Abstract
We used an in vitro technique to investigate blood volumes required to detect bacteremia and fungemia with low concentrations of an organism. At 1 to 10 CFU/ml, Escherichia coli, Staphylococcus epidermidis, Staphylococcus aureus, Listeria monocytogenes, Candida albicans, and Candida parapsilosis isolates were detected in volumes as low as 0.5 ml. Detection of Streptococcus agalactiae and detection of bacteremia at <1 CFU/ml were unreliable.
TEXT
Neonatal sepsis continues to be a serious condition. While incidence of early-onset neonatal Streptococcus agalactiae infections has decreased, late-onset disease and Enterobacteriaceae sepsis rates remain stable (1–3). Additionally, the frequent use of central venous catheters in premature and ill newborns places them at risk of developing central line-associated bloodstream infections. Coagulase-negative Staphylococcus species are a leading causative agent. Staphylococcus aureus, Enterobacteriaceae, and Candida species are reported less frequently (4).
There is evidence to suggest that the likelihood of a blood culture testing positive in a bacteremic neonate is dependent on the volume of blood drawn (5). Accordingly, the College of American Pathologists requires that laboratories have a system to monitor blood culture volumes submitted. To provide guidance around appropriate blood draw volumes in pediatric patients, there are currently four sets of published recommendations. However, when applied to neonates, the recommended volumes vary considerably (Fig. 1) (6–9).
FIG 1.
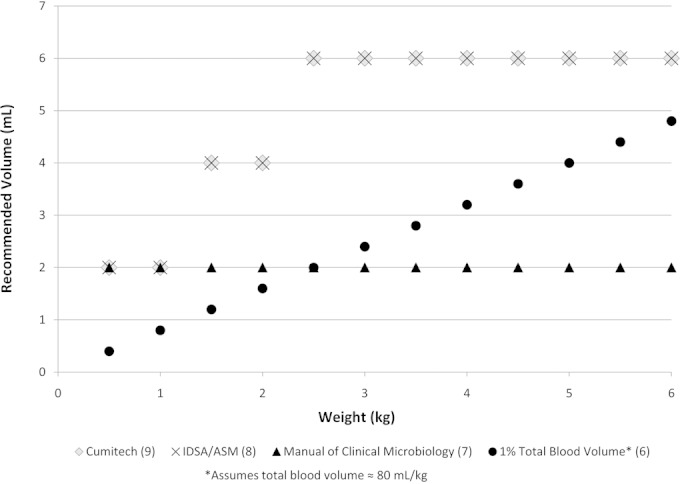
Blood culture volume recommendations by weight in kilograms.
In this study, we used in vitro methods to investigate the blood volumes necessary to detect common agents of neonatal sepsis at low levels (1 to 10 CFU/ml) and ultralow levels (<1 CFU/ml). Also, we compared the sensitivity of pediatric Bactec Peds Plus/F media and adult Bactec Plus Aerobic/F media (BD Diagnostics, Sparks, MD).
We used American Type Culture Collection (ATCC) organisms, as summarized in Table 1. These organisms were chosen to represent common agents of neonatal sepsis (10, 11). S. agalactiae serotypes Ia and III were chosen, as they are the most prevalent serotypes involved in invasive neonatal S. agalactiae infections in the United States (12). After overnight culture on 5% sheep's blood agar and a subsequent subculture, each organism was serially diluted using a digital pipettor to achieve concentrations of 1 to 10 CFU/ml and <1 CFU/ml. Specifically, for each bacterial isolate, a stock suspension was prepared starting with a 1.0 McFarland standard suspension (≈3 × 108 CFU/ml). Three successive 100-fold dilutions were prepared using sterile saline (Remel, Lenexa, KS) to produce a stock suspension concentration of ≈3 × 102 CFU/ml. With Candida species, we started with a 0.5 McFarland standard (≈3 × 106 CFU/ml) and performed two successive 100-fold dilutions using sterile saline to produce a stock suspension concentration of ≈3 × 102 CFU/ml (13). To produce an experimental source of blood with 1 to 10 CFU/ml, 1 ml of the stock suspension was added to 49 ml of donated, banked, citrated, adult human whole blood that had been collected within the previous 48 h (≈6 CFU/ml). To produce blood with <1 CFU/ml of organism, a 1:10 dilution was performed on the stock solution, then 1 ml of this solution was added to 49 ml of blood, generating a bacterial concentration of ≈0.6 CFU/ml. At each step, suspensions were either vortexed or swirled to ensure homogeneity.
TABLE 1.
Organism suspension colony counts
| Organism | ATCC strain | Organism suspension, CFU/ml |
|
|---|---|---|---|
| <1 | 1–10 | ||
| E. coli | 25922 | 0.4 | 4.8 |
| S. agalactiae | 12386 | 0.2 | 3.4 |
| S. agalactiae (serotype 1a) | BAA 1138 | 0.4 | 4.6 |
| S. agalactiae (serotype III) | 12403 | 0.2 | 4.2 |
| S. aureus (MSSA)a | 25923 | 0.8 | 6.2 |
| S. aureus (MRSA)b | BAA 1556 | 0.2 | 6.4 |
| S. epidermidis | 12228 | 0.6 | 3.0 |
| L. monocytogenes | BAA 751 | 0.2 | 3.0 |
| C. albicans | 60193 | 0.6 | 3.2 |
| C. parapsilosis | 22019 | 0.8 | 6.6 |
MSSA, methicillin-susceptible S. aureus.
MRSA, methicillin-resistant S. aureus.
Once the organism suspensions were prepared, colony counts were verified as follows. A 1-ml aliquot of each of the final suspensions of 1 to 10 CFU/ml and <1 CFU/ml was inoculated on to a Mueller-Hinton agar plate with 5% sheep's blood and streaked for quantitation. None of the <1 CFU/ml suspensions yielded growth at this volume. Colony counts are summarized in Table 1. We also inoculated 100 μl of the penultimate suspensions of the <1 CFU/ml preparations (≈30 CFU/ml) to ensure that bacterial cells were present prior to the final 1:50 dilution.
Organism suspensions were then inoculated into Bactec Peds Plus/F and Bactec Plus Aerobic/F bottles at volumes of 0.5, 1, 1.5, 2, and 3 ml in triplicate. A Peds Plus/F and a Plus Aerobic/F bottle each were inoculated with 3 ml of banked blood only (no organism) to serve as negative controls. All bottles were incubated in a Bactec FX instrument for a maximum of 5 days. Positive blood cultures were subcultured to 5% Columbia sheep's blood agar to confirm growth of the appropriate organism. Terminal subcultures were performed on all negative bottles using 200 μl of blood culture broth.
A total of 600 bottles were inoculated with organism suspension. Negative control bottles were negative at 5 days. Terminal subcultures did not yield growth. Organism recovery is summarized in Table 2 . A total of 219 of 300 (73%) Peds Plus/F bottles and 194 of 300 (65%) Plus Aerobic/F bottles were positive (P = 0.03). Among the organism/bacterial load/volume triplicate groups in which organism was recovered in 3 of 3 bottles in both bottle types (n = 234 bottles qualified), the mean time to detection was 20.4 h for Peds Plus/F and was 21.1 h for Plus Aerobic/F (P = 0.02).
TABLE 2.
Blood culture bottle yield by organism, volume of blood, level of bacteremia, and bottle type
| Organism (ATCC strain) | Volume (ml) | Yieldc with: |
|||
|---|---|---|---|---|---|
| Bactec Peds Plus/F |
Bactec Plus Aerobic/F |
||||
| <1 CFU/ml | 1–10 CFU/ml | <1 CFU/ml | 1–10 CFU/ml | ||
| E. coli (25922) | 0.5 | 1/3 | 3/3 | 1/3 | 3/3 |
| 1 | 2/3 | 3/3 | 1/3 | 3/3 | |
| 1.5 | 3/3 | 3/3 | 2/3 | 3/3 | |
| 2 | 3/3 | 3/3 | 2/3 | 3/3 | |
| 3 | 3/3 | 3/3 | 2/3 | 3/3 | |
| Total | 12/15 | 15/15 | 8/15 | 15/15 | |
| S. agalactiae (12386) | 0.5 | 0/3 | 2/3 | 1/3 | 2/3 |
| 1 | 0/3 | 1/3 | 0/3 | 2/3 | |
| 1.5 | 2/3 | 2/3 | 1/3 | 3/3 | |
| 2 | 2/3 | 3/3 | 1/3 | 3/3 | |
| 3 | 2/3 | 3/3 | 2/3 | 3/3 | |
| Total | 6/15 | 11/15 | 5/15 | 13/15 | |
| S. agalactiae (BAA 1138) | 0.5 | 1/3 | 3/3 | 0/3 | 2/3 |
| 1 | 2/3 | 3/3 | 0/3 | 2/3 | |
| 1.5 | 2/3 | 3/3 | 1/3 | 0/3 | |
| 2 | 2/3 | 2/3 | 0/3 | 2/3 | |
| 3 | 0/3 | 2/3 | 0/3 | 2/3 | |
| Total | 7/15 | 13/15 | 1/15 | 8/15 | |
| S. agalactiae (12403) | 0.5 | 0/3 | 3/3 | 0/3 | 1/3 |
| 1 | 0/3 | 3/3 | 0/3 | 0/3 | |
| 1.5 | 1/3 | 3/3 | 0/3 | 1/3 | |
| 2 | 2/3 | 3/3 | 2/3 | 0/3 | |
| 3 | 1/3 | 3/3 | 0/3 | 2/3 | |
| Total | 4/15 | 15/15 | 2/15 | 4/15 | |
| MSSAa (25923) | 0.5 | 1/3 | 3/3 | 2/3 | 3/3 |
| 1 | 3/3 | 3/3 | 2/3 | 3/3 | |
| 1.5 | 3/3 | 3/3 | 3/3 | 3/3 | |
| 2 | 3/3 | 3/3 | 3/3 | 3/3 | |
| 3 | 3/3 | 3/3 | 3/3 | 3/3 | |
| Total | 13/15 | 15/15 | 13/15 | 15/15 | |
| MRSAb (BAA 1556) | 0.5 | 0/3 | 3/3 | 0/3 | 3/3 |
| 1 | 1/3 | 3/3 | 0/3 | 3/3 | |
| 1.5 | 1/3 | 3/3 | 0/3 | 3/3 | |
| 2 | 1/3 | 3/3 | 1/3 | 3/3 | |
| 3 | 2/3 | 3/3 | 2/3 | 3/3 | |
| Total | 5/15 | 15/15 | 3/15 | 15/15 | |
| S. epidermidis (12228) | 0.5 | 1/3 | 3/3 | 2/3 | 3/3 |
| 1 | 3/3 | 3/3 | 2/3 | 3/3 | |
| 1.5 | 3/3 | 3/3 | 2/3 | 3/3 | |
| 2 | 2/3 | 2/3 | 3/3 | 3/3 | |
| 3 | 3/3 | 3/3 | 2/3 | 3/3 | |
| Total | 12/15 | 14/15 | 11/15 | 15/15 | |
| L. monocytogenes (BAA 751) | 0.5 | 0/3 | 3/3 | 0/3 | 3/3 |
| 1 | 0/3 | 3/3 | 0/3 | 3/3 | |
| 1.5 | 0/3 | 3/3 | 2/3 | 3/3 | |
| 2 | 1/3 | 3/3 | 2/3 | 3/3 | |
| 3 | 0/3 | 3/3 | 2/3 | 3/3 | |
| Total | 1/15 | 15/15 | 6/15 | 15/15 | |
| C. albicans (60193) | 0.5 | 1/3 | 3/3 | 0/3 | 3/3 |
| 1 | 0/3 | 3/3 | 0/3 | 3/3 | |
| 1.5 | 1/3 | 3/3 | 0/3 | 3/3 | |
| 2 | 1/3 | 3/3 | 2/3 | 3/3 | |
| 3 | 3/3 | 3/3 | 2/3 | 3/3 | |
| Total | 6/15 | 15/15 | 4/15 | 15/15 | |
| C. parapsilosis (22019) | 0.5 | 2/3 | 3/3 | 2/3 | 3/3 |
| 1 | 1/3 | 3/3 | 2/3 | 3/3 | |
| 1.5 | 2/3 | 3/3 | 3/3 | 3/3 | |
| 2 | 2/3 | 3/3 | 2/3 | 3/3 | |
| 3 | 3/3 | 3/3 | 2/3 | 3/3 | |
| Total | 10/15 | 15/15 | 11/15 | 15/15 | |
MSSA, methicillin-susceptible S. aureus.
MRSA, methicillin-resistant S. aureus.
Number of bottles positive per triplicate.
At a bacterial load of 1 to 10 CFU/ml, Escherichia coli, Staphylococcus aureus (methicillin-sensitive Staphylococcus aureus [MSSA] and methicillin-resistant Staphylococcus aureus [MRSA]), Listeria monocytogenes, Candida albicans, and Candida parapsilosis were recovered in all bottles tested. S. agalactiae was detected in 39 of 45 (87%) Peds Plus/F bottles and in 25 of 45 (56%) Plus Aerobic/F bottles. We failed to recover S. agalactiae ATCC BAA 1138 consistently, even with 3 ml of blood.
At <1 CFU/ml, the Peds Plus/F recovered E. coli in bottles inoculated with ≥1.5 ml of organism suspension; ≥1.0 ml was required for recovery of MSSA, and 3 ml was required for C. albicans and C. parapsilosis. With Peds Plus/F, the detection of S. agalactiae, L. monocytogenes, and MRSA was unreliable at all volumes tested at this organism load. The Plus Aerobic/F detected MSSA consistently at volumes of ≥1.5 ml but had poor sensitivity for all other organisms at this organism load.
There is a paucity of data supporting recommended draw volumes in neonates. To ensure that blood culturing does not contribute to the development of iatrogenic anemia, care is required when extrapolating concepts of ideal blood draw volumes from older children to younger ones. In this study, we sought to determine the blood volumes required to reliably detect low levels and ultralow levels of organisms commonly implicated in neonatal bacteremia.
Our data suggest that, if detection of bacteremia at 1 to 10 CFU/ml is the clinical goal, blood draw volumes as low as 0.5 ml may reliably detect most organisms implicated in early-onset neonatal bacteremia. However, detection of certain strains of S. agalactiae may require >3 ml. At simulated bacteremia levels of <1 CFU/ml, Peds Plus/F detected the organism with volumes as low as 1 to 1.5 ml for E. coli, MSSA, S. aureus, and Staphylococcus epidermidis. Three milliliters appeared to suffice for the detection of Candida species. S. agalactiae, MRSA, and L. monocytogenes were difficult to detect reliably, even at volumes as high as 3 ml. The Plus Aerobic/F bottles were unreliable in detecting any organism at any volume except for MSSA. Overall, we failed to recover organism in 86 of 120 blood cultures (72%) inoculated with 0.5 ml or 1 ml of blood with an organism load of <1 CFU/ml, suggesting that bacteremia at this level may be missed in significant numbers when small volumes of blood are drawn for culture.
It was previously suggested that neonatal bacteremia is associated with high bacterial loads (14, 15). Kellogg et al. refuted this concept in a study that used a pediatric isolator system and large-volume blood collections (4.5% of total blood volume) to quantify bacteremia in infants at 0 to 2 months of age. They reported that 7 of 15 (47%) cases of S. agalactiae and 4 of 11 (36%) cases of E. coli bacteremia had bacterial loads of <1 CFU/ml (16). These results are somewhat difficult to interpret, however, as other studies have suggested that bacterial recovery from pediatric isolator tubes may be inferior to broth-based media, such as those used in our study (5, 17). Bacterial inhibition by saponin (the cell lysing agent present in pediatric isolator systems) has been suggested as the culprit (14). Further studies confirming the concept of ultralow level bacteremia in neonates and older children, therefore, would be beneficial. Nevertheless, if detection of bacteremia levels of <1 CFU/ml is desired, blood volumes of >3 ml need to be considered and balanced against the hazard of drawing larger volumes in small neonates.
Brown et al. published a study in 1995 based on the Bactec NR-660 system, which was introduced in 1984 (18). This system used infrared spectrophotometry to detect carbon dioxide produced by growing organisms. E. coli and S. agalactiae bacteremia was simulated at levels of 0.4, 4, and 40 CFU/ml. Bactec NR-6A bottles were inoculated with volumes of 0.25, 0.5, and 1 ml. At 1 to 10 CFU/ml, E. coli was detected in 26 of 26 bottles with 0.5 ml or 1 ml and in 12 of 15 bottles inoculated with 0.25 ml. S. agalactiae at 1 to 10 CFU/ml was detected in 10 of 15, 13 of 15, and 15 of 15 bottles inoculated with 0.25, 0.5, and 1 ml of seeded blood. Both E. coli and S. agalactiae grew inconsistently in bottles inoculated with any volume at levels of <1 CFU/ml. Our findings corroborated these results.
Using in vitro methods, we observed superior sensitivity and shorter time to positivity in Peds Plus/F bottles than in adult Plus Aerobic/F media. We are not aware of any other data in the published literature documenting this finding. This may be relevant for pediatric hospital settings that use the Bactec blood culture system.
This study has a number of limitations. First, banked, citrated, whole adult human blood was used as a proxy for fresh blood from a neonate. Citrate may have a subtle inhibitory effect on bacterial growth, which may have led to an overestimation of the volume of blood required to detect bacteremia. Also, plasma proteins, including coagulation factors, complement, and immunoglobulin, may degrade with prolonged storage. Additionally, there have been anecdotal observations of impaired organism recovery in seeded blood cultures when old, expired units of blood are used. We minimized this effect by using blood that was received within 48 h of donation, immediately upon receipt. Second, our findings apply strictly to the Bactec FX system. In a recent study, we demonstrated superior sensitivity and swifter time to detection of bacteremia in Bactec Peds Plus/F media compared to BacT/Alert Pediatric FAN media (19). Required blood volumes, therefore, may vary according to the blood culture system in use. Third, colony counts were performed on the penultimate dilution for blood prepared with bacterial concentrations of <1 CFU/ml. We used this approach, as we could not find an accurate or reliable method to perform colony counts using large volumes of blood, which we anticipated would be required for testing for S. agalactiae. Fourth, actual bacterial loads in neonatal bacteremia are unknown. Our decision to examine organism recovery in simulated bloodstream infections with <1 CFU/ml and 1 to 10 CFU/ml was based on the findings of Kellogg et al. (16). Finally, this study used simulation techniques to estimate the volume of blood required to detect bacteremia in neonates. Multicenter, randomized clinical studies involving bacteremic neonates that examine the yield of incrementally increasing blood culture draw volumes in neonates would be ideal and are much needed. While this study does not determine the adequacy or inadequacy of current published pediatric blood draw volume recommendations, our data can potentially be used in conjunction with studies that investigate magnitude of bacteremia in neonates to better understand appropriate blood draw volumes in this population.
In summary, published recommendations for pediatric blood culture volumes vary, and we sought to determine the volumes required to detect organisms implicated in neonatal bacteremia. In an in vitro setting, with the Bactec Peds Plus/F system, 0.5 ml appeared sufficient to detect all organisms at a load of 1 to 10 CFU/ml with the exception of S. agalactiae. Recovery of S. agalactiae varied considerably according to strain. Larger volumes would likely need to be collected if detection of bacteremia at levels of <1 CFU/ml was desired, but this would need to be considered in tandem with the risks associated with increased draw volumes. The Peds Plus/F media recovered organism with superior sensitivity and faster time to detection than the adult Plus Aerobic/F media. Large-scale clinical studies examining recommended volumes using the various blood culture systems would be beneficial.
ACKNOWLEDGMENTS
We thank Deborah Blecker-Shelly for her input into the study protocol. We also thank Pamala Blair and Maria Dirkes from transfusion medicine at CHOP for their assistance in procuring appropriate banked whole blood.
We have no conflicts of interest to declare.
This study was not funded.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2009. QuickStats: infant, neonatal, and postneonatal mortality rates—United States, 1940 to 2006. Morbid Mortal Wkly Rep 58:646. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2013. CDC grand rounds: public health approaches to reducing US infant mortality. Morbid Mortal Wkly Rep 62:625–628. [PMC free article] [PubMed] [Google Scholar]
- 3.Stoll BJ, Hansen NI, Sánchez PJ, Faix RG, Poindexter BB, Van Meurs KP, Bizzarro MJ, Goldberg RN, Frantz ID 3rd, Hale EC, Shankaran S, Kennedy K, Carlo WA, Watterberg KL, Bell EF, Walsh MC, Schibler K, Laptook AR, Shane AL, Schrag SJ, Das A, Higgins RD. 2011. Early onset neonatal sepsis: the burden of group B streptococcal and E. coli disease continues. Pediatrics 127:817–826. doi: 10.1542/peds.2010-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudeck MA, Horan TC, Peterson KD, Allen-Bridson K, Morrell G, Pollock DA, Edwards JR. 2011. National Healthcare Safety Network (NHSN) report, data summary for 2010, device-associated module. Am J Infect Control 39:798–816. doi: 10.1016/j.ajic.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Isaacman DJ, Karasic RB, Reynolds EA, Kost SI. 1996. Effect of number of blood cultures and volume of blood on detection of bacteremia in children. J Pediatr 128:190–195. doi: 10.1016/S0022-3476(96)70388-8. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2007. Principles and procedures for blood cultures. CLSI document M47A Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Baron EJ, Thomson RB Jr. 2011. Specimen collection, transport, and processing: bacteriology, p 228–271. In Versalovic J, Carroll KC, Jorgensen JH, Funke G, Landry ML, Warnock DW (ed), Manual of clinical microbiology. 10th ed, vol 2 Washington DC, ASM Press. [Google Scholar]
- 8.Baron EJ, Miller JM, Weinstein MP, Richter SS, Gilligan PH, Thomson RB Jr, Bourbeau P, Carroll KC, Kehl SC, Dunne WM, Robinson-Dunn B, Schwartzman JD, Chapin KC, Snyder JW, Forbes BA, Patel R, Rosenblatt JE, Pritt BS. 2013. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin Infect Dis 57:e22–e121. doi: 10.1093/cid/cit278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baron EJ, Weinstein MP, Dunne WM, Yagupsky P, Welch DF, Wilson DM. 2005. Cumitech 1C, blood cultures IV. Coordinating ed, Baron EJ. Washington, DC, ASM Press. [Google Scholar]
- 10.Bizzarro MJ, Raskind C, Baltimore RS, Gallagher PG. 2005. Seventy-five years of neonatal sepsis at Yale: 1928 to 2003. Pediatrics 116:595–602. doi: 10.1542/peds.2005-0552. [DOI] [PubMed] [Google Scholar]
- 11.Simonsen KA, Anderson-Berry AL, Delair SF, Davies HD. 2014. Early-onset neonatal sepsis. Clin Microbiol Rev 27:21–47. doi: 10.1128/CMR.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrieri P, Lynfield R, Creti R, Flores AE. 2013. Serotype IV and invasive group B Streptococcus disease in neonates, Minnesota, USA, 2000 to 2010. Emerg Infect Dis 19:551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard— 3rd ed CLSI document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.Yagupsky P, Nolte FS. 1990. Quantitative aspects of septicemia. Clin Microbiol Rev 3:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietzman DE, Fischer GW, Schoenknecht FD. 1974. Neonatal Escherichia coli septicemia: bacterial counts in blood. J Pediatr 85:128–130. doi: 10.1016/S0022-3476(74)80308-2. [DOI] [PubMed] [Google Scholar]
- 16.Kellogg JA, Ferrentino FL, Goodstein MH, Liss J, Shapiro SL, Bankert DA. 1997. Frequency of low level bacteremia in infants from birth to two months of age. Pediatr Infect Dis J 16:381–385. doi: 10.1097/00006454-199704000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Eisenach K, Dyke J, Boehme M, Johnson B, Cook MB. 1992. Pediatric blood culture evaluation of the Bactec Peds Plus and the DuPont Isolator 1.5 systems. Diagn Microbiol Infect Dis 15:225–231. doi: 10.1016/0732-8893(92)90117-C. [DOI] [PubMed] [Google Scholar]
- 18.Brown DR, Kutler D, Bellipady R, Chan T, Cohen M. 1995. Bacterial concentration and blood volume required for a positive blood culture. J Perinatol 15:157–159. [PubMed] [Google Scholar]
- 19.Sullivan KV, Turner NN, Lancaster DP, Shah AR, Chandler LJ, Friedman DF, Blecker-Shelly DL. 2013. Superior sensitivity and decreased time to detection with the Bactec Peds Plus/F system compared to the BacT/Alert pediatric FAN blood culture system. J Clin Microbiol 51:4083–4086. doi: 10.1128/JCM.02205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


