the exception of ciprofloxacin, contrasting with other reported N. thailandica isolates (7), which exhibited completely different patterns, thus suggesting important interspecies variability and/or methodological differences.
While there have been no randomized controlled trials to allow the recommendation of an optimal therapy for pulmonary nocardiosis, in general, the agents most commonly used include trimethoprim-sulfamethoxazole, amikacin, carbapenems, and third-generation cephalosporins (5, 13), which were used successfully in our patient.
In conclusion, Nocardia species are primarily opportunist organisms, and infection occurs more frequently in patients with underlying immune suppression (14). To date, N. thailandica has only been reported from wound and eye infections in Japan and India (4, 7). Thus, this is the first reported case of N. thailandica in the United States and the first report of pulmonary infection, suggesting an increasing role of N. thailandica as an opportunistic pathogen in immunocompromised patients and the broadening of its disease spectrum. In addition, this case highlights the importance of using a combined approach in Nocardia identification, the ever-increasing utility of novel diagnostic platforms like MALDI-TOF MS for its identification, and the need for microbial databases for advanced diagnostic methods to be continually checked and updated to include rare, novel, geographically limited, and emerging pathogens.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of the isolate studied was deposited in GenBank under accession number KP966094.
REFERENCES
- 1.Edwards U, Rogal T, Blöcker H, Emde M, Böttger EC. 1989. Isolation and direct sequencing of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conville PS, Zelazny AM, Witebsky FG. 2006. Analysis of secA1 gene sequences for identification of Nocardia species. J Clin Microbiol 44:2760–2766. doi: 10.1128/JCM.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, Hotta K, Shiba T, Hattori M. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc Natl Acad Sci U S A 101:14925–14930. doi: 10.1073/pnas.0406410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kageyama A, Poonwan N, Yazawa K, Suzuki S-I, Kroppenstedt RM, Mikami Y. 2004. Nocardia vermiculata sp. nov. and Nocardia thailandica sp. nov. isolated from clinical specimens. Actinomycetologica 18:27–33. doi: 10.3209/saj.18_27. [DOI] [Google Scholar]
- 5.Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev 19:259–282. doi: 10.1128/CMR.19.2.259-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2011. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes. Approved standard, 2nd ed CLSI document M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 7.Reddy AK, Garg P, Kaur I. 2010. Speciation and susceptibility of Nocardia isolated from ocular infections. Clin Microbiol Infect 16:1168–1171. doi: 10.1111/j.1469-0691.2009.03079.x. [DOI] [PubMed] [Google Scholar]
- 8.Minero MV, Marín M, Cercenado E, Rabadán PM, Bouza E, Muñoz P. 2009. Nocardiosis at the turn of the century. Medicine 88:250–261. doi: 10.1097/MD.0b013e3181afa1c8. [DOI] [PubMed] [Google Scholar]
- 9.Kurahara Y, Tachibana K, Tsuyuguchi K, Akira M, Suzuki K, Hayashi S. 2014. Pulmonary nocardiosis: a clinical analysis of 59 cases. Respir Investig 52:160–166. doi: 10.1016/j.resinv.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Blackmon KN, Ravenel JG, Gomez JM, Ciolino J, Wray DW. 2011. Pulmonary nocardiosis: computed tomography features at diagnosis. J Thorac Imaging 26:224–229. doi: 10.1097/RTI.0b013e3181f45dd5. [DOI] [PubMed] [Google Scholar]
- 11.Schlaberg R, Fisher MA, Hanson KE. 2014. Susceptibility profiles of Nocardia isolates based on current taxonomy. Antimicrob Agents Chemother 58:795–800. doi: 10.1128/AAC.01531-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhde KB, Pathak S, McCullum I Jr, Jannat-Khah DP, Shadomy SV, Dykewicz CA, Clark TA, Smith TL, Brown JM. 2010. Antimicrobial-resistant nocardia isolates, United States, 1995-2004. Clin Infect Dis 51:1445–1448. doi: 10.1086/657399. [DOI] [PubMed] [Google Scholar]
- 13.Tripodi MF, Durante-Mangoni E, Fortunato R, Cuccurullo S, Mikami Y, Farina C, Utili R. 2011. In vitro activity of multiple antibiotic combinations against Nocardia: relationship with a short-term treatment strategy in heart transplant recipients with pulmonary nocardiosis. Transpl Infect Dis 13:335–343. doi: 10.1111/j.1399-3062.2010.00588.x. [DOI] [PubMed] [Google Scholar]
- 14.Lebeaux D, Morelon E, Suarez F, Lanternier F, Scemla A, Frange P, Mainardi JL, Lecuit M, Lortholary O. 2014. Nocardiosis in transplant recipients. Eur J Clin Microbiol Infect Dis 33:689–702. doi: 10.1007/s10096-013-2015-5. [DOI] [PubMed] [Google Scholar]