Abstract
The association between human papillomavirus 31 (HPV31) DNA loads and the risk of cervical intraepithelial neoplasia grades 2 and 3 (CIN2–3) was evaluated among women enrolled in the atypical squamous cells of undetermined significance (ASCUS) and low-grade squamous intraepithelial lesion (LSIL) triage study (ALTS), who were monitored semiannually over 2 years and who had HPV31 infections detected at ≥1 visit. HPV31 DNA loads in the first HPV31-positive samples and in a random set of the last positive samples from women with ≥2 HPV31-positive visits were measured by a real-time PCR assay. CIN2–3 was histologically confirmed at the same time as the first detection of HPV31 for 88 (16.6%) of 530 women. After adjustment for HPV31 lineages, coinfection with other oncogenic types, and the timing of the first positive detection, the odds ratio (OR) per 1-log-unit increase in viral loads for the risk of a concurrent diagnosis of CIN2–3 was 1.5 (95% confidence interval [CI], 1.2 to 1.9). Of 373 women without CIN2–3 at the first positive visit who had ≥1 later visit, 44 had subsequent diagnoses of CIN2–3. The initial viral loads were associated with CIN2–3 diagnosed within 6 months after the first positive visit (adjusted OR, 1.5 [95% CI, 1.0 to 2.4]) but were unrelated to CIN2–3 diagnosed later. For a random set of 49 women who were tested for viral loads at the first and last positive visits, changes in viral loads were upward and downward among women with and without follow-up CIN2–3 diagnoses, respectively, although the difference was not statistically significant. Results suggest that HPV31 DNA load levels at the first positive visit signal a short-term but not long-term risk of CIN2–3.
INTRODUCTION
Cervical infection with oncogenic human papillomavirus (HPV) is a prerequisite for cervical cancer and its precursor lesions, which have typically been defined for clinical purposes as cervical intraepithelial neoplasia grades 2 and 3 (CIN2–3) (1). However, most infections are transient, with only a small proportion leading to the development of treatable cervical precancerous lesions (2, 3). It is still largely undetermined why, for a given type of HPV, some infections progress while others do not. The HPV DNA load, as measured in an exfoliated cervical scraping sample, reflects the number of free virions, the number of infected cells with either productive infection or clonal expansion, and the numbers of viral copies in individual cells. Whether HPV DNA load levels are associated with the risk of CIN2–3 deserves consideration.
Studies of the clinical relevance of HPV DNA loads have focused mainly on HPV16 (4–8), the type that possesses the greatest oncogenic potential (9–11). Data on oncogenic types other than HPV16 are much less common. Some reports suggest that the viral-load-related risk of CIN2–3 may be HPV type specific (12, 13); others show that differences in slopes (daily changes in viral loads) between transient infections and infections leading to CIN3 are constant across types (14, 15). HPV31 is one of the oncogenic types and is phylogenetically closely related to HPV16 (16). Results from recent meta-analyses indicated that the average prevalence of HPV31 infections ranked sixth (after HPV16, HPV18, HPV58, HPV33, and HPV45 infections) among 30,848 invasive cervical cancer cases (9) and second (after HPV16 infections) among 7,094 women with high-grade squamous intraepithelial lesions (HSILs) (referring to both cytological diagnoses of high-grade lesions and histological diagnoses of CIN2–3 or carcinoma in situ in the study) (10).
Few studies have examined the relationship between HPV31 DNA loads and the risk of cervical lesions, with inconsistent findings being reported (17–24). Most previous studies were limited by small sample sizes and a cross-sectional study design. To date, published data on longitudinal changes in HPV31 DNA loads, stratified according to whether there was a subsequent diagnosis of CIN2–3, are even more rare (14, 15). The profile of changes in viral loads over time may help to distinguish infections that tend to resolve from those that lead to the development of cervical lesions.
We recently demonstrated the association of variant lineages of the HPV31 genome with the likelihood of viral persistence (25) and disease progression (26) in a large-scale longitudinal study. The present study is an extension of our previous observations, designed to examine whether HPV31 DNA loads at the first positive visit were associated with concurrent and/or subsequent diagnoses of CIN2–3 in the same study population. Furthermore, we explored the clinical implications of viral load fluctuations by comparing the HPV31 DNA levels at the first and last positive visits for a random set of women with or without a subsequent diagnosis of CIN2–3.
MATERIALS AND METHODS
Study subjects.
Study subjects were women enrolled in the atypical squamous cells of undetermined significance (ASCUS) and low-grade squamous intraepithelial lesion (LSIL) triage study (ALTS), a multicenter randomized trial designed to evaluate three strategies for management of women with equivocal or mild abnormal cervical cytological findings. A detailed description of the ALTS design and study population is presented elsewhere (27). Briefly, between January 1997 and December 1998, a total of 5,060 women with diagnoses of ASCUS or LSILs, as established through liquid-based cytology within the previous 6 months, were enrolled in the trial. At enrollment, participants were randomly assigned to one of three management arms, i.e., immediate colposcopy (referral of all women to colposcopy), HPV triage (referral to colposcopy if the testing result at enrollment was oncogenic HPV positive or cytological evidence of a HSIL), or conservative management (referral to colposcopy if the cytological findings at enrollment indicated a HSIL). All participants, regardless of arm, were scheduled for liquid-based cytology and HPV testing semiannually for 2 years. A colposcopic examination was performed again if follow-up cytological evidence of a HSIL was identified. At study exit, all participants were requested to undergo an exit procedure that included cytology, HPV testing, and colposcopic examination with biopsy of any visible lesions.
An ALTS participant was eligible for this study if she had HPV31 DNA detectable by a PCR-based reverse line blot assay (28) in her cervical swab sample at ≥1 visit. For each infection, we chose to use the first HPV31-positive sample for viral load measurements. Of a total of 557 HPV31-positive women, 232 had HPV31 DNA detected at ≥2 visits. We also selected the last HPV31-positive sample (n = 58) for a random set of 25% of the women with ≥2 HPV31-positive visits, for a comparison of viral loads in paired samples. The first positive sample was not available for 27 women, leaving 530 women in analyses of viral loads at the first positive visit. Of the 58 women who were randomly selected for testing of viral loads in paired positive samples, 5 were excluded due to lack of either the first positive sample (n = 4) or the last positive sample (n = 1). We also excluded 4 women with paired positive specimens who had a diagnosis of CIN2–3 at the first positive visit, leaving 49 women for the analysis of changes in viral loads for women with or without a subsequent diagnosis of CIN2–3.
Data on HPV genotyping, cervical lesions, and characteristics of the study subjects were obtained from the ALTS database. The ALTS protocol was approved by the institutional review boards at the National Cancer Institute and all four clinical centers involved in the trial. The protocol for this study was approved by the University of Washington institutional review board.
Clinical endpoint.
In the ALTS, cervical lesions were initially diagnosed by the clinical center pathologists, and then the findings were reviewed by a panel of expert pathologists for quality control and safety monitoring. The endpoint used in this study was the first episode of CIN2–3 (unless otherwise specified) histologically confirmed by the panel of expert pathologists. Cervical tissues were obtained by biopsy, endocervical curettage, and/or excision procedures. If more than one tissue block was examined at a single visit, then the most severe diagnosis was assigned to the patient. CIN2–3 was considered to be HPV31 related only if HPV31 DNA was detected in the cervical swab sample collected at the screening visit immediately preceding the colposcopic visit that yielded the histological diagnosis.
Quantification of HPV31 DNA loads.
DNAs were extracted with a QIAamp DNA Blood minikit (Qiagen, Valencia, CA) from aliquots of 50- to 100-μl cervical swab samples in specimen transport medium and then were resuspended in 25 μl AE buffer, as described previously (29). HPV31 copy numbers and cellular DNA amounts (evaluated by testing for the β-actin gene) were measured with a multiplex real-time PCR assay. The sequences of the primers and probe for the HPV31 E7 gene were as follows: forward primer (nucleotide positions 808 to 827), 5′-AATGGGCTCATTTGGAATCG; reverse primer (nucleotide position 873 to 848), 5′-TGGATCAGCCATTGTAGTTACAGTCT; fluorescent probe (nucleotide positions 830 to 845), 5′-VIC-TGCCCCAACTGTTCTA-minor groove binder (MGB). The primers and probe for the human β-actin gene are commercially available (Applied Biosystems, Foster City, CA). Each sample was assayed in triplicate. The assay was set up with the TaqMan Universal PCR master mix kit (Applied Biosystems, Foster City, CA), in a final reaction volume of 5 μl containing a 0.5-μl DNA sample input. Amplification was performed with an Applied Biosystems 7900HT sequence detection system, with a cycling program of holding at 50°C for 2 min and then at 95°C for 10 min followed by a two-step cycle of 10 s at 95°C and 1 min at 60°C for 40 cycles.
Two logarithmic 5-point standard curves, one for HPV31 and the other for cellular DNA, were implemented for each assay set for absolute quantification. Positive and negative controls were included in each assay run from sample preparation through all PCR steps. The numbers of viral copies were normalized according to the input amounts of cellular DNA (β-actin) and log10 transformed. The mean values of three measurements (expressed as log10 HPV31 copy number per nanogram of cellular DNA) were used for analysis.
HPV31 DNA was undetectable by real-time PCR in 46 samples that were positive by the initial PCR-based reverse line blot assay. The negative results were not explained by a lack of sufficient sample input or the presence of potential inhibitors, because the amounts of cellular DNA were similar for samples with and without detectable HPV31 DNA (data not shown). Also, repeated testing of these samples with double or even triple amounts of sample input did not yield positive signals (data not shown). Considering that the negative finding might result from a tiny amount of HPV31 DNA, a value of 1 viral copy/ng of cellular DNA was arbitrarily assigned to each of these samples. Results remained similar when these 46 samples were excluded from the analysis; therefore, for simplicity, results obtained with these samples excluded are not presented.
Statistical analyses.
The main exposure of interest was HPV31 DNA load, which was treated as a continuous variable for all analyses. Odds ratios (ORs) and 95% confidence intervals (CIs) for the association of the HPV31 DNA load at the first positive visit with a concurrent diagnosis of CIN2–3 were estimated using unconditional logistic regression (30). Factors potentially associated with the risk of CIN2–3, such as age, race, sexual behavior, smoking status, timing of the initial HPV31 detection, lineages of HPV31 variants, and coinfection with other oncogenic types, were initially examined by univariate analyses. Variables with P values of <0.20 were entered into multivariate models as covariates. We used backward stepwise regression to establish the final model, with a P value of <0.20 being the criterion for entering and removing variables. Covariates included in the final model were lineages of HPV31 variants (A, B, or C), coinfection with other oncogenic types (yes or no for infection with HPV16/18/33/35/39/45/51/52/56/58/59), and timing of the initial HPV31 detection (at enrollment or during follow-up monitoring). Since hierarchically HPV16 confers greater risk of CIN2–3 than does HPV31, the results are presented separately for all HPV31-positive women and for women without HPV16 coinfection at the first HPV31-positive visit.
Among women without CIN2–3 at the first HPV31-positive visit, we further examined the association between the initial HPV31 DNA load and the risk of a subsequent diagnosis of CIN2–3 during follow-up monitoring, using unconditional logistic regression. In this analysis, the time of the first HPV31-positive test was rescaled as month 0, the next visit as month 6, and the visit after that as month 12. Data from the last 2 visits were combined because of a limited number of CIN2–3 cases at months 18 and 24. Women with a previous diagnosis of CIN2–3 were removed from risk sets for analyses of the initial viral-load-related risk of CIN2–3 at subsequent visits. Again, the ORs were adjusted for lineages of HPV31 variants, coinfection with other oncogenic types, and the timing of the initial HPV31 detection.
A trend of increasing HPV31 DNA loads at the first positive visit with increasing severity of concurrent cervical cytological findings was examined with a trend test. For a random set of 49 women, a paired t test was used for pairwise comparison of HPV31 DNA loads for the first and last positive samples, and Student's t test was used to compare the lengths of time from the first positive visit to the last positive visit for women with versus without a subsequent diagnosis of CIN2–3. The significance of changes in viral loads in paired samples for women with versus without a subsequent diagnosis of CIN2–3 was assessed by linear regression with adjustment for the time period from the first positive visit to the last positive visit. All statistical tests were conducted at the 5% two-sided significance level.
RESULTS
Of the 530 HPV31-positive women included, 321 were initially positive at enrollment and 209 during follow-up monitoring. Abnormal cervical cytological findings at the first HPV31-positive visit were detected for 355 (67.2%) of the 528 women (2 with Pap smears insufficient for cytological diagnosis), including 153 (29.0%) with ASCUS, 151 (28.6%) with LSILs, and 51 (9.7%) with HSILs. As shown in Fig. 1, the mean values of log10-transformed HPV31 copy numbers per nanogram of cellular DNA increased from 2.55 log10 copies/ng (95% CI, 2.34 to 2.76 log10 copies/ng) among women with normal cytological findings to 3.32 log10 copies/ng (95% CI, 3.10 to 3.56 log10 copies/ng) among women with ASCUS and 3.96 log10 copies/ng (95% CI, 3.72 to 4.19 log10 copies/ng) among women with LSILs and then decreased slightly to 3.75 log10 copies/ng (95% CI, 3.39 to 4.10 log10 copies/ng) among women with HSILs (P for trend of <0.001).
FIG 1.
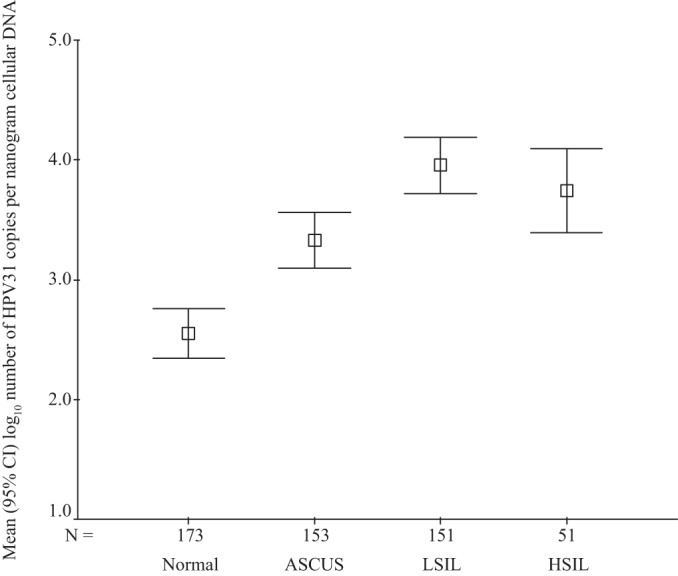
Means (squares) and 95% confidence intervals (upper and lower bounds) of log10-transformed HPV31 copy number per nanogram of cellular DNA at the first positive visit, stratified according to cervical cytological findings. Excluded were two women whose Pap smears were insufficient for cytological evaluation. HPV, human papillomavirus; ASCUS, atypical squamous cells of undetermined significance; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion.
CIN2–3 was histologically confirmed at the first HPV31-positive visit for 88 (16.6%) of the 530 women. The mean ± standard deviation (SD) values of log10-transformed HPV31 copy number per nanogram of cellular DNA at the first positive visit were 3.89 ± 1.27 and 3.17 ± 1.54 log10 copies/ng for women with and without a concurrent diagnosis of CIN2–3, respectively (Table 1). After adjustment for the lineages of HPV31 variants, coinfection with other oncogenic types, and the timing of the initial HPV31 detection, the OR for the association of 1-log-unit increases in HPV31 DNA loads with the risk of a concurrent diagnosis of CIN2–3 was 1.5 (95% CI, 1.2 to 1.9). Higher viral loads remained associated with a concurrent diagnosis of CIN2–3 across cytological categories, although a statistically significant difference was seen only among women with LSILs (adjusted OR, 1.4 [95% CI, 1.0 to 2.1]).
TABLE 1.
Association of 1-log-unit increases in log10-transformed HPV31 DNA loads at the first positive visit with risk of concurrent diagnosis of CIN2–3
| Subjects and cervical cytological results at first positive visita | Women with <CIN2 |
Women with CIN2–3 |
Crude OR (95% CI) | Adjusted OR (95% CI)c | ||
|---|---|---|---|---|---|---|
| No. | Viral load (log10 copies/ng) (mean ± SD)b | No. | Viral load (log10 copies/ng) (mean ± SD) | |||
| All women | ||||||
| Overall | 442 | 3.17 ± 1.54 | 88 | 3.89 ± 1.27 | 1.4 (1.2–1.7) | 1.5 (1.2–1.9) |
| Within normal limits | 165 | 2.51 ± 1.40 | 8 | 3.33 ± 0.75 | 1.6 (0.9–2.9) | 1.5 (0.5–4.4) |
| ASCUS | 128 | 3.27 ± 1.48 | 25 | 3.63 ± 1.30 | 1.2 (0.9–1.7) | 1.3 (0.8–2.1) |
| LSIL | 125 | 3.89 ± 1.47 | 26 | 4.31 ± 1.43 | 1.2 (0.9–1.7) | 1.4 (1.0–2.1) |
| HSIL | 23 | 3.54 ± 1.36 | 28 | 3.91 ± 1.15 | 1.3 (0.8–2.0) | 1.5 (0.8–2.8) |
| Women without HPV16 coinfection at first positive visit | ||||||
| Overall | 360 | 3.20 ± 1.53 | 62 | 4.04 ± 1.18 | 1.5 (1.2–1.9) | 1.6 (1.2–2.1) |
| Within normal limits | 140 | 2.54 ± 1.35 | 7 | 3.32 ± 0.81 | 1.6 (0.9–3.1) | 1.8 (0.5–6.1) |
| ASCUS | 103 | 3.34 ± 1.49 | 16 | 3.99 ± 1.07 | 1.5 (0.9–2.3) | 1.6 (0.9–3.0) |
| LSIL | 100 | 3.93 ± 1.49 | 20 | 4.37 ± 1.20 | 1.3 (0.9–1.8) | 1.2 (0.8–1.9) |
| HSIL | 16 | 3.62 ± 1.22 | 18 | 4.01 ± 1.36 | 1.3 (0.7–2.2) | 2.1 (1.0–4.7) |
HPV, human papillomavirus; CIN, cervical intraepithelial neoplasia; ASCUS, atypical squamous cells of undetermined significance; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; SD, standard deviation; OR, odd ratio; CI, confidence interval.
Viral loads are presented as log10-transformed HPV31 copy number per nanogram of cellular DNA.
Adjusted for lineages of HPV31 variants (A, B, or C), coinfection with other oncogenic types (yes or no), and the timing of the initial HPV31 detection (at enrollment or during follow-up monitoring).
A total of 108 women (26 with and 82 without CIN2–3) had HPV16 coinfections at the first HPV31-positive visit. The mean ± SD values of log10-transformed HPV31 DNA copy number per nanogram of cellular DNA were 3.56 ± 1.43 and 3.04 ± 1.59 log10 copies/ng for HPV16-positive women with and without a concurrent diagnosis of CIN2–3, respectively (P = 0.14). When the analysis was restricted to women without HPV16 coinfection (n = 422), the associations between the risk of a concurrent diagnosis of CIN2–3 and 1-log-unit increases in viral loads remained similar except for data stratified according to cytological findings, with a statistically significant association being seen among women with HSILs (Table 1). A total of 207 women (24 with and 183 without CIN2–3) did not have coinfections with other oncogenic types at the first HPV31-positive visit. Among women without other oncogenic types detected, the mean ± SD values of log10-transformed HPV31 DNA copy number per nanogram of cellular DNA were 4.26 ± 1.02 and 3.12 ± 1.50 log10 copies/ng for women with and without a concurrent diagnosis of CIN2–3, respectively; the adjusted OR associating the risk of a concurrent diagnosis of CIN2–3 with 1-log-unit increases in HPV31 DNA loads was 1.6 (95% CI, 1.1 to 2.4). Infection with HPV31 alone at the first positive visit was detected for 114 women (16 with and 98 without CIN2–3). Among women with single-type infections, increased viral loads remained associated with CIN2–3 (i.e., 4.46 ± 0.93 and 3.34 ± 1.51 log10 copies/ng for women with and without a concurrent diagnosis of CIN2–3, respectively), although the association was not statistically significant (adjusted OR, 1.5 [95% CI, 0.9 to 2.6]).
Thirty-nine women had a diagnosis of CIN3 at the first HPV31-positive visit. Mean ± SD values of log10-transformed HPV31 copy number per nanogram of cellular DNA were 3.74 ± 1.27 log10 copies/ng for CIN3 cases overall, 3.85 ± 1.23 log10 copies/ng for 25 cases without HPV16 coinfection, and 3.84 ± 0.39 log10 copies/ng for 5 cases without coinfection with other oncogenic types at the first HPV31-positive visit. After adjustment for the lineages of HPV31 variants, coinfection with other oncogenic types, and the timing of the initial HPV31 detection, the ORs for the association of 1-log-unit increases in HPV31 DNA loads at the first positive visit with the risk of a concurrent diagnosis of CIN3 were 1.4 (95% CI, 1.0 to 1.9) for women overall and 1.5 (95% CI, 1.0 to 2.2) for women without HPV16 coinfection.
We next examined whether HPV31 DNA load levels at the first positive visit played a role in the subsequent development of CIN2–3. Eighty-eight women who had a diagnosis of CIN2–3 at the first positive visit were excluded from this analysis. We also excluded 69 women who were not in the risk set for subsequent development of CIN2–3 because they had HPV31 infections that were initially detected at the exit visit (n = 41) or they were lost to follow-up monitoring after the first positive visit (n = 28). Overall, CIN2–3 was histologically confirmed for 44 (11.8%) of the 373 women who had ≥1 visit since the first HPV31 detection; it was seen in 16 (4.7%) of 341 women at month 6, 12 (4.2%) of 284 women at month 12, and 16 (6.3%) of 256 women at month 18 or 24. After adjustment for the lineages of HPV31 variants, coinfection with other oncogenic types, and the timing of the initial HPV31 detection, the viral loads at the first positive visit were statistically significantly associated with a risk of CIN2–3 diagnosed 6 months after the first HPV31 detection (adjusted OR, 1.5 [95% CI, 1.0 to 2.4]) but were unrelated to CIN2–3 that occurred after that time (Table 2).
TABLE 2.
Association of 1-log-unit increases in log10-transformed HPV31 DNA loads at the first positive visit with risk of subsequent diagnosis of CIN2–3
| Time from first HPV31-positive visit (mo)a | No. of women followedb | Women with <CIN2 |
Women with CIN2–3 |
Crude OR (95% CI) | Adjusted OR (95% CI)d | ||
|---|---|---|---|---|---|---|---|
| No. | Viral load (log10 copies/ng) (mean ± SD)c | No. | Viral load (log10 copies/ng) (mean ± SD) | ||||
| 6 | 341 | 325 | 3.17 ± 1.49 | 16 | 3.87 ± 0.97 | 1.4 (1.0–2.2) | 1.5 (1.0–2.4) |
| 12 | 284 | 272 | 3.25 ± 1.48 | 12 | 3.44 ± 1.13 | 1.1 (0.7–1.6) | 1.0 (0.6–1.6) |
| 18 or 24 | 256 | 240 | 3.25 ± 1.54 | 16 | 3.35 ± 0.85 | 1.0 (0.7–1.4) | 0.9 (0.6–1.5) |
HPV, human papillomavirus; CIN, cervical intraepithelial neoplasia; SD, standard deviation; OR, odd ratio; CI, confidence interval.
Actual number of women seen at visits subsequent to the first HPV31-positive test who did not have a diagnosis of CIN2–3 at previous visits. Thirty-two women did not return for follow-up visits at 6 months but did return for later visits. Thus, the number of women seen at 6 months was 341 (not 373).
Viral loads are presented as log10-transformed HPV31 copy number per nanogram of cellular DNA.
Adjusted for lineages of HPV31 variants (A, B, or C), coinfection with other oncogenic types (yes or no), and the timing of the initial HPV31 detection (at enrollment or during follow-up monitoring).
Forty-nine women were included in a pairwise comparison of HPV31 DNA loads between the first and last positive visits, including 11 with and 38 without a diagnosis of CIN2–3 at the last positive visit. The lengths of time (mean ± SD) between the two visits were 13.3 ± 8.0 and 10.9 ± 6.2 months for women with and without a diagnosis of CIN2–3, respectively (P = 0.29). In contrast to a slight decrease in HPV31 DNA loads, from 3.32 log10 copies/ng at the first positive visit to 3.05 log10 copies/ng at the last positive visit, among women without CIN2–3, the viral loads increased from 3.98 to 4.43 log10 copies/ng among women with CIN2–3, although the difference was not statistically significant (P = 0.23) (Table 3).
TABLE 3.
Pairwise comparison of HPV31 DNA loads between first and last positive visits, stratified according to diagnosis of CIN2–3 at last positive visit
| CIN2–3 at last positive visita | No. of sample pairs | Viral load (log10 copies/ng) (mean ± SD)b |
Paired difference (log10 copies/ng) (mean [95% CI]) | P for first vs last visitc | P for with vs without CIN2–3d | |
|---|---|---|---|---|---|---|
| First positive visit | Last positive visit | |||||
| No | 38 | 3.32 ± 1.20 | 3.05 ± 1.39 | 0.28 (−0.20 to 0.75) | 0.24 | 0.23 |
| Yes | 11 | 3.98 ± 1.30 | 4.43 ± 1.42 | −0.44 (−1.36 to 0.48) | 0.31 | |
HPV, human papillomavirus; CIN, cervical intraepithelial neoplasia; SD, standard deviation; CI, confidence interval.
Viral loads are presented as log10-transformed HPV31 copy number per nanogram of cellular DNA.
Testing for differences in viral loads between the first and last positive visits, by paired t test.
Testing for differences in changes in viral loads in paired samples between women with versus without a diagnosis of CIN2–3 at the last positive visit, by linear regression with adjustment for the time from the first positive visit to the last positive visit.
DISCUSSION
In this study of the clinical relevance of HPV31 DNA loads among ALTS participants, we found that HPV31 DNA load levels at the first positive visit were significantly associated with a concurrent diagnosis of CIN2–3. The association could not be explained by other factors shown to predict risk for cervical lesions, including the lineages of HPV variants, coinfection with other oncogenic HPV types, and the timing of the initial HPV31 detection. Also, the association was independent of the role of HPV16, the most oncogenic type of HPV. Because testing for viral loads was performed without knowledge of any clinical information and cervical lesions were diagnosed by the panel of expert pathologists prior to the viral load measurements, biases in ascertainment of exposures and outcomes were minimized.
In agreement with previous reports regarding HPV DNA loads according to cervical cytological findings (18, 24), we observed that HPV31 DNA loads increased significantly from women with normal cytological findings to those with cytological diagnoses of LSILs; DNA loads were slightly lower among women with HSILs than among those with LSILs. This trend may in part reflect differences in the numbers of cells featured with productive infections and/or viral copies in individual cells among women with various cytological manifestations. Cervical specimens obtained by scraping contain mixed cell populations. The slightly higher viral loads in women with cytological diagnoses of LSILs versus those with HSILs may result from a large number of ASCUS/LSIL cells obtained from women with LSILs, compared to those with HSILs. Considering a correlation of HPV DNA loads and cervical cytological findings and the possibility that some cytological abnormalities might represent an intermediate step (or an indicator) in the HPV-induced pathogenesis of CIN2–3, we treated this variable as a stratifier, rather than a confounder, in our analyses. The tendency for increased risk of CIN2–3 with increasing HPV31 DNA loads was maintained across cytological categories.
The published data regarding the risk of cervical lesions according to HPV31 DNA loads have been inconsistent (17–24). The discrepancy may be in part due to limited sample sizes and/or differences in study designs and laboratory methods. Our results are consistent with some previous reports that showed a positive relationship between higher HPV31 DNA loads and concurrent detection of cervical lesions of greater severities (17, 19).
It should be noted that CIN2–3 cells are usually featured with clonal expansion of the virus and contain many fewer viral copes than cells featured with productive infections, such as LSILs. Thus, the increased viral loads detected in cervical swab samples from CIN2–3 cases should not be contributed by the lesions. As reported previously (31), pathological features in the mucosa surrounding CIN3 lesions, rather than the lesions themselves, are the main determinants of HPV DNA loads. This is because specimens obtained by scraping consist mainly of surface epithelium and favor the collection of maturing cells (cytologically appearing as ASCUS/LSIL); CIN3 lesions contribute only a small fraction of the cells removed by scraping. Accordingly, the finding of the significant (albeit modest) association of increasing HPV31 DNA loads with concurrent diagnoses of CIN2–3 could be explained by the possibility that specimens from women with versus without CIN2–3 contain more cells featured with productive HPV infections and/or more viral copies in individual cells.
Longitudinal data on the clinical relevance of HPV31 DNA loads are rare; Hesselink et al. showed a relative risk of 1.7 (95% CI, 1.1 to 2.7) for the association of 10-fold changes in HPV31 DNA loads with an 18-month cumulative risk of CIN2–3 in a population-based cervical screening cohort (19). The present study extended previous reports by showing a time-dependent, viral-load-related risk of CIN2–3. We observed that the initial viral loads were significantly associated with CIN2–3 that occurred within 6 months after the first positive detection but were unrelated to CIN2–3 that occurred later. This finding somewhat agrees with our previous reports, indicating that the initially detected viral loads were associated with short-term but not long-term persistence of HPV infection (32).
Another interesting observation of the present study is the different changes in HPV31 DNA loads between women with versus without a follow-up diagnosis of CIN2–3. In contrast to a slight decrease among women who did not develop CIN2–3 under observation, the viral loads were higher and tended to increase even further among women who had a diagnosis of CIN2–3 at the last positive visit. This agrees with previous reports on clinical outcomes with respect to changes in HPV16 DNA loads (6, 14), suggesting that sustained higher viral loads may signal the progression of cervical lesions.
Several limitation of this study should be addressed. First, there are currently no universal HPV DNA standards for viral load quantification. Although the standard used for our real-time PCR assay was carefully calibrated, the absolute values of viral loads determined by cervical cytology and histology may not be generalizable to other studies. However, this lack of generalizability does not affect the validity of relative comparisons of viral loads that were derived from the same standard. Second, viral loads are known to fluctuate during the natural course of infections. As noted, this study included a large fraction of HPV31 infections with unknown onset times, due to “left censoring” with the first positive detection at enrollment. To minimize confounding effects possibly introduced by the uncertainty of the onset times for these infections, we included a variable for infection initially detected at enrollment versus during follow-up monitoring as a covariate in analyses of the risk association. Related to this limitation is the fact that not all HPV31-positive samples were tested for viral loads. Thus, we were unable to examine the effects of dynamic changes in viral loads on the development of CIN2–3. Third, CIN2–3 was considered to be HPV31 related only if HPV31 DNA was detected in the cervical swab sample obtained at the screening visit immediately preceding the lesion diagnosis. It is possible that not all infections seen in swab samples exist in tissue samples; lesions could be caused by types other than HPV31. While the present study was unable to ascertain such misclassified cases (because HPV testing was not performed on tissue samples in the ALTS), the comparable results derived from analyses restricted to women without coinfection with HPV16 or other oncogenic types argue against the idea that the observed association would be substantially disturbed by this potential misclassification. Lastly, our findings pertain to women who had a cytological diagnosis of ASCUS or LSIL within 6 months prior to enrollment into the ALTS and who underwent an intensive procedure for follow-up monitoring and clinical examination. Thus, the frequency of CIN2–3 diagnoses seen in this study might not be generalizable to all clinical populations. However, this does not affect the validity of the assessment of the viral-load-associated risk of CIN2–3.
In summary, data from this study suggest that HPV31 DNA load levels at the first positive visit signal a short-term but not long-term risk of CIN2–3; sustained higher or increasing viral loads over time may represent a biomarker for progression. This does not suggest that this biomarker has clinical utility, as the differences are modest and the data are limited to one HPV genotype; rather, we present this as another clue to the pathogenesis of cervical cancer following HPV infection.
ACKNOWLEDGMENTS
The research reported in this publication was supported by the National Cancer Institute, National Institutes of Health (grant CA133569). X.L. was financially supported by a Plan of Cultivating Future Talents award from Shengjing Hospital, China Medical University.
The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The study funder had no role in the study design, data collection, analysis, or interpretation, the writing of the manuscript, or the decision to submit the manuscript for publication.
This study was part of a project ancillary to the ASCUS-LSIL Triage Study but does not represent the ALTS Group. We thank the ALTS Group for providing cervical samples and ALTS data for this study.
We declare no commercial or other associations that might pose a conflict of interest.
REFERENCES
- 1.Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. 2002. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 55:244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez AC, Schiffman M, Herrero R, Wacholder S, Hildesheim A, Castle PE, Solomon D, Burk R. 2008. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst 100:513–517. doi: 10.1093/jnci/djn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med 338:423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 4.Trevisan A, Schlecht NF, Ramanakumar AV, Villa LL, Franco EL. 2013. Human papillomavirus type 16 viral load measurement as a predictor of infection clearance. J Gen Virol 94:1850–1857. doi: 10.1099/vir.0.051722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xi LF, Kiviat NB, Galloway DA, Zhou XH, Ho J, Koutsky LA. 2008. Effect of cervical cytologic status on the association between human papillomavirus type 16 DNA load and the risk of cervical intraepithelial neoplasia grade 3. J Infect Dis 198:324–331. doi: 10.1086/589715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xi LF, Hughes JP, Castle PE, Edelstein ZR, Wang C, Galloway DA, Koutsky LA, Kiviat NB, Schiffman M. 2011. Viral load in the natural history of human papillomavirus type 16 infection: a nested case-control study. J Infect Dis 203:1425–1433. doi: 10.1093/infdis/jir049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Duin M, Snijders PJ, Schrijnemakers HF, Voorhorst FJ, Rozendaal L, Nobbenhuis MA, van den Brule AJ, Verheijen RH, Helmerhorst TJ, Meijer CJ. 2002. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer 98:590–595. doi: 10.1002/ijc.10232. [DOI] [PubMed] [Google Scholar]
- 8.Sundstrom K, Ploner A, Dahlstrom LA, Palmgren J, Dillner J, Adami HO, Ylitalo N, Sparen P. 2013. Prospective study of HPV16 viral load and risk of in situ and invasive squamous cervical cancer. Cancer Epidemiol Biomarkers Prev 22:150–158. doi: 10.1158/1055-9965.EPI-12-0953-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. 2011. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int J Cancer 128:927–935. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- 10.Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, Winer R, Clifford GM. 2007. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer 121:621–632. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- 11.Guan P, Howell-Jones R, Li N, Bruni L, de Sanjose S, Franceschi S, Clifford GM. 2012. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer 131:2349–2359. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 12.Gravitt PE, Kovacic MB, Herrero R, Schiffman M, Bratti C, Hildesheim A, Morales J, Alfaro M, Sherman ME, Wacholder S, Rodriguez AC, Burk RD. 2007. High load for most high risk human papillomavirus genotypes is associated with prevalent cervical cancer precursors but only HPV16 load predicts the development of incident disease. Int J Cancer 121:2787–2793. doi: 10.1002/ijc.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Rio-Ospina L, Soto-De Leon SC, Camargo M, Moreno-Perez DA, Sanchez R, Perez-Prados A, Patarroyo ME, Patarroyo MA. 2015. The DNA load of six high-risk human papillomavirus types and its association with cervical lesions. BMC Cancer 15:100. doi: 10.1186/s12885-015-1126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depuydt CE, Criel AM, Benoy IH, Arbyn M, Vereecken AJ, Bogers JJ. 2012. Changes in type-specific human papillomavirus load predict progression to cervical cancer. J Cell Mol Med 16:3096–3104. doi: 10.1111/j.1582-4934.2012.01631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depuydt CE, Jonckheere J, Berth M, Salembier GM, Vereecken AJ, Bogers JJ. 2015. Serial type-specific human papillomavirus (HPV) load measurement allows differentiation between regressing cervical lesions and serial virion productive transient infections. Cancer Med 4:1294–1302. doi: 10.1002/cam4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 17.Moberg M, Gustavsson I, Gyllensten U. 2004. Type-specific associations of human papillomavirus load with risk of developing cervical carcinoma in situ. Int J Cancer 112:854–859. doi: 10.1002/ijc.20480. [DOI] [PubMed] [Google Scholar]
- 18.Marongiu L, Godi A, Parry JV, Beddows S. 2014. Human papillomavirus 16, 18, 31 and 45 viral load, integration and methylation status stratified by cervical disease stage. BMC Cancer 14:384. doi: 10.1186/1471-2407-14-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hesselink AT, Berkhof J, Heideman DA, Bulkmans NW, van Tellingen JE, Meijer CJ, Snijders PJ. 2009. High-risk human papillomavirus DNA load in a population-based cervical screening cohort in relation to the detection of high-grade cervical intraepithelial neoplasia and cervical cancer. Int J Cancer 124:381–386. doi: 10.1002/ijc.23940. [DOI] [PubMed] [Google Scholar]
- 20.Moberg M, Gustavsson I, Wilander E, Gyllensten U. 2005. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br J Cancer 92:891–894. doi: 10.1038/sj.bjc.6602436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swan DC, Tucker RA, Tortolero-Luna G, Mitchell MF, Wideroff L, Unger ER, Nisenbaum RA, Reeves WC, Icenogle JP. 1999. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J Clin Microbiol 37:1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson S, Safari H, Mints M, Lewensohn-Fuchs I, Gyllensten U, Johansson B. 2005. Type distribution, viral load and integration status of high-risk human papillomaviruses in pre-stages of cervical cancer (CIN). Br J Cancer 92:2195–2200. doi: 10.1038/sj.bjc.6602648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreuter A, Brockmeyer NH, Pfister H, Altmeyer P, Wieland U. 2007. Increased human papillomavirus type 31 DNA load in a verrucous high-grade intraepithelial neoplasia of a human immunodeficiency virus-infected patient with extensive bowenoid papulosis. Br J Dermatol 156:596–598. doi: 10.1111/j.1365-2133.2007.07690.x. [DOI] [PubMed] [Google Scholar]
- 24.Snijders PJ, Hogewoning CJ, Hesselink AT, Berkhof J, Voorhorst FJ, Bleeker MC, Meijer CJ. 2006. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int J Cancer 119:1102–1107. doi: 10.1002/ijc.21956. [DOI] [PubMed] [Google Scholar]
- 25.Xi LF, Schiffman M, Koutsky LA, He Z, Winer RL, Hulbert A, Lee SK, Ke Y, Kiviat NB. 2013. Persistence of newly detected human papillomavirus type 31 infection, stratified by variant lineage. Int J Cancer 132:549–555. doi: 10.1002/ijc.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xi LF, Schiffman M, Koutsky LA, Hulbert A, Lee SK, Defilippis V, Shen Z, Kiviat NB. 2012. Association of human papillomavirus type 31 variants with risk of cervical intraepithelial neoplasia grades 2–3. Int J Cancer 131:2300–2307. doi: 10.1002/ijc.27520. [DOI] [PubMed] [Google Scholar]
- 27.Schiffman M, Adrianza ME. 2000. ASCUS-LSIL Triage Study: design, methods and characteristics of trial participants. Acta Cytol 44:726–742. [DOI] [PubMed] [Google Scholar]
- 28.Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlee F, Hildesheim A, Schiffman MH, Scott DR, Apple RJ. 2000. Improved amplification of genital human papillomaviruses. J Clin Microbiol 38:357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winer RL, Xi LF, Shen Z, Stern JE, Newman L, Feng Q, Hughes JP, Koutsky LA. 2014. Viral load and short-term natural history of type-specific oncogenic human papillomavirus infections in a high-risk cohort of midadult women. Int J Cancer 134:1889–1898. doi: 10.1002/ijc.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosmer DW Jr, Wang CY, Lin IC, Lemeshow S. 1978. A computer program for stepwise logistic regression using maximum likelihood estimation. Comput Programs Biomed 8:121–134. doi: 10.1016/0010-468X(78)90047-8. [DOI] [PubMed] [Google Scholar]
- 31.Sherman ME, Wang SS, Wheeler CM, Rich L, Gravitt PE, Tarone R, Schiffman M. 2003. Determinants of human papillomavirus load among women with histological cervical intraepithelial neoplasia 3: dominant impact of surrounding low-grade lesions. Cancer Epidemiol Biomarkers Prev 12:1038–1044. [PubMed] [Google Scholar]
- 32.Xi LF, Hughes JP, Edelstein ZR, Kiviat NB, Koutsky LA, Mao C, Ho J, Schiffman M. 2009. Human papillomavirus (HPV) type 16 and type 18 DNA loads at baseline and persistence of type-specific infection during a 2-year follow-up. J Infect Dis 200:1789–1797. doi: 10.1086/647993. [DOI] [PMC free article] [PubMed] [Google Scholar]


