Abstract
Salmonella enterica serovar Typhimurium is an important foodborne human pathogen that often causes self-limiting but severe gastroenteritis. Prolonged excretion of S. Typhimurium after the infection can lead to secondary transmissions. However, little is known about within-host genomic variation in bacteria associated with asymptomatic shedding. Genomes of 35 longitudinal isolates of S. Typhimurium recovered from 11 patients (children and adults) with culture-confirmed gastroenteritis were sequenced. There were three or four isolates obtained from each patient. Single nucleotide polymorphisms (SNPs) were analyzed in these isolates, which were recovered between 1 and 279 days after the initial diagnosis. Limited genomic variation (5 SNPs or fewer) was associated with short- and long-term carriage of S. Typhimurium. None of the isolates was shown to be due to reinfection. SNPs occurred randomly, and the majority of the SNPs were nonsynonymous. Two nonsense mutations were observed. A nonsense mutation in flhC rendered the isolate nonmotile, whereas the significance of a nonsense mutation in yihV is unknown. The estimated mutation rate is 1.49 × 10−6 substitution per site per year. S. Typhimurium isolates excreted in stools following acute gastroenteritis in children and adults demonstrated limited genomic variability over time, regardless of the duration of carriage. These findings have important implications for the detection of possible transmission events suspected by public health genomic surveillance of S. Typhimurium infections.
INTRODUCTION
Nontyphoid Salmonella (NTS) infections generally result in a short-term, self-limiting gastroenteritis. However, NTS can be excreted continually and asymptomatically in stools for many weeks, even after the initial diarrheal episode has been resolved. Children are the more common carriers, especially children under the age of 3 years (1, 2). Fecal shedding of NTS after an intestinal infection can last for up to 4 weeks in adults and 7 weeks in children (3). In a very small proportion of cases, carriage can last for a year after the initial onset of the disease (3). Carriers excrete large numbers of bacteria in their feces and can facilitate the transmission of Salmonella to other hosts by contaminating water and food sources. The persistence of fecal shedding in asymptomatic patients can have a duration similar to that for patients with clinical disease (4). Antibiotic treatment of NTS disease is rarely indicated, as it does not assist in clearance of infection but may increase the duration of asymptomatic shedding (4).
Salmonella enterica serovar Typhimurium is one of the leading causes of NTS gastroenteritis in humans in Australia and other countries. Salmonellosis is a notifiable disease in all Australian states and territories. In the state of New South Wales (NSW), the notification rate has been around 50 cases per 100,000 population (5). Sequencing of S. Typhimurium genomes or analyses of tandem repeats within the genome have increasingly been employed for tracking community outbreaks and transmission chains (6, 7). However, the magnitude of genomic variation in isolates associated with carriage remains unknown. Although they are relatively infrequent, we identified 11 cases of prolonged carriage based on data from the NSW Enteric Reference Laboratory, Pathology West, Sydney, Australia. In this study, isolates from these 11 patients, who had samples positive for S. Typhimurium over different periods following gastroenteritis, were compared using multilocus variable-number tandem-repeat analysis (MLVA) and high-throughput genome sequencing. We aimed to establish the genetic relatedness of the longitudinal isolates recovered from patients and to determine if there was any reinfection by other endemic strains.
MATERIALS AND METHODS
Ethics statement.
The isolates used in this study were obtained during routine diagnostic testing and follow-up and were submitted to the NSW Enteric Reference Laboratory, Pathology West, Sydney, New South Wales, Australia, for confirmatory testing.
Selection of patients and samples.
A total of 35 isolates collected from 11 different patients were selected for sequencing. Each isolate came from a sample collected at a given time point for purposes of laboratory diagnosis or bacterial clearance testing. Fecal samples were subcultured on xylose-lysine-deoxycholate agar and their identification confirmed. Serotyped cultures were then stored on STGG (skim milk powder, tryptone soy broth, glucose, and glycerol) medium at −80°C. Prior to genomic experiments, stored isolates were plated out on nutrient agar, and DNA were extracted from randomly selected single colonies. For each patient, there were three or four isolates analyzed. Each isolate differed by 1 to 182 days from the isolate collected earlier from the same patient (Table 1). The ages of the patients ranged from 1 to 70 years. All isolates were subjected to MLVA at the NSW Enteric Reference Laboratory, using a protocol compatible with the European MLVA-5 scheme (8). Genomic DNA was extracted by phenol-chloroform extraction using the method described by Octavia and Lan (9).
TABLE 1.
Isolates analyzed in this study
| Patient no. | MLVA type | Isolate | Date collected (day/mo/yr) | No. of days aparta | No. of days from first isolation | No. of reads | Fold coverage |
|---|---|---|---|---|---|---|---|
| 1 | 2-8-13-11-112 | A1 | 14/12/2010 | 0 | 939,662 | 31.96 | |
| 2-8-13-11-112 | A2 | 2/02/2011 | 50 | 50 | 960,460 | 21.93 | |
| 2-8-13-11-112 | A3 | 12/03/2011 | 38 | 98 | 1,247,482 | 34.72 | |
| 2 | 2-14-10-10-212 | B1 | 6/01/2013 | 0 | 766,736 | 23.84 | |
| 2-14-10-10-212 | B2 | 30/01/2013 | 24 | 24 | 934,536 | 19.62 | |
| 2-14-10-10-212 | B3 | 26/02/2013 | 27 | 51 | 1,217,706 | 38.18 | |
| 3 | 2-8-13-11-112 | C1 | 20/02/2012 | 0 | 1,598,820 | 43.67 | |
| 2-8-13-11-112 | C2 | 2/04/2012 | 42 | 42 | 1,062,126 | 33.33 | |
| 2-8-13-11-112 | C3 | 1/05/2012 | 29 | 71 | 906,012 | 26.59 | |
| 2-8-13-11-112 | C4 | 31/05/2012 | 30 | 101 | 1,098,896 | 34.37 | |
| 4 | 2-7-6-13-212 | D1 | 10/02/2011 | 0 | 1,556,456 | 59.34 | |
| 2-7-6-13-212 | D2 | 8/03/2011 | 26 | 26 | 1,091,980 | 40.11 | |
| 2-7-6-13-212 | D3 | 3/05/2011 | 56 | 82 | 1,380,158 | 48.92 | |
| 5 | 3-17-0-0-311 | E1 | 27/06/2011 | 0 | 1,423,720 | 48.62 | |
| 3-17-0-0-311 | E2 | 26/10/2011 | 121 | 121 | 1,206,680 | 41.31 | |
| 3-17-0-0-311 | E3 | 27/10/2011 | 1 | 122 | 877,310 | 33.76 | |
| 6 | 2-8-7-8-212 | F1 | 18/12/2010 | 0 | 986,098 | 34.32 | |
| 2-7-6-12-212 | F2-1b | 7/01/2011 | 20 | 20 | 1,211,578 | 41.38 | |
| 2-7-6-12-212 | F2-2b | 7/01/2011 | 0 | 20 | 2,167,602 | 76.01 | |
| 7 | 4-13-7-0-211 | G1 | 6/11/2010 | 0 | 1,082,482 | 34.89 | |
| 3-12-7-0-211 | G2 | 15/11/2010 | 9 | 9 | 1,207,848 | 37.58 | |
| 3-12-7-0-211 | G3 | 30/11/2010 | 15 | 24 | 1,481,814 | 49.41 | |
| 3-12-7-0-211 | G4 | 31/05/2011 | 182 | 206 | 2,190,794 | 41.5 | |
| 8 | 2-7-6-14-212 | H1 | 4/01/2011 | 0 | 971,980 | 36.78 | |
| 2-7-6-14-212 | H2 | 14/04/2011 | 100 | 100 | 625,940 | 23.58 | |
| 2-7-6-14-212 | H3 | 10/10/2011 | 179 | 279 | 1,474,180 | 57.72 | |
| 9 | 4-8-11-8-211 | I1 | 30/04/2012 | 0 | 1,147,288 | 42.81 | |
| 4-8-11-9-211 | I2 | 26/09/2012 | 149 | 149 | 907,912 | 34.24 | |
| 4-8-11-9-211 | I3 | 8/10/2012 | 12 | 161 | 1,230,354 | 48.1 | |
| 10 | 2-7-6-12-212 | J1 | 2/12/2011 | 0 | 1,074,820 | 40.38 | |
| 2-7-6-12-212 | J2 | 6/12/2011 | 4 | 4 | 1,777,186 | 60.26 | |
| 2-7-6-12-212 | J3 | 23/01/2012 | 48 | 52 | 1,620,496 | 63.97 | |
| 11 | 2-11-10-8-212 | K1 | 12/05/2012 | 0 | 2,559,340 | 98.02 | |
| 2-11-10-8-212 | K2 | 13/05/2012 | 1 | 1 | 1,374,252 | 53.14 | |
| 2-11-10-8-212 | K3 | 6/07/2012 | 54 | 55 | 1,839,088 | 72.94 |
Number of days since the collection of the prior isolate.
F2-1 and F2-2 were two different isolates from the same day.
Genome sequencing and assembly.
A 250-bp paired-end library was constructed for each purified DNA sample by using a NexteraXT kit (Illumina) and was sequenced on a MiSeq (Illumina) platform. Reads were assembled using VelvetOptimiser (version 2.2.5) and velvet (version 1.2.10) (10). Contigs were then compared to the S. Typhimurium LT2 reference genome (accession no. NC_003197) and reordered using progressiveMauve (version 2.3.1) (11).
Identification of SNPs.
Single nucleotide polymorphisms (SNPs) were determined using read mapping as well as alignments of de novo-assembled sequences, based on the approach we applied previously (12). Reads were mapped to the S. Typhimurium LT2 chromosomal genome (accession no. NC_003197) by using Burrows-Wheeler alignment (BWA) (version 0.7.5a) (13). Raw SNP calls were filtered for quality scores of ≥20, with a cutoff of 20 reads covering the SNP site and ≥70% of the reads supporting the SNP. SNPs were also validated by comparison of de novo-assembled genomes to the genome of strain LT2 by using progressiveMauve. This was done to eliminate the problem of reads that may be mapped to repeats or homologous regions with mismatches being called SNPs. The final list of SNPs was made up of the SNPs identified by both methods. These SNPs were separated into the following three categories: nonsynonymous SNPs (nsSNPs), synonymous SNPs (sSNPs), and intergenic (IG) regions.
Prophage and plasmid.
Prophage sequences were identified using PHAST (14). Reads were mapped to S. Typhimurium LT2 plasmid pSLT (accession no. NC_003277.1) by using BWA. Mapped reads were extracted using bamtools (15) and were then assembled using VelvetOptimiser and velvet. Contigs were then compared to the complete pSLT genome and reordered using progressiveMauve.
Phylogenetic analysis.
Identified SNPs that were located in repeat regions, insertion sequences, or prophage sequences were excluded from phylogenetic analysis. The remaining SNPs were concatenated, and a maximum parsimony tree was generated using the PAUP package (16), with heuristic searches based on tree bisection and the reconnection swap method. S. enterica serovar Enteritidis PT4 strain NCTC13349 (accession no. AM933172) and S. enterica serovar Choleraesuis strain SC-B67 (accession no. AE017220) were used as the outgroup.
Estimation of in vivo mutation rate.
The mutation rate was estimated from the number of SNPs observed in isolates from all patients. We assumed that both the mutation rate and the growth rate can be averaged for the life span of S. Typhimurium in the host. To estimate the mutation rate and 95% confidence interval (CI), the poissfit function in Matlab (MathWorks) was used, where the total number of SNPs observed was divided by the total time. Mutation was modeled as a Poisson process, which assumes that the mutation rate per unit time is constant.
Nucleotide sequence accession number.
The raw sequencing data from this study were submitted to GenBank under BioProject no. PRJNA285421.
RESULTS AND DISCUSSION
Genome sequencing and overview of the genome contents.
The 35 isolates from the 11 patients were sequenced using Illumina paired-end sequencing with a read length of 250 bp. The strains were multiplexed in batches of 24 for sequencing. The average number of reads generated per genome was 1,282,583, and the coverage depth for all genomes, on average, was 45×. The reads were mapped to LT2 as a reference and also assembled de novo for SNP discovery and determining the presence of novel genes. On average, 96% of the LT2 genome was covered by reads from each sequenced genome. Overall, there were 26 SNPs detected in the 35 isolates obtained from these patients (see Table S1 in the supplemental material). No isolates carried mutator mutations that may increase the mutation rate (17). We did not search for insertions and deletions. The isolates from two patients (patients 5 and 7) did not contain the virulence plasmid pSLT (18). Several virulence factors are known to play a role in fecal shedding or the carrier state, including genes encoding thin aggregative fimbriae (19), genes encoding the secreted effectors ShdA and MisL (20, 21), and genes located on Salmonella pathogenicity island 16 that are responsible for O-antigen glycosylation (22). The isolates analyzed in this study were all found to carry these genes, but none of these genes carried mutations in isolates from the same patient, suggesting that no further adaptive changes in these genes occurred during carriage.
SNP differences between serial isolates from the same patient.
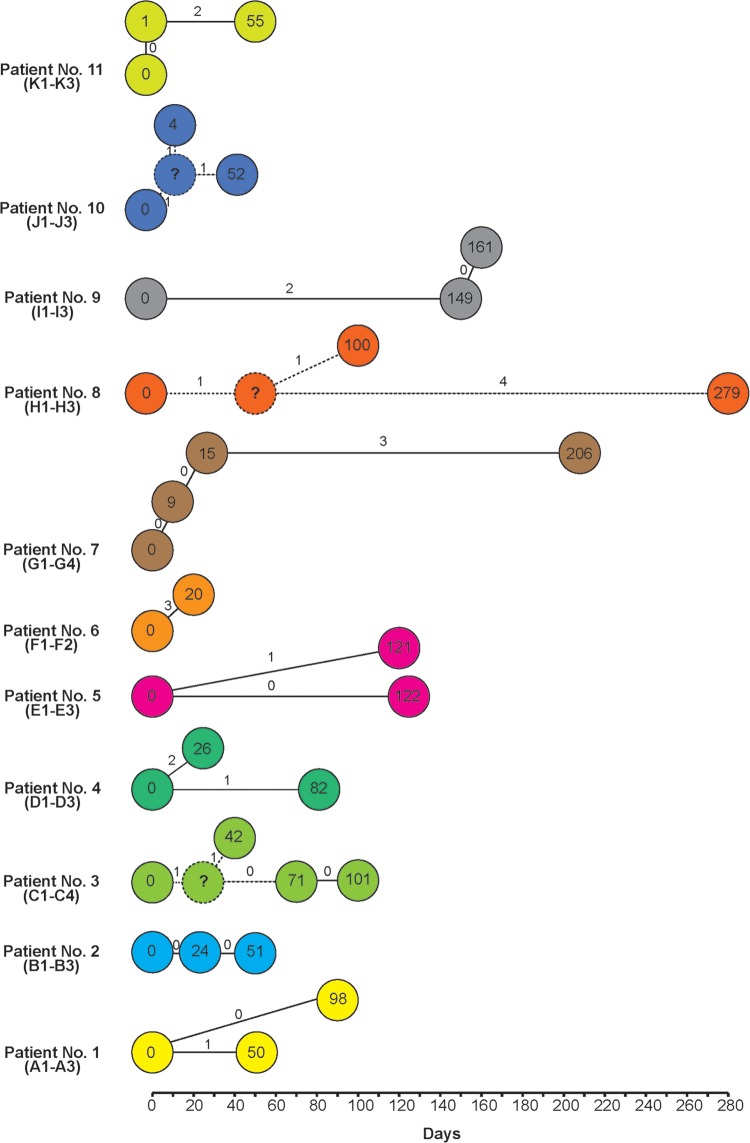
To determine the genetic variation between isolates from the same patient over different periods, we analyzed the SNPs (Fig. 1). The serial isolates showed 0 to 5 SNP differences, suggesting that in all cases the initial infecting isolate persisted in the patient between episodes of disease. SNPs between serial isolates (J1 and J2) were observed as few as 4 days apart. Identical serial isolates were found as long as 122 days apart (E1 and E3). There were two cases of isolates obtained 1 day apart, and these isolates were treated as being from the same episode.
FIG 1.
Numbers of single nucleotide polymorphism differences (on the connecting lines) in isolates from the same patients plotted over time. The connections are based on the assumption that the isolates within the same patient had mutations through a sequence of steps. The numbers in circles show numbers of days since the first isolate was collected. For patients 3 and 10, the symbol “?” is used to represent a postulated intermediate. Hypothetical connections are indicated by dashed lines.
For patient 1, one SNP was detected in the day 50 (A2) isolate, but the day 88 isolate (A3) was identical to the day 0 isolate (A1). For patient 2, all three isolates, from day 0, day 24, and day 51, were identical. For patient 3, two SNPs were observed in the day 42 isolate, one of which was present in the day 71 and day 101 isolates, while the other SNP was not found in the latter isolates. For patient 4, the day 26 isolate (D2) differed by two SNPs from the day 0 (D1) isolate. These SNPs were not observed in the day 56 isolate (D3). Instead, this isolate had another SNP. For patient 5, one SNP was detected in the day 121 isolate (E2). Interestingly, this SNP was not observed in the day 122 isolate (E3), even though the two isolates were collected only 1 day apart. For patient 6, two isolates were collected 20 days apart, and three SNPs were found. For patient 7, no SNPs were detected from the day 9 and day 24 isolates. Three SNPs were detected in the day 206 isolate (G4). For patient 8, two SNPs were detected in the day 100 isolate (H2). However, one of these SNPs was still observed in the day 279 isolate (H3). Additionally, H3 had four other SNPs. For patient 9, the day 149 (I2) and day 161 (I3) isolates were identical and differed from the day 0 (I1) isolate by two SNPs. For patient 10, the day 4 isolate (J2) had two SNPs, one of which was still observed in the day 52 isolate (J3), which carried an additional SNP. For patient 11, the day 0 and day 1 isolates were identical and the day 55 isolate had two SNPs.
Nature of the SNPs identified.
Most of the SNPs observed were located in coding regions, and the majority of them were nsSNPs (Table 2). The nsSNPs were located in 20 different genes, which were mostly in the metabolism category. There were only three synonymous SNPs, which affected three different genes, two of which also belonged to the metabolism category. Multiple SNPs from the same isolate were in different genes which are distantly located, suggesting that they resulted from mutation rather than recombination.
TABLE 2.
Nonsynonymous SNPs in the isolates analyzed
| Isolatea | Gene | Locus | Product | COGb | Amino acid change |
|---|---|---|---|---|---|
| A2 | ttrA | STM1383 | Tetrathionate reductase complex subunit A | Metabolism | A → T |
| C2 | mukB | STM0994 | Cell division protein MukB | Cellular processes and signaling | E → G |
| C2 | nifJ | STM1651 | Pyruvate-flavodoxin oxidoreductase | Metabolism | R → C |
| D2 | STM1787 | Hydrogenase 1 large subunit | Metabolism | R → C | |
| D2 | hisC | STM2073 | Histidinol-phosphate aminotransferase | Metabolism | P → L |
| D3 | masA | STM1076 | Methylglyoxal synthase | Metabolism | T → I |
| E2 | topB | STM1298 | DNA topoisomerase III | Information storage and processing | Q → L |
| F2-1 | malT | STM3515 | Transcriptional regulator MalT | Information storage and processing | E → K |
| G4 | rpoS | STM2924 | RNA polymerase sigma factor RpoS | Information storage and processing | R → C |
| H2 | STM1618 | Transcriptional repressor of sgc operon | Information storage and processing, metabolism | E → K | |
| H2 | recG | STM3744 | ATP-dependent DNA helicase RecG | Information storage and processing | R → S |
| H3 | btuB | STM4130 | Vitamin B12/cobalamin outer membrane transporter | Metabolism | R → H |
| H3 | dcuA | STM4325 | Anaerobic C4-dicarboxylate transporter | Poorly characterized | P → S |
| H3 | yjiE | STM4511 | DNA-binding transcriptional regulator | Information storage and processing | F → L |
| I2 | STM3833 | Mandelate racemase | Cellular processes and signaling, poorly characterized | E → K | |
| I2 | yihV | STM4024.S | Sugar kinase | Metabolism | Stop → E |
| J2 | STM3021 | Inner membrane protein | Poorly characterized | T → P | |
| J2 | rfaK | STM3714 | Hexose transferase | Cellular processes and signaling | T → I |
| K3 | flhC | STM1924.S | Transcriptional activator FlhC | Cellular processes and signaling | Q → Stop |
| K3 | yhdG | STM3384 | tRNA-dihydrouridine synthase B | Information storage and processing | R → S |
The isolate in which the SNP was observed. See Table 1 for isolate details.
COG, cluster of orthologous groups.
Two SNPs resulted in a stop codon. One of these SNPs was observed in the day 55 isolate collected from patient 11 (K3) and affected flhC, a master regulator of the biogenesis of flagella in Salmonella (23). Using triphenyltetrazolium chloride motility agar and a swimming motility assay, we demonstrated that K3 was not motile, while K1 and K2 were both motile. The second nonsense SNP was found in yihV in the day 0 isolate from patient 9 (I1). Interestingly, two subsequent isolates from this patient (I2 and I3) had another SNP in the same position that resulted in a different amino acid from that in the reference strain S. Typhimurium LT2. I2 could have been a direct descendant of I1, which meant that the nonsense mutation was subsequently reversed. However, it is also possible that I2 was present when I1 was sampled and that the two SNPs were independent mutational changes. YihV plays a role in O-antigen capsule assembly and translocation and biofilm formation (24, 25). However, we did not test whether the SNPs observed in yihV affected capsule production and biofilm formation. Considering the functions of the genes involved, these SNPs may have an adaptive advantage.
The majority of the nsSNPs were not maintained in the subsequent isolates. They are likely to have been transient, although it is also possible that the nsSNPs were maintained for a time but were not sampled, as only a single isolate was obtained at any given time point. However, isolates from two patients (C2 to C4 and H2-H3) each contained one nsSNP that was maintained for 59 and 179 days, respectively: one was found in the mukB gene, which plays a role in chromosomal partitioning, and the other was found in the STM1618 gene, encoding a DeoR family transcriptional repressor. It is unknown whether these SNPs have any adaptive value.
There was no overlap in the sets of genes harboring SNPs between isolates from different patients. This suggests that none of these genes was a hot spot for mutation and that the SNPs observed were due to random mutational events. In addition, there was no evidence of an increased mutation rate after the initial infection, as there was no burst of SNPs in any of the subsequent isolates. Only a few SNPs appeared to be passed on in the subsequent isolations, but their significance is unknown.
Estimation of mutation rate from genome data.
By summing the total number of SNPs observed and the total number of days between isolations, we obtained an approximate estimate of the mutation rate in vivo. In total, there were 29 SNPs in 1,579 days, which equated to a total of 0.018 SNP/day or 6.70 SNPs/year (95% CI, 4.49 to 9.63 SNPs/year). Using the genome size of 4,487,272 bp, the mutation rate is 1.49 × 10−6 substitution per site per year. There are three different mutation rates reported for S. Typhimurium. The lowest rate is 1.9 × 10−7 substitution per site per year, estimated for ST313, causing invasive infections in Africa (26); the intermediate rate is 3.4 × 10−7 substitution per site per year, obtained from the epidemic DT104 infections (27); and the highest rate is 1.2 ×10−6 substitution per site per year, obtained from a DT135a outbreak (28). Our estimate of the mutation rate is the highest but is only 12% higher than the previous highest estimate. S. Typhimurium appears to evolve a little faster in vivo. To our knowledge, this is the first estimate of the in vivo mutation rate for S. Typhimurium in a carrier state. We caution that this mutation rate estimate should be regarded as an upper bound, because the total time available for mutation may have been longer than the 1,579 days measured here. For example, in a few instances, isolates obtained close in time differed by one or more SNPs, but these SNPs may have arisen earlier than the collection time of the earlier isolate.
Comparison of MLVA and genome data.
The isolates were initially typed by MLVA to determine whether they were related by using the MLVA-5 scheme (8). All but three patients had identical MLVA profiles for all subsequent isolates, suggesting carriage. For patient 7, the first three isolates (G1 to G3) had identical MLVA profiles, while the last isolate, collected at day 206 (G4), differed by one repeat at STTR9. The day 206 (G4) isolate also differed from the others by three SNPs. For patient 9, the day 149 (I2) and day 161 (I3) isolates differed from the first isolate (I1) by one repeat each, at STTR10pl and STTR6, respectively. There was no SNP distinguishing I2 and I3, and both differed from I1 by two SNPs. Isolates from the same patients with identical MLVA profiles may differ by up to 5 SNPs (patient 8).
In addition, two sets of two patients each had the same MLVA profile. Patient 1 and patient 3 isolates had the same profile but differed by 8 SNPs, while patient 6 and patient 10 isolates had the same profile but differed by 19 SNPs. We previously also observed that isolates with identical MLVA types had one or more SNP difference (6). The genome data offered a much higher resolution than the MLVA data and demonstrated that MLVA lacks the power to reveal the true relationships of the isolates from carriers.
We obtained a variable-number tandem repeat (VNTR) mutation rate by using the same approach as that used for estimation of the SNP mutation rate. There were five occurrences of VNTR repeat changes in a total of 1,227 days observed. This equates to a VNTR mutation rate of 1.49 per year (95% CI, 0.48 to 3.47 per year). This estimate is higher than that reported in a previous study using a mouse model in which S. Typhimurium was passaged in vivo (29), with an estimated VNTR mutation rate of 0.84 per year. The VNTR mutation rate difference could be due to the different hosts or to the mode of infection, as the mouse model used was a systemic infection through intravenous inoculation (29).
Phylogenetic analysis of serial S. Typhimurium isolates.
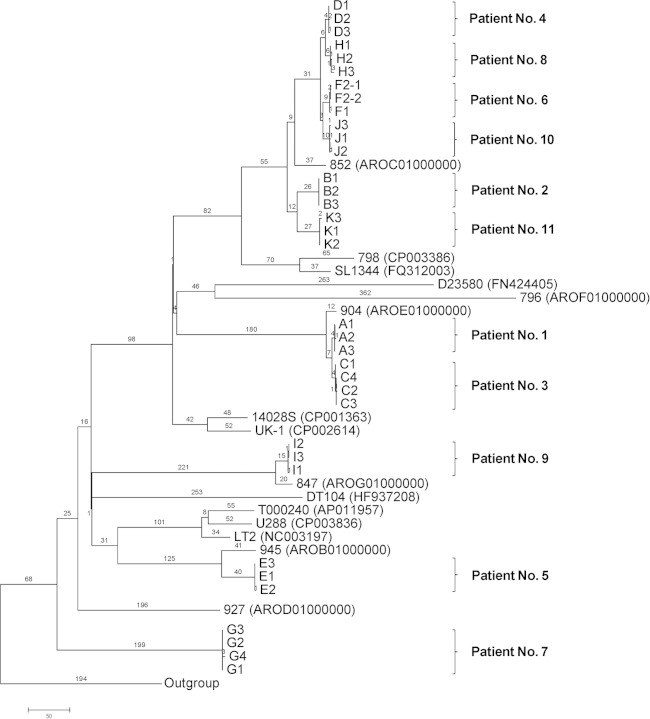
To determine the phylogenetic relationships among the isolates from carriers and other S. Typhimurium isolates, a genomic tree based on SNPs from the 35 isolates and from 16 publicly available genomes from GenBank was generated using the maximum parsimony method (Fig. 2). All the serial S. Typhimurium isolates from the same patient were grouped together. Isolates from patients 4, 6, 8, and 10 clustered together, and based on their relationship with the DT170 isolates, it is likely that they belonged to phage type DT170, the most common phage type in NSW, Australia, where the patients resided (7). The phylogenetic analysis showed that the isolates from carriers did not belong to a single clone. Thus, the present study is unlike the case of invasive S. Typhimurium infections in Africa, where one clone (ST313) was causing the infections (30).
FIG 2.
Maximum parsimony tree of S. Typhimurium genomes based on SNPs identified by mapping to the reference genome of S. Typhimurium LT2. The numbers on branches correspond to numbers of SNP differences. GenBank accession numbers for the publicly available genomes are given in parentheses. The unit of the scale bar is number of SNPs. S. Enteritidis and S. Choleraesuis were used as the outgroup.
Concluding comments.
Our analysis of genomic variation demonstrated a limited genomic variation within S. Typhimurium isolates excreted at the time of symptomatic illness and during the carriage stage of infection. None of the longitudinal isolates was due to reinfection. The genomic changes during carriage were mostly random, and the estimated mutation rate is 1.49 × 10−6 substitution per site per year. Our data fill a gap in the knowledge of Salmonella microevolution during long-term human carriage, which can be used for case definition using whole-genome sequencing.
A limitation of this study is that only a single isolate was obtained from each patient at any time point, as this has been the general clinical microbiology practice for diagnostic purposes. The isolate sampled was likely to be the predominant isolate in the population at a given time point. However, it is likely that there are other variants present at any given time. A metagenomic approach or in-depth sampling would be required to fully assess the population diversity during chronic carriage.
In a previous study, a significant proportion of patients with recurrence or carriage of NTS infection had an underlying disease (31). However, we were unable to determine if the patients whose isolates were analyzed in this study had any underlying diseases. HIV infection puts patients at high risk for having recurrent salmonellosis (32), and in recent years, there has been an increase in invasive S. Typhimurium infections in HIV patients in Africa. By genome sequencing of 47 serial isolates from 14 patients, Okoro et al. (30) showed that recurrence of invasive S. Typhimurium infection in HIV patients can be due to either recrudescence, reinfection, or a combination of both. Recrudescence accounted for 78% of recurring infections (30). However, recrudescence and multiple infections in the same patient were not uncommon (30).
In uncomplicated NTS gastroenteritis, the NTS infections tend to be self-limiting, and the median duration of NTS excretion is 4 to 5 weeks, based on previous studies. In the 11 patients we examined, all but 1 excreted NTS for longer than 35 days, with the longest duration being 279 days after the initial infection. Carriers pose potential risks as sources of infection. However, studies on the effects of prolonged shedding and carriage of NTS are scarce. In previous studies, the risks of household transmission of Salmonella ranged from 6% to 39% (33, 34). The prolonged shedding of NTS postinfection is an important issue, as evidenced by sporadic outbreaks traced back to asymptomatic carriers as the source of infection (35–37). These findings have important implications for the detection of possible transmission events suspected by public health genomic surveillance of S. Typhimurium infections.
ACKNOWLEDGMENTS
This study was supported by a grant from the National Health and Medical Research Council (grant APP1050227).
We thank the enteric team from the Communicable Disease Branch, the NSW Ministry of Health, and Peter Howard from the NSW Enteric Reference Laboratory, ICPMR-Pathology West, Westmead, Australia, for their assistance in the study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01733-15.
REFERENCES
- 1.Devi S, Murray CJ. 1991. Salmonella carriage rate amongst school children—a three year study. Southeast Asian J Trop Med Public Health 22:357–361. [PubMed] [Google Scholar]
- 2.Kazemi M, Gumpert G, Marks MI. 1974. Clinical spectrum and carrier state of nontyphoidal Salmonella infections in infants and children. Can Med Assoc J 110:1253–1257. [PMC free article] [PubMed] [Google Scholar]
- 3.Buchwald DS, Blaser MJ. 1984. A review of human salmonellosis. II. Duration of excretion following infection with nontyphi Salmonella. Rev Infect Dis 6:345–356. [DOI] [PubMed] [Google Scholar]
- 4.Murase T, Yamada M, Muto T, Matsushima A, Yamai S. 2000. Fecal excretion of Salmonella enterica serovar Typhimurium following a food-borne outbreak. J Clin Microbiol 38:3495–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Notifiable Diseases Surveillance System. 28 April 2014, accession date Australia's notifiable disease status: annual report of the National Notifiable Diseases Surveillance System. Department of Health and Ageing, Canberra, Australia: http://www9.health.gov.au/cda/source/rpt_2.cfm. [Google Scholar]
- 6.Octavia S, Wang Q, Tanaka MM, Kaur S, Sintchenko V, Lan R. 2015. Delineating community outbreaks of Salmonella enterica serovar Typhimurium by use of whole-genome sequencing: insights into genomic variability within an outbreak. J Clin Microbiol 53:1063–1071. doi: 10.1128/JCM.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sintchenko V, Wang Q, Howard P, Ha CW, Kardamanidis K, Musto J, Gilbert GL. 2012. Improving resolution of public health surveillance for human Salmonella enterica serovar Typhimurium infection: 3 years of prospective multiple-locus variable-number tandem-repeat analysis (MLVA). BMC Infect Dis 12:78. doi: 10.1186/1471-2334-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindstedt B-A, Torpdahl M, Vergnaud G, Le Hello S, Weill F, Tietze E, Malorny B, Prendergast D, Ní Ghallchóir E, Lista R, Schouls LM, Söderlund R, Börjesson S, Åkerström S. 2013. Use of multilocus variable-number tandem repeat analysis (MLVA) in eight European countries, 2012. Euro Surveill 18:20385. [DOI] [PubMed] [Google Scholar]
- 9.Octavia S, Lan R. 2006. Frequent recombination and low level of clonality within Salmonella enterica subspecies I. Microbiology 152:1099–1108. doi: 10.1099/mic.0.28486-0. [DOI] [PubMed] [Google Scholar]
- 10.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang S, Octavia S, Feng L, Liu B, Reeves PR, Lan R, Wang L. 2013. Genomic diversity and adaptation of Salmonella enterica serovar Typhimurium from analysis of six genomes of different phage types. BMC Genomics 14:718. doi: 10.1186/1471-2164-14-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT. 2011. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Inc, Publishers, Sunderland, MA. [Google Scholar]
- 17.LeClerc JE, Li B, Payne WL, Cebula TA. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 18.Ahmer BM, Tran M, Heffron F. 1999. The virulence plasmid of Salmonella typhimurium is self-transmissible. J Bacteriol 181:1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner C, Hensel M. 2011. Adhesive mechanisms of Salmonella enterica. Adv Exp Med Biol 715:17–34. doi: 10.1007/978-94-007-0940-9_2. [DOI] [PubMed] [Google Scholar]
- 20.Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Baumler AJ. 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol 57:196–211. doi: 10.1111/j.1365-2958.2005.04666.x. [DOI] [PubMed] [Google Scholar]
- 21.Kingsley RA, van Amsterdam K, Kramer N, Baumler AJ. 2000. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect Immun 68:2720–2727. doi: 10.1128/IAI.68.5.2720-2727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogomolnaya LM, Santiviago CA, Yang HJ, Baumler AJ, Andrews-Polymenis HL. 2008. ‘Form variation’ of the O12 antigen is critical for persistence of Salmonella Typhimurium in the murine intestine. Mol Microbiol 70:1105–1119. doi: 10.1111/j.1365-2958.2008.06461.x. [DOI] [PubMed] [Google Scholar]
- 23.Ikebe T, Iyoda S, Kutsukake K. 1999. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet Syst 74:179–183. doi: 10.1266/ggs.74.179. [DOI] [PubMed] [Google Scholar]
- 24.Amarasinghe JJ, D'Hondt RE, Waters CM, Mantis NJ. 2013. Exposure of Salmonella enterica serovar Typhimurium to a protective monoclonal IgA triggers exopolysaccharide production via a diguanylate cyclase-dependent pathway. Infect Immun 81:653–664. doi: 10.1128/IAI.00813-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson DL, White AP, Snyder SD, Martin S, Heiss C, Azadi P, Surette M, Kay WW. 2006. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J Bacteriol 188:7722–7730. doi: 10.1128/JB.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, Kariuki S, Msefula CL, Gordon MA, de Pinna E, Wain J, Heyderman RS, Obaro S, Alonso PL, Mandomando I, MacLennan CA, Tapia MD, Levine MM, Tennant SM, Parkhill J, Dougan G. 2012. Intracontinental spread of human invasive Salmonella typhimurium pathovariants in sub-Saharan Africa. Nat Genet 44:1215–1221. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mather AE, Reid SW, Maskell DJ, Parkhill J, Fookes MC, Harris SR, Brown DJ, Coia JE, Mulvey MR, Gilmour MW, Petrovska L, de Pinna E, Kuroda M, Akiba M, Izumiya H, Connor TR, Suchard MA, Lemey P, Mellor DJ, Haydon DT, Thomson NR. 2013. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341:1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawkey J, Edwards DJ, Dimovski K, Hiley L, Billman-Jacobe H, Hogg G, Holt KE. 2013. Evidence of microevolution of Salmonella typhimurium during a series of egg-associated outbreaks linked to a single chicken farm. BMC Genomics 14:800. doi: 10.1186/1471-2164-14-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimovski K, Cao H, Wijburg OL, Strugnell RA, Mantena RK, Whipp M, Hogg G, Holt KE. 2014. Analysis of Salmonella enterica serovar Typhimurium variable-number tandem-repeat data for public health investigation based on measured mutation rates and whole-genome sequence comparisons. J Bacteriol 196:3036–3044. doi: 10.1128/JB.01820-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okoro CK, Kingsley RA, Quail MA, Kankwatira AM, Feasey NA, Parkhill J, Dougan G, Gordon MA. 2012. High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella typhimurium disease. Clin Infect Dis 54:955–963. doi: 10.1093/cid/cir1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musher DM, Rubenstein AD. 1973. Permanent carriers of nontyphosa salmonellae. Arch Intern Med 132:869–872. [PubMed] [Google Scholar]
- 32.Rubino S, Spanu L, Mannazzu M, Schiaffino A, Mura MS, Cappuccinelli P, Aceti A. 1999. Molecular typing of non-typhoid Salmonella strains isolated from HIV-infected patients with recurrent salmonellosis. AIDS 13:137–139. [PubMed] [Google Scholar]
- 33.Ethelberg S, Olsen KE, Gerner-Smidt P, Molbak K. 2004. Household outbreaks among culture-confirmed cases of bacterial gastrointestinal disease. Am J Epidemiol 159:406–412. doi: 10.1093/aje/kwh049. [DOI] [PubMed] [Google Scholar]
- 34.Wilson R, Feldman RA, Davis J, LaVenture M. 1982. Salmonellosis in infants: the importance of intrafamilial transmission. Pediatrics 69:436–438. [PubMed] [Google Scholar]
- 35.Kumar A, Nath G, Bhatia BD, Bhargava V, Loiwal V. 1995. An outbreak of multidrug resistant Salmonella typhimurium in a nursery. Indian Pediatr 32:881–885. [PubMed] [Google Scholar]
- 36.Medus C, Smith KE, Bender JB, Besser JM, Hedberg CW. 2006. Salmonella outbreaks in restaurants in Minnesota, 1995 through 2003: evaluation of the role of infected foodworkers. J Food Prot 69:1870–1878. [DOI] [PubMed] [Google Scholar]
- 37.Todd EC, Greig JD, Bartleson CA, Michaels BS. 2008. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 4. Infective doses and pathogen carriage. J Food Prot 71:2339–2373. [DOI] [PubMed] [Google Scholar]