Abstract
Macrolide-resistant Mycoplasma pneumoniae (MRMP) is rapidly emerging in Asia, but information on the temporal relationship between the increase in macrolide resistance and changes in strain types is scarce. Between 2011 and 2014, M. pneumoniae infection was diagnosed by PCR as part of routine care in a health care region in Hong Kong. Testing was initiated by clinicians, mainly in patients with suspected M. pneumoniae pneumonia. Specimens positive for M. pneumoniae were retrospectively investigated by macrolide resistance genotyping and a four-locus (Mpn13 to -16) multilocus variable-number tandem-repeat analysis (MLVA) scheme. The overall percentage of M. pneumoniae-positive specimens was 17.9%, with annual rates ranging from 9.8% to 27.2%. The prevalence of MRMP had rapidly increased from 13.6% in 2011 to 30.7% in 2012, 36.6% in 2013, and 47.1% in 2014 (P = 0.038). Two major MLVA types, 4-5-7-2 and 3-5-6-2, accounted for 75% to 85% of the infections each year. MLVA types 4-5-7-2 and 3-5-6-2 predominated among macrolide-resistant and macrolide-sensitive groups, respectively. The increase in MRMP was mainly caused by increasing macrolide resistance in the prevalent MLVA type 4-5-7-2, changing from 25.0% in 2011 to 59.1% in 2012, to 89.7% in 2013, and to 100% in 2014 (P < 0.001). In conclusion, increasing MRMP in Hong Kong was linked to a single MLVA type, which was both prevalent and increasingly resistant to macrolides.
INTRODUCTION
Mycoplasma pneumoniae is a common cause of community-acquired pneumonia and other respiratory tract infections (1). Community epidemics occur at intervals of 3 to 7 years. Infections develop in persons of all ages, but it is primarily a disease of children and teenagers (2). When treatment is indicated, a macrolide is usually the drug of choice (1, 2). However, macrolide-resistant M. pneumoniae (MRMP) has become increasingly prevalent worldwide, and high rates of infection (>80%) have been found in certain parts of the world (3–6). MRMP infections have been associated with persistence of symptoms, slower reduction in bacterial load, longer hospital stays, requirement of alternative therapy, and higher frequency of complications (1, 7, 8). Strain typing is important for understanding changes in disease epidemiology and for investigations of outbreaks. In 2009, a multilocus variable-number tandem-repeat analysis (MLVA) scheme based upon five loci (Mpn1 and Mpn13 to -16) was developed for the molecular typing of M. pneumoniae (9). It was initially used for an investigation of isolates but was later modified for directly typing M. pneumoniae in respiratory specimens (10–12). An amended 4-locus MLVA scheme was later proposed after studies raised concerns on the instability of the Mpn1 locus (13, 14). In clinical laboratories, culture and characterization of M. pneumoniae are seldom performed. Therefore, M. pneumoniae typing was usually carried out on isolates collected from sporadic cases and outbreaks (9, 13, 15), limiting the inferences that can be made about trends in M. pneumoniae infections. In addition, information on the temporal relationship between the increase in macrolide resistance and changes in strain types is scarce (15). Here, MLVA was used to investigate the M. pneumoniae strain type and macrolide resistance genotype in respiratory specimens collected consecutively from patients in a health care region in Hong Kong over a 4-year period.
MATERIALS AND METHODS
Study design.
This retrospective study was conducted in a health care region in Hong Kong comprising one university-affiliated hospital with 1,600 beds, three extended-care hospitals with a total of 1,600 beds, and one pediatric hospital with 160 beds. A diagnostic PCR assay for M. pneumoniae was provided as a routine service for inpatients by a clinical microbiology laboratory (7, 16). Testing was initiated by clinicians, mainly in patients with features suspected to be due to M. pneumoniae pneumonia (2, 17). Nasopharyngeal aspirate samples were collected in viral transport medium (18). Sputum and other respiratory specimens were collected using standard techniques (16). Patients were included if their respiratory specimens were obtained for M. pneumoniae testing by PCR between January 2011 and December 2014. During the study period, a total of 1,657 respiratory specimens from 1,433 patients were investigated by a real-time PCR test for the presence of M. pneumoniae. Overall, 257 (17.9%) patients, including 274 (16.5%) specimens, were positive for M. pneumoniae. The 274 M. pneumoniae-positive specimens comprised 264 nasopharyngeal aspirates/swabs, five pleural specimens, and five other respiratory specimens (sputum and bronchial aspirate). The data were analyzed by five age groups: 0 to 1 year (infants, n = 11), 2 to 11 years (children, n = 195), 12 to 17 years (teenagers, n = 33), 18 to 64 years (adults, n = 16), and ≥65 years (seniors, n = 2). The patients were diagnosed with pneumonia (n = 231), upper respiratory tract infection (n = 7), nonspecific respiratory illness (n = 9), and acute bronchiolitis (n = 1). In nine patients, no information on the syndromic diagnosis was available. Clinical features and macrolide resistance genotyping results for 101 of the patients were reported previously (7, 16). Nucleic acid extracts from the 257 patients with positive M. pneumoniae results were retrospectively retrieved for further testing. Only one specimen from each patient was included.
Nucleic acid extraction.
Nucleic acid extraction was performed by using the NucliSENS easyMAG extraction system (bioMérieux, France) and stored at −80°C, as described previously (16). All testing was performed on nucleic acid extracted from the clinical specimens. Culture for M. pneumoniae was not performed.
Real-time qPCR for detection of M. pneumoniae.
Real-time quantitative (qPCR) was conducted to detect M. pneumoniae using TaqMan universal PCR master mix (Applied Biosystems) in a StepOnePlus instrument (Applied Biosystems, Foster City, CA), as previously described (16). A series of 6 log10 dilutions equivalent to 10 to 1 × 106 copies per reaction mixture were prepared from a plasmid (pC-RII-TOPO vector; Invitrogen, CA) containing the corresponding target bacterial sequence to generate calibration curves; these were run in parallel with the test specimens. The detection limit of the qPCR assay was approximately 10 copies per reaction mixture (16).
MRMP genotype detection.
Simple-Probe real-time PCR coupled to melting curve analysis (Simple-Probe PCR) was performed on the extracted nucleic acid from specimens to identify MRMP. The MRMP assay was done using the LightCycler FastStart DNA master HybProbe kit (Roche Diagnostics, Germany), according to a previously published protocol (16). The detection limit of Simple-Probe PCR for both wild-type and mutants was 103 copies per reaction. A randomly chosen subset of the specimens was subjected to Sanger sequencing for confirmation.
MLVA typing.
Previously published primers were used to amplify four variable-number tandem-repeat (VNTR) loci (Mpn13 to -16) (12). Our initial testing showed that nonspecific bands were commonly observed if all loci were amplified together in one multiplex reaction. After optimization, good results were obtained through amplification of the loci in two duplex reactions, with one for Mpn13 and Mpn15, and one for Mpn14 and Mpn16. The PCR was performed using 2 μl of nucleic acid and 15 μl of 2× Qiagen multiplex master mixes, and the concentrations of each primer were 0.2 μM for Mpn13 and Mpn15, 0.4 μM for Mpn14, and 0.08 μM for Mpn16. The total reaction mixture volume was made up to 30 μl with nuclease-free water. A Veriti 96-well thermal cycler (Applied Biosystems) was used for amplification. The cycling conditions were a denaturation step of 15 min at 95°C, and an amplification step of 40 cycles of 30 s at 95°C, 30 s at 62°C, and 45 s at 72°C. The products were then pooled into one tube for product size determination in one lane by capillary electrophoresis using an ABI 3130 genetic analyzer (Applied Biosystems), and the data were analyzed using the GeneMapper software (version 4.0; Applied Biosystems). The primers were fluorescently labeled at the 5′ end with VIC (green, Mpn13 and Mpn15), NED (yellow, Mpn14), or 6-carboxyfluorescein (6-FAM) (blue, Mpn16) (Applied Biosystems). The fluorescent labels for the targets together with the expected product sizes of each locus allowed the amplicons for all four loci to be sized in one reaction mixture. The number of repeats for each locus was calculated according to the PCR fragment size. The MLVA type was designated by the numeric combination of the number of tandem repeats at four loci (Mpn13 to -16), as suggested previously (13, 15). The number of repeats was rounded up to an integer value (9, 12). The number of repeats in each locus (1 to 2 specimens for each product size) was confirmed by Sanger sequencing.
Statistical analysis.
Statistical analysis was performed using SPSS Statistics version 23 for Windows. Chi-square tests were used to compare categorical variables. A P value of <0.05 was considered statistically significant.
RESULTS
Prevalence of M. pneumoniae.
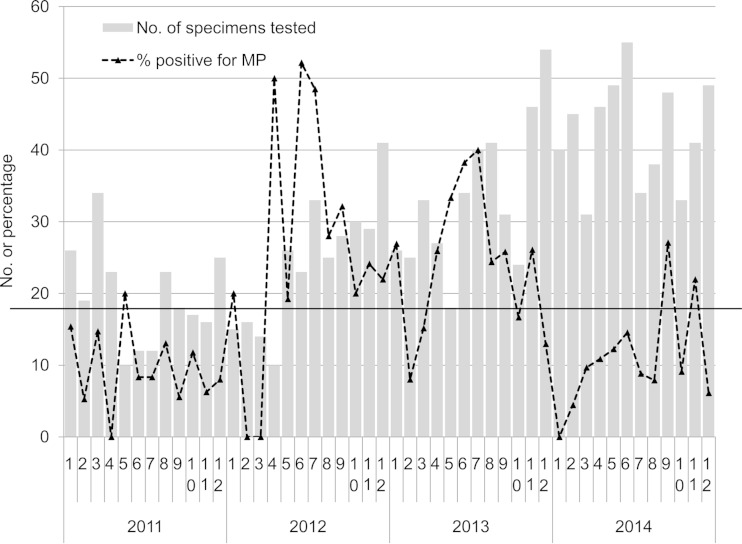
The percentages of test-positive patients per month from 2011 to 2014 are shown in Fig. 1. Higher positive rates (>1 standard deviation above average for the entire period) were observed in 2012 (April, June, July, and September) and 2013 (May to July). The percentages of test-positive patients by year were 9.8% in 2011, 27.2% in 2012, 24.3% in 2013, and 11.4% in 2014 (P < 0.001).
FIG 1.
M. pneumoniae-positive rate in respiratory specimens by date of request for a health care region in Hong Kong, January 2011 to December 2014. The histogram shows the monthly number of specimens tested. The dotted line shows the percentage of specimens positive for M. pneumoniae in each month. The horizontal line shows the average positive percentage for the entire period. Only one specimen per patient was included.
The M. pneumoniae-positive rate was highest among children age 2 to 11 years (33.2%) and teenagers age 12 to 17 years (30.6%), and then in infants age 0 to 1 year (7.6%); it was lowest in adults age 18 to 64 years (4.0%) and seniors age ≥65 years (1.0%) (P < 0.001). The positive rate was higher in females than in males (21.8% versus 14.5%, respectively; P < 0.001).
Prevalence of macrolide-resistant genotype.
The M. pneumoniae-positive specimens for 16 patients were of an insufficient amount and not investigated further. Macrolide resistance genotyping was successfully carried out on all specimens from the remaining 241 patients. Simple-Probe real-time PCR coupled to melting curve analysis identified 34.9% (84/241) of the unique patient specimens as having the MRMP genotype. The A2063G transition was the only mutation identified. A subset of 88 specimens, including 61 with the macrolide-sensitive M. pneumoniae (MSMP) genotype and 27 with the MRMP genotype, were further analyzed by Sanger sequencing. The results were 100% concordant with the melting curve analysis. The annual prevalence of MRMP among all M. pneumoniae-positive patients had significantly increased from 13.6% (3/22) in 2011 to 30.7% (23/75) in 2012, 36.6% (34/93) in 2013, and 47.1% (24/51) in 2014 (P = 0.038). The prevalence of the MRMP genotype was higher among children (age 0 to 1 year, 30.0%; age 2 to 11 years, 36.1%; age 12 to 17 years, 39.7%) than in adults (age 18 to 64 years, 20.0%, age ≥65 years, 0%), but the difference was not statistically significant (P = 0.122). The MRMP prevalence rates among males (33.7%) and females (35.7%) were similar (P = 0.742).
Temporal changes in macrolide resistance rate and MLVA types.
Specimens from the 241 patients with sufficient DNA extracts were further investigated by MLVA typing, and successful results were obtained for 205 (85.1%) patients. The number of repeats in the four loci were 3 to 5 for Mpn13, 4 to 6 for Mpn14, 6 to 7 for Mpn15, and 2 to 3 for Mpn16, giving seven distinct MLVA types. The major types were 3-5-6-2 (44.4%), 4-5-7-2 (36.6%), and 4-5-7-3 (14.1%). Other rare types, including 3-6-6-2 (n = 4), 5-5-7-2 (n = 3), 4-6-7-3 (n = 2), and 4-4-7-3 (n = 1), accounted for only 4.9% of the total.
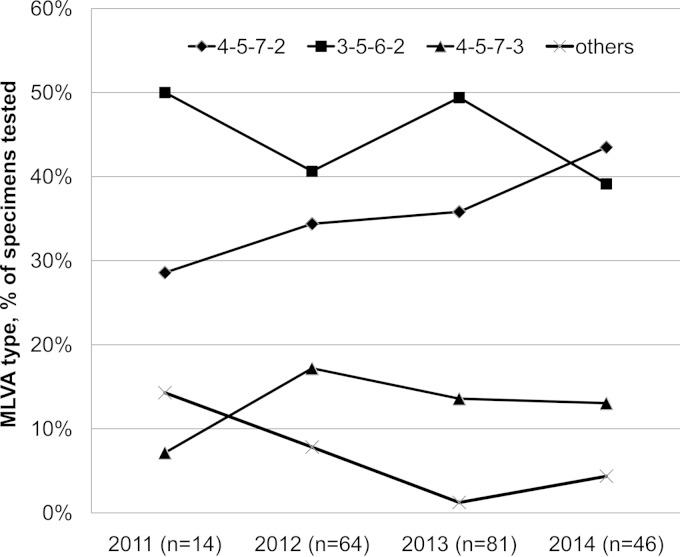
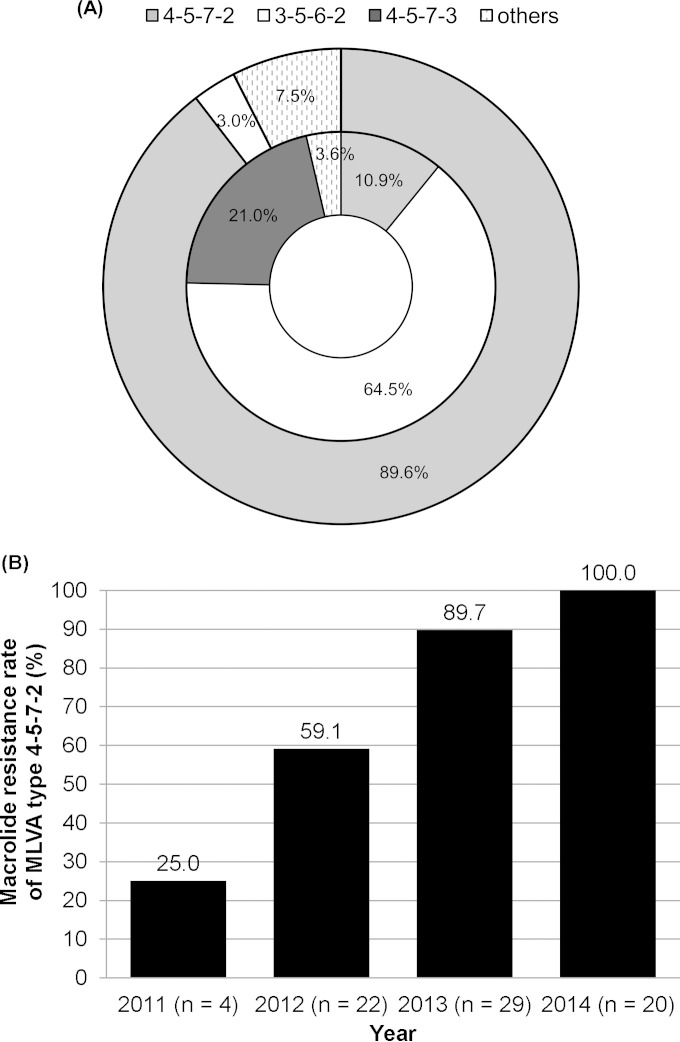
During the 4-year period, types 4-5-7-2 and 3-5-6-2 were predominant (Fig. 2). Together, the two types comprised 75% to 85% of all positive specimens in each year. The proportion of type 4-5-7-2 had an increasing trend from 29% in 2011 to 34% in 2012, 36% in 2013, and 43% in 2014, but the difference was not statistically significant (P = 0.686). Of the seven MLVA types, five and four MLVA types were found among specimens with the macrolide-sensitive M. pneumoniae (MSMP) and MRMP genotypes, respectively (Fig. 3A). The two major MLVA types (3-5-6-2 and 4-5-7-2) occurred in the MRMP and MSMP groups at different frequencies. The prevalence of MLVA type 4-5-7-2 was substantially higher in the MRMP group than that in the MSMP group (89.6% versus 10.9%, respectively; P < 0.001). In contrast, MLVA type 3-5-6-2 was more prevalent in the MSMP group than in the MRMP group (64.5% versus 3.0%, respectively; P < 0.001). The other five MLVA types were found at low frequencies in the MRMP group (for types 5-5-7-2 and 4-6-7-3) and the MSMP group (for types 4-5-7-3, 4-4-7-3, and M3-6-6-2). Stratification by year revealed that the macrolide resistance rate of MLVA type 4-5-7-2 had significantly increased from 25.0% in 2011 to 100% in 2014 (Fig. 3B) (P < 0.001).
FIG 2.
MLVA type in M. pneumoniae specimens in a health care region in Hong Kong, 2011 to 2014. The percentage of each MLVA type for each year is shown. The number of patients in each year is shown within parentheses.
FIG 3.
Macrolide resistance in M. pneumoniae specimens in a health care region in Hong Kong, 2011 to 2014. (A) MLVA type according to macrolide resistance genotype. The proportions of MLVA types for MRMP (n = 67) and MSMP (n = 138) groups are shown in the outer and inner rings, respectively. Others in the MRMP group included types 4-6-7-3 and 5-5-7-2. Others in the MSMP group included types 3-6-6-2 and 4-4-7-3. (B) Changes in macrolide resistance rate of MLVA type 4-5-7-2 during 2011 to 2014. The number of patients in each year is shown within parentheses.
DISCUSSION
In this study, an increase in the rate of M. pneumoniae infection was noted in 2012 and 2013, suggesting that there was an epidemic outbreak during this period. During the entire period, the changes in the annual cycle of positive rates were irregular, with peaks in early summer (in 2012), mid-summer (in 2013), and early autumn (in 2014). This is in line with reports describing more M. pneumoniae infections with increased relative humidity and ambient temperature (19, 20). In our neighborhood areas, recent epidemic outbreaks of M. pneumoniae infections were also noted in South Korea from 2010 to 2011, in Japan from 2011 to 2012, and in Beijing and Shanghai, China, in 2012 (4, 21, 22). Notably, a substantial increase in the prevalence of MRMP infection was noted in the areas during or shortly after those epidemics (3, 4, 21–23). Our data revealed that the MRMP infection rate had increased by >3-fold from 13.6% in 2011 to 47.1% in 2014. Reported rates of MRMP infection range from 62.9% in South Korea (23) and >80% to 90% in China and Japan (3, 8), compared to ≤10% in Europe and the United States (1, 15). The relationship between M. pneumoniae epidemics and MRMP emergence is likely to be complex, involving selection pressure from the widespread administration of macrolides (23).
Two MLVA types (3-5-6-2 [44.4%] and 4-5-7-2 [36.6%]) accounted for 81.0% of the infections during the study period. Both types occur worldwide and were among the predominant types found in many studies. Among international collections of M. pneumoniae isolates collected over decades, 14.0% to 20.8% and 50.6% to 55.1% were of MLVA types 3-5-6-2 and 4-5-7-2, respectively (9, 13). During the periods with a high proportion of M. pneumoniae-positive specimens, the major genotypes did not change (Fig. 2A). This suggests that the increase in diagnosis of M. pneumoniae infections is likely a result of increased transmission of cocirculating MLVA types rather than the introduction of new types into the community.
This study found that the increase in MRMP was predominantly a result of increasing resistance in MLVA type 4-5-7-2. The macrolide resistance rate of this type had drastically increased from 25% in 2011 to 100% in 2014. All other MLVA types, including the prevalent 3-5-6-2 type, remained largely macrolide sensitive. Qu et al. (22) previously reported the same significant correlation between macrolide resistance and susceptibility with types 4-5-7-2 and 3-5-6-2, respectively. In the two international M. pneumoniae collections described by Dégrange et al. (9) and Benitez et al. (13), four (33.3%) of 12 and nine (90%) of 10 MRMP isolates, respectively, had MLVA type 4-5-7-2. In the United States, 13 (68.4%) of 19 MRMP isolates identified during CDC-assisted investigations across the country between 2006 and 2013 belonged to MLVA type 4-5-7-2 (15). In China, this MLVA type accounted for >90% of the MRMP isolates identified in Beijing from 2010 to 2013 (12, 22, 24). Of the 21 distinct types that can be distinguished by the amended four-locus MLVA scheme (5, 6, 9–15, 22, 24–26) (see Table S1 in the supplemental material), MRMP has been detected in 12 types. Among the published reports, the prevalences of the other 11 types among MRMP were low, and their occurrences were sporadic (5, 6, 9, 12–15, 22, 24, 26).
As far as we know, this is the first report to demonstrate a link between changes in MRMP prevalence and increasing resistance within a single MLVA type. The inclusion of consecutive specimens from a 4-year period and a relatively large sample size are the strengths of this study. Given that this retrospective analysis examined specimens from inpatients only, of whom the majority were diagnosed with pneumonia, the findings may not be representative of mild M. pneumoniae infections in the community. M. pneumoniae is a genetically conserved organism. A pairwise comparison of the four published M. pneumoniae genomes (strains M29, M129, 309, and FH) revealed that the difference between strains of different MLVA types (4-5-7-2 versus 3-5-6-2) was 0.3% to 0.5%, while the difference between strains of the same MLVA types was 0.08% (see Table S2 in the supplemental material). Therefore, whole-genome sequencing may be the ultimate approach for resolving whether there is any MRMP subclone within type 4-5-7-2.
In summary, we demonstrated a link between increasing macrolide resistance and the expansion of MRMP strains of the MLVA type 4-5-7-2 in Hong Kong during 2011 to 2014. It is worrying that type 4-5-7-2 may be associated with more severe disease (22). Increasing public awareness, enhancing access to rapid diagnostics, and improving surveillance for M. pneumoniae infection and macrolide resistance are necessary to inform case and outbreak management and to understand the burden of disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Health and Medical Research Fund of the Food and Health Bureau of the Government of the Hong Kong Special Administrative Region.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01983-15.
REFERENCES
- 1.Principi N, Esposito S. 2013. Macrolide-resistant Mycoplasma pneumoniae: its role in respiratory infection. J Antimicrob Chemother 68:506–511. doi: 10.1093/jac/dks457. [DOI] [PubMed] [Google Scholar]
- 2.Bradley JS, Byington CL, Shah SS, Alverson B, Carter ER, Harrison C, Kaplan SL, Mace SE, McCracken GH Jr, Moore MR, St. Peter SD, Stockwell JA, Swanson JT, Pediatric Infectious Diseases Society, Infectious Diseases Society of. 2011. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin Infect Dis 53:e25–e76. doi: 10.1093/cid/cir531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Ye X, Zhang H, Xu X, Wang M. 2012. Multiclonal origin of macrolide-resistant Mycoplasma pneumoniae isolates as determined by multilocus variable-number tandem-repeat analysis. J Clin Microbiol 50:2793–2795. doi: 10.1128/JCM.00678-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada T, Morozumi M, Tajima T, Hasegawa M, Sakata H, Ohnari S, Chiba N, Iwata S, Ubukata K. 2012. Rapid effectiveness of minocycline or doxycycline against macrolide-resistant Mycoplasma pneumoniae infection in a 2011 outbreak among Japanese children. Clin Infect Dis 55:1642–1649. doi: 10.1093/cid/cis784. [DOI] [PubMed] [Google Scholar]
- 5.Pereyre S, Touati A, Petitjean-Lecherbonnier J, Charron A, Vabret A, Bébéar C. 2013. The increased incidence of Mycoplasma pneumoniae in France in 2011 was polyclonal, mainly involving M. pneumoniae type 1 strains. Clin Microbiol Infect 19:E212–E217. doi: 10.1111/1469-0691.12107. [DOI] [PubMed] [Google Scholar]
- 6.Xue G, Wang Q, Yan C, Jeoffreys N, Wang L, Li S, Gilbert GL, Sun H. 2014. Molecular characterizations of PCR-positive Mycoplasma pneumoniae specimens collected from Australia and China. J Clin Microbiol 52:1478–1482. doi: 10.1128/JCM.03366-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong KN, Chiu SS, Chan BW, To KK, Chan EL, Ho PL. 2014. Severe macrolide-resistant Mycoplasma pneumoniae pneumonia associated with macrolide failure. J Microbiol Immunol Infect doi: 10.1016/j.jmii.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, Zhang Y, Sheng Y, Zhang L, Shen Z, Chen Z. 2014. More complications occur in macrolide-resistant than in macrolide-sensitive Mycoplasma pneumoniae pneumonia. Antimicrob Agents Chemother 58:1034–1038. doi: 10.1128/AAC.01806-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dégrange S, Cazanave C, Charron A, Renaudin H, Bébéar C, Bébéar CM. 2009. Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae. J Clin Microbiol 47:914–923. doi: 10.1128/JCM.01935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalker V, Stocki T, Mentasti M, Fleming D, Harrison T. 2011. Increased incidence of Mycoplasma pneumoniae infection in England and Wales in 2010: multiocus [sic]variable number tandem repeat analysis typing and macrolide susceptibility. Euro Surveill 16:pii=19865 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19865. [PubMed] [Google Scholar]
- 11.Dumke R, Jacobs E. 2011. Culture-independent multi-locus variable-number tandem-repeat analysis (MLVA) of Mycoplasma pneumoniae. J Microbiol Methods 86:393–396. doi: 10.1016/j.mimet.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Yan C, Sun H, Xue G, Zhao H, Wang L, Feng Y, Li S. 2014. A single-tube multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical specimens by use of multiplex PCR-capillary electrophoresis. J Clin Microbiol 52:4168–4171. doi: 10.1128/JCM.02178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benitez AJ, Diaz MH, Wolff BJ, Pimentel G, Njenga MK, Estevez A, Winchell JM. 2012. Multilocus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical isolates from 1962 to the present: a retrospective study. J Clin Microbiol 50:3620–3626. doi: 10.1128/JCM.01755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H, Xue G, Yan C, Li S, Cao L, Yuan Y, Zhao H, Feng Y, Wang L, Fan Z. 2013. Multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical specimens and proposal for amendment of MLVA nomenclature. PLoS One 8:e64607. doi: 10.1371/journal.pone.0064607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz MH, Benitez AJ, Winchell JM. 2015. Investigations of Mycoplasma pneumoniae infections in the United States: trends in molecular typing and macrolide resistance from 2006 to 2013. J Clin Microbiol 53:124–130. doi: 10.1128/JCM.02597-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan KH, To KK, Chan BW, Li CP, Chiu SS, Yuen KY, Ho PL. 2013. Comparison of pyrosequencing, Sanger sequencing, and melting curve analysis for detection of low-frequency macrolide-resistant Mycoplasma pneumoniae quasispecies in respiratory specimens. J Clin Microbiol 51:2592–2598. doi: 10.1128/JCM.00785-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM Jr, Musher DM, Niederman MS, Torres A, Whitney CG, Infectious Diseases Society of America, American Thoracic Society. 2007. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 44(Suppl 2):S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu SS, Ho PL, Peiris MJ, Chan KH, Chan EL. 2014. Population-based hospitalization incidence of respiratory viruses in community-acquired pneumonia in children younger than 5 years of age. Influenza Other Respir Viruses 8:626–627. doi: 10.1111/irv.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onozuka D, Hashizume M, Hagihara A. 2009. Impact of weather factors on Mycoplasma pneumoniae pneumonia. Thorax 64:507–511. doi: 10.1136/thx.2008.111237. [DOI] [PubMed] [Google Scholar]
- 20.Xu YC, Zhu LJ, Xu D, Tao XF, Li SX, Tang LF, Chen ZM. 2011. Epidemiological characteristics and meteorological factors of childhood Mycoplasma pneumoniae pneumonia in Hangzhou. World J Pediatr 7:240–244. doi: 10.1007/s12519-011-0318-0. [DOI] [PubMed] [Google Scholar]
- 21.Kim EK, Youn YS, Rhim JW, Shin MS, Kang JH, Lee KY. 2015. Epidemiological comparison of three Mycoplasma pneumoniae pneumonia epidemics in a single hospital over 10 years. Korean J Pediatr 58:172–177. doi: 10.3345/kjp.2015.58.5.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu J, Yu X, Liu Y, Yin Y, Gu L, Cao B, Wang C. 2013. Specific multilocus variable-number tandem-repeat analysis genotypes of Mycoplasma pneumoniae are associated with diseases severity and macrolide susceptibility. PLoS One 8:e82174. doi: 10.1371/journal.pone.0082174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong KB, Choi EH, Lee HJ, Lee SY, Cho EY, Choi JH, Kang HM, Lee J, Ahn YM, Kang YH, Lee JH. 2013. Macrolide resistance of Mycoplasma pneumoniae, South Korea, 2000–2011. Emerg Infect Dis 19:1281–1284. doi: 10.3201/eid1908.121455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao F, Liu G, Cao B, Wu J, Gu Y, He L, Meng F, Zhu L, Yin Y, Lv M, Zhang J. 2013. Multiple-locus variable-number tandem-repeat analysis of 201 Mycoplasma pneumoniae isolates from Beijing, China, from 2008 to 2011. J Clin Microbiol 51:636–639. doi: 10.1128/JCM.02567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalker V, Stocki T, Litt D, Bermingham A, Watson J, Fleming DM, Harrison TG. 2012. Increased detection of Mycoplasma pneumoniae infection in children in England and Wales, October 2011 to January 2012. Euro Surveill 17:pii=20081 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20081. [PubMed] [Google Scholar]
- 26.Dumke R, Schnee C, Pletz MW, Rupp J, Jacobs E, Sachse K, Rohde G, CAPNETZ Study Group . 2015. Mycoplasma pneumoniae and Chlamydia spp. infection in community-acquired pneumonia, Germany, 2011–2012. Emerg Infect Dis 21:426–434. doi: 10.3201/eid2103.140927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.