Abstract
Resistance of Bordetella pertussis to erythromycin has been increasingly reported. We developed an allele-specific PCR method for rapid detection of erythromycin-resistant B. pertussis directly from nasopharyngeal (NP) swab samples submitted for diagnostic PCR. Based on the proven association of erythromycin resistance with the A2047G mutation in the 23S rRNA of B. pertussis, four primers, two of which were designed to be specific for either the wild-type or the mutant allele, were used in two different versions of the allele-specific PCR assay. The methods were verified with results obtained by PCR-based sequencing of 16 recent B. pertussis isolates and 100 NP swab samples submitted for diagnostic PCR. The detection limits of the two PCR assays ranged from 10 to 100 fg per reaction for both erythromycin-susceptible and -resistant B. pertussis. Two amplified fragments of each PCR, of 286 and 112 bp, respectively, were obtained from a mutant allele of the isolates and/or NP swab samples containing B. pertussis DNAs. For the wild-type allele, only a 286-bp fragment was visible when the allele-specific PCR assay 1 was performed. No amplification was found when a number of non-Bordetella bacterial pathogens and NP swab samples that did not contain the DNAs of B. pertussis were examined. This assay can serve as an alternative for PCR-based sequencing, especially for local laboratories in resource-poor countries.
INTRODUCTION
Pertussis has resurged in many countries. Macrolides, especially erythromycin, are considered the first-choice antibiotics for treatment of pertussis and postexposure prophylaxis (1). The first erythromycin-resistant Bordetella pertussis was discovered in 1994 (2). Although resistant strains are still rare, they have been reported in several countries, including China (3–7), where macrolide-resistant B. pertussis has become prevalent (7).
B. pertussis is a fastidious bacterium. The culture positivity rate is low in immunized populations. The PCR method has been developed and used for diagnosis of pertussis in many laboratories all over the world. However, it is possible that resistant B. pertussis strains are missed due to failure of the culture and when culture is not performed (8).
Several studies have proven the association between macrolide resistance and the A2047G mutation in the 23S rRNA of B. pertussis (3–7). Based on the molecular mechanism identified, methods have been developed for detection of the point mutation in 23S rRNA of B. pertussis in order to study the susceptibility to macrolides when B. pertussis isolates are available (3, 5). In a previous study, we developed a PCR-based sequencing method for identification of the A2047G mutation in cultured B. pertussis isolates and clinical nasopharyngeal (NP) specimens (7).
Although the sequencing is widely used, it is not available or accessible everywhere because it takes longer to obtain the results in many of the local laboratories, especially in resource-poor countries. In this study, we aimed to develop a simple allele-specific PCR method for direct detection of erythromycin-resistant B. pertussis from clinical specimens submitted for diagnostic PCR.
MATERIALS AND METHODS
Bacteria strains and clinical samples.
Fourteen erythromycin-resistant B. pertussis strains with the confirmed mutation A2047G and 2 susceptible strains without the mutation were used for this study (9). The 16 clinical strains were isolated between 2012 and 2013 during a prospective study conducted in Xi'an, China (7, 9). They were isolated throughout the study period and not from local small outbreaks. Erythromycin MICs ranged from 0.023 to >256 μg/ml, and all 14 erythromycin-resistant strains had MICs of >256 μg/ml (9). During the prospective study, NP swab samples were taken from 313 patients with suspected pertussis infection and tested for culture and PCRs (targeting IS481 and ptx-Pr). The age of these patients was 6 days to 11 years (median, 3 months). Of the 313 NP samples, 16 (5.1%) and 168 (53.7%) were positive for culture and IS481 PCR, respectively. Of the 168 samples positive for IS481 PCR, 122 and 100 were positive for ptx-Pr and 23S rRNA PCRs, respectively. All 100 samples positive for 23S rRNA PCR were also positive for ptx-Pr PCR. The primers designed for the 23S rRNA PCR were based on the sequence of domain V of the 23S rRNA gene of the Chinese B. pertussis vaccine strain CS (GenBank accession number CP002695.1). The sequences of the primers were previously reported (7), and the sequencing was performed for the PCR-positive products using the forward primer with the BigDye Terminator v31 cycle sequencing kit on ABI3730xl (Applied Biosystems, Carlsbad, CA) from Life Technology Corporation (Shanghai, China). Of the 100 NP samples tested by sequencing, 15 had the wild-type allele and 85 had the mutant allele A2047G. In addition, 100 NP swab samples testing negative for culture, IS481 PCR, and ptx-Pr PCR were selected and used to evaluate the performance of the 2 allele-specific PCR methods. All DNA samples were stored at −20°C.
The ATCC reference strains and clinical isolates B. pertussis ATCC 9797, Bordetella parapertussis ATCC 15311, Bordetella bronchiseptica ATCC 4613, Klebsiella pneumonia ATCC 70060, Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 29213, the erythromycin-resistant B. pertussis strain bp12152, Neisseria meningitidis, and Haemophilus influenzae were used to determine the specificity and sensitivity of the PCR assays.
Allele-specific PCR assays.
The sequences of the four primers used for the two allele-specific PCR assays are shown in Table 1 and Fig. 1. Primers MP and WP contained a specific mismatch A at the 3′ end that did not complement the published sequence of 23S rRNA of B. pertussis (Fig. 1). The G at the 3′ end of the MP primer was complementary to 2047G of erythromycin-resistant B. pertussis, whereas the A at the 3′ end of the WP primer was complementary to A2047 of erythromycin-sensitive B. pertussis. In each of the two PCRs, three primers were used. For assay 1, primers FP, MP, and RP were included; for assay 2, primers FP, WP, and RP were included. The length of the PCR products based on the forward primer FP and reverse primer RP was 286 bp, whereas the length of the PCR products based on the forward primer MP or WP and reverse primer RP was 121 bp. The working principle of the two PCR assays is described below. In assay 1, the two mismatches at the 3′ end of primer MP would guarantee the absence of PCR amplification if the sequence of erythromycin-sensitive B. pertussis DNA is used as the target. Therefore, there would be only one PCR product of 286 bp. However, if the sequence of erythromycin-resistant B. pertussis DNA is used as the target, PCR amplification of 121 bp would also be present, since there was only one mismatch at the 3′ end of primer MP. In assay 2, the two mismatches at the 3′ end of primer WP would guarantee the absence of PCR amplification if the sequence of erythromycin-resistant B. pertussis DNA is used as the target. Therefore, there would only be one PCR product of 286 bp. However, if the sequence of erythromycin-sensitive B. pertussis DNA is used as the target, PCR amplification of 121 bp would also be present, since there was only one mismatch at the 3′ end of the primer WP. DNA extraction of the reference strains was performed by using the DNA minikit (Qiagen). For allele-specific PCR assay 1, the 20 μl total volume of PCR mixture contained 10 μl of HotStar Taq master mix; 0.5 μM each of primers FP, MP, and RP; and 2 μl of DNA extracts. Amplification was performed in PTC200 thermal cycler (Bio-Rad, CA) with the following conditions: 95°C for 15 min, 35 cycles of 94°C for 1 min, 60°C for 30 s, 72°C for 30 s, and the final extension with 72°C for 10 min. PCR products were resolved on 2% agarose gel, stained with GelRed, and photographed with GelDox XR (Bio-Rad).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Source or reference |
|---|---|---|
| FP | GTGATGGGGTGCAAGCTCTT | This study |
| RP | TCTGGCGACTCGAGTTCTGC | 7 |
| MPa | ATCTACCCGCGGCTAGACAGG | This study |
| WPa | ATCTACCCGCGGCTAGACAGA | This study |
The primers MP and WP contained a specific mismatch A (underlined) at the 3′end that did not complement the published sequence of 23S rRNA of B. pertussis (see Fig. 1). The G (boldfaced) at the 3′ end of the MP primer was complementary to 2047G of erythromycin-resistant B. pertussis, whereas the A (boldfaced) at the 3′ end of the WP primer was complementary to A2047 of erythromycin-sensitive B. pertussis.
FIG 1.
Partial sequence of the 23S rRNA gene of Bordetella pertussis, showing the positions of primers FP, MP, and RP used in the allele-specific PCR. Consensus bases are shown with dashes, and the mismatched bases in the primer are indicated by box and the underline. The position of primer WP is the same as that of primer MP, and the sequence of primer WP is shown in Table 1.
For allele-specific PCR assay 2, the total volume, reaction mixture, and conditions were the same as those of PCR assay 1, except for primer MP, which was replaced by primer WP.
The work flow for determining the A to G mutation of the 2047 nucleotide of 23S rRNA from the clinical NP samples submitted for diagnostic PCR is illustrated in Fig. 2. It is known that B. pertussis carries three copies of the 23S rRNA gene. Heterozygous erythromycin-resistant strains have been reported (3). If the information on heterogeneous susceptibility to erythromycin or coinfection with erythromycin-susceptible B. pertussis is needed, the allele-specific PCR assay 2 should be performed.
FIG 2.
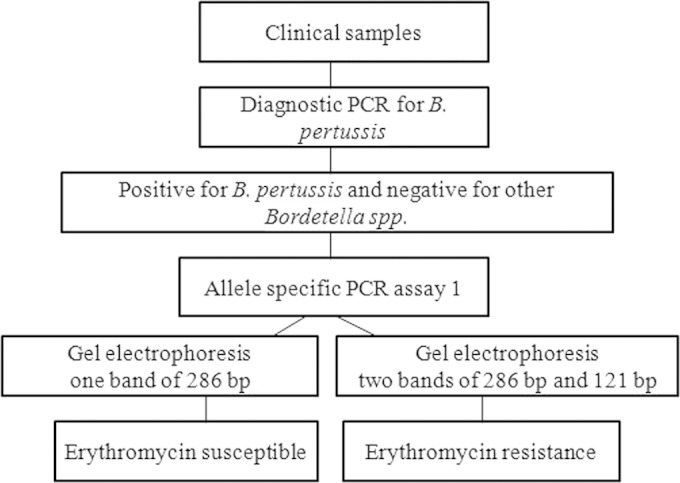
Work flow for determining the A to G mutation of the 2047 nucleotide of 23S rRNA from clinical samples submitted for diagnostic PCR used.
Detection limits and specificity assay.
The detection limits of the two allele-specific PCR assays were measured using serially diluted DNA solutions of an erythromycin-susceptible strain of B. pertussis ATCC 9797 and an erythromycin-resistant strain of bp12152. The concentrations tested ranged from 1 fg/μl to 1 ng/μl.
The specificity of the two PCR assays was determined by using DNA extracts from other Bordetella spp., including B. parapertussis ATCC 15311, B. bronchiseptica ATCC 4613, B. holmesii FR4020, B. petrii FR3497, B. avium FR3815, and B. hinzii FR3756 and from other bacterial species mentioned above. The concentrations used were 1 ng/μl.
RESULTS
Allele-specific PCR assays. Allele-specific PCR assay 1.
As designed, a single band of a 286-bp fragment was visible when the PCR was tested with the DNA extracts from erythromycin-susceptible B. pertussis (Fig. 3). In contrast, two bands of 286 and 121 bp were obtained with the DNA extracts from erythromycin-resistant B. pertussis.
FIG 3.
Gel electrophoresis of the amplified products by allele-specific PCR assays 1 and 2. Lanes: M, 100-bp ladder; WT, ATCC 9797 B. pertussis reference strain (erythromycin sensitive); MT, clinical B. pertussis isolate (erythromycin resistant); S, clinical NP swab; N, negative control of PCR.
Allele-specific PCR assay 2.
For erythromycin-susceptible B. pertussis, two bands of 286 and 121 bp were obtained. Only one band of 286 bp was obtained when erythromycin-resistant B. pertussis was tested (Fig. 3).
Detection limits and specificity.
The detection limits of the two PCR assays ranged from 10 to 100 fg per reaction for erythromycin-susceptible and -resistant B. pertussis. No amplification was detected when DNAs isolated from K. pneumoniae, E. coli, S. aureus, N. meningitidis, and H. influenzae were tested. However, except for B. holmesii, all Bordetella spp. tested produced the 286-bp fragment.
Evaluation of the allele-specific PCR assays by clinical samples submitted for diagnostic PCR.
Of the 100 NP samples tested by PCR-based sequencing, 15 were found to have a wild-type allele, and 85 were found to have the mutant allele A2047G (Table 2). Ninety-five of these samples were positive according to allele-specific PCR assays, and they contained 16 culture-positive samples. The reasons the 5 remaining samples were negative by the allele-specific PCR assays are unknown. One explanation may be that these DNA samples contained a limited amount of B. pertussis DNA. Another explanation may be that the samples were already used several times for different diagnostic PCR tests and PCR-based sequencing. In addition, we cannot exclude the possibility that there might be competitive binding of the three primers used in one PCR, especially when a limited amount of the target DNAs were included.
TABLE 2.
Comparison of the results from PCR-based sequencing and the allele-specific PCR assay in determination of the A2047G mutation of 23S rRNA in B. pertussis from 200 nasopharyngeal swab samples submitted for diagnostic PCRa
| Allele-specific PCR assay | PCR sequencing assay |
Total | ||
|---|---|---|---|---|
| Wild type | Mutation type | Negative | ||
| Wild type | 14b | 0 | 0 | 14 |
| Mutation type | 0 | 81c | 0 | 81 |
| Negative | 1 | 4 | 100 | 105 |
| Total | 15 | 85 | 100 | 200 |
The 200 nasopharyngeal (NP) swab samples included 100 positive and 100 negative samples tested by diagnostic ptx-Pr PCR (7). Of the 100 ptx-Pr PCR positive samples, 16 were also culture positive. Of the 16 B. pertussis strains, 14 were erythromycin resistant and 2 were erythromycin sensitive.
Containing the 2 NP samples from which B. pertussis was isolated and proven to be erythromycin sensitive.
Containing the 14 NP samples from which B. pertussis was isolated and proven to be erythromycin resistant.
Moreover, when the other 100 NP samples that were negative for the diagnostic (ptx-Pr) PCR were tested with the allele-specific PCR assays, they all tested negative.
DISCUSSION
Macrolides, especially erythromycin, are used as the first-choice antibiotics for treatment and prevention of pertussis transmission. However, the susceptibility test cannot be done without bacterial isolates. In this study, we developed a simple PCR method for detection of erythromycin-resistant B. pertussis directly from clinical NP samples. This method is specific, rapid, and easily undertaken compared to currently available methods. Moreover, it is suitable for certain local laboratories, especially those in resource-poor countries where the sequencing technology is not available or where it takes a longer time to obtain the sequencing results. It is well known that the correct and early use of antibiotics is important for treatment and prevention, in addition to saving resources.
Antibiotic resistance plays a key role in the (re)emergence of infectious diseases (10). After the first erythromycin-resistant B. pertussis was reported, researchers paid more attention to the resistant strains, especially when pertussis resurgence occurred. However, as far as we can discern, antibiotic-resistant B. pertussis was only found occasionally in the United States and France (4). Studies carried out in Australia, Taiwan, the United Kingdom, Romania, and Canada showed that B. pertussis isolated in these countries was sensitive to erythromycin (11–15). According to recent research in Iran, 2 of 11 B. pertussis strains isolated between 2009 and 2010 were macrolide resistant (16). However, in China, macrolide-resistant B. pertussis emerged and became common when strains in different regions were tested (5, 17).
In the antibiotic era, bacteria face selective pressure from antibiotics and always evolve to adapt. To our knowledge, there is no molecular mechanism other than the A2047G substitution in the 23S rRNA of B. pertussis that has been linked to erythromycin resistance. Our previous study based on the sequencing of 23S rRNA of B. pertussis also found no other mutations in the gene studied (7). Therefore, we considered that detection of the specific A to G mutation can provide the same information as that obtained from phenotypic analysis of B. pertussis isolates.
In the allele-specific PCR assay, when primers designed to be specific for either wild-type or mutant allele are used, results depend on the absence or presence of amplification. In this study, we found that primer MP contains two mismatches at its 3′ end, and this guarantees the absence of PCR amplification when the sequence of erythromycin-sensitive B. pertussis DNA is used as the target. In contrast, primer WP was found to contain two mismatches at its 3′ end, and this guarantees the absence of PCR amplification when the sequence of erythromycin-resistant B. pertussis DNA is used as the target. The sequences of the two primers are almost identical, except for the last nucleotide at their 3′ ends. Our results confirm that the last nucleotides of the 3′ ends of primers are crucial for primer binding (18).
So far, the molecular mechanism of B. pertussis resistance to erythromycin has been associated with the A2047G substitution in domain V of 23S rRNA (3). It remains to be shown if there is an alternative mechanism that can confer erythromycin resistance. Azithromycin is another commonly used macrolide and more active than erythromycin against B. pertussis. In our previous study, we analyzed the MICs of azithromycin against 14 erythromycin-resistant and 2 erythromycin-sensitive strains (9). Although the MICs of azithromycin against the 16 tested strains were generally lower than those of erythromycin, all strains except the 2 that were erythromycin sensitive were also resistant to azithromycin, suggesting that the A2047G mutation in domain V of 23S rRNA of B. pertussis might be implicated in azithromycin resistance, at least among the clinical isolates analyzed in this study. Further studies are needed to confirm the association.
The study had some limitations. First, the sequences in 23S rRNA are similar among the Bordetella species. As shown in the present study, the allele-specific PCRs may also amplify all except B. holmesii of the Bordetella species. However, this should not be a problem in clinical practice, since this PCR will be performed after the diagnostic PCR specific for B. pertussis. Second, it is known that there are 3 copies of 23S rRNA in the genome of B. pertussis. The heterozygote and homozygote variants of 23S rRNA in B. pertussis found can confer the heterogeneous susceptibility to erythromycin (3). However, we tested the 100 clinical samples using this allele-specific PCR and PCR-based sequencing and did not find any heterozygote variants, suggesting that the heterozygote variants of 23S rRNA in B. pertussis are rare in this study area. In order to determine whether the heterozygote variant of 23S rRNA in B. pertussis or the coinfection of erythromycin-susceptible B. pertussis exists (19), we suggest performing an additional allele-specific PCR assay 2 when allele-specific PCR assay 1 produces two different fragments.
In conclusion, we developed a simple PCR assay for rapid identification of erythromycin-resistant B. pertussis directly from clinical specimens submitted for diagnostic PCR for pertussis. The assay serves as an alternative for PCR-based sequencing, especially for local laboratories in resource-poor countries, where sequencing is difficult to access.
ACKNOWLEDGMENTS
This study was supported by the Natural Science Foundation of Shaanxi Province (2015JM8391).
We thank Nicole Guiso and Sophie Guillot for kindly providing us DNAs of B. holmesii, B. petrii, B. avium, and B. hinzii.
We thank Tom Hamilton for language revision of the manuscript.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Tiwari T, Murphy TV, Moran J, National Immunization Program, CDC . 2005. Recommended antimicrobial agents for the treatment and postexposure prophylaxis of pertussis: 2005 CDC guidelines. MMWR Recomm Rep 54:1–16. [PubMed] [Google Scholar]
- 2.Anonymous. 1994. Erythromycin-resistant Bordetella pertussis–Yuma County, Arizona, May–October 1994. MMWR Morb Mortal Wkly Rep 43:807–810. [PubMed] [Google Scholar]
- 3.Bartkus JM, Juni BA, Ehresmann K, Miller CA, Sanden GN, Cassiday PK, Saubolle M, Lee B, Long J, Harrison AR Jr, Besser JM. 2003. Identification of a mutation associated with erythromycin resistance in Bordetella pertussis: implications for surveillance of antimicrobial resistance. J Clin Microbiol 41:1167–1172. doi: 10.1128/JCM.41.3.1167-1172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillot S, Descours G, Gillet Y, Etienne J, Floret D, Guiso N. 2012. Macrolide-resistant Bordetella pertussis infection in newborn girl, France. Emerg Infect Dis 18:966–968. doi: 10.3201/eid1806.120091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Li M, Wang L, Xin T, He Q. 2013. High-resolution melting analysis for the detection of two erythromycin-resistant Bordetella pertussis strains carried by healthy schoolchildren in China. Clin Microbiol Infect 19:E260–E262. doi: 10.1111/1469-0691.12161. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Li Y, Hou T, Liu X, Liu Y, Yu T, Chen Z, Gao Y, Li H, He Q. 2013. Appearance of macrolide-resistant Bordetella pertussis strains in China. Antimicrob Agents Chemother 57:5193–5194. doi: 10.1128/AAC.01081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Cui Z, Li Y, Hou T, Liu X, Xi Y, Liu Y, Li H, He Q. 2014. High prevalence of erythromycin-resistant Bordetella pertussis in Xi'an, China. Clin Microbiol Infect 20:O825–O830. doi: 10.1111/1469-0691.12671. [DOI] [PubMed] [Google Scholar]
- 8.Mattoo S, Cherry JD. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18:326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Liu X, Zhang B, He Q, Wang Z. 2015. Where macrolide resistance is prevalent. APMIS 123:361–363. doi: 10.1111/apm.12357. [DOI] [PubMed] [Google Scholar]
- 10.Morens DM, Folkers GK, Fauci AS. 2004. The challenge of emerging and re-emerging infectious diseases. Nature 430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sintchenko V, Brown M, Gilbert GL. 2007. Is Bordetella pertussis susceptibility to erythromycin changing? MIC trends among Australian isolates 1971–2006. J Antimicrob Chemother 60:1178–1179. [DOI] [PubMed] [Google Scholar]
- 12.Yao SM, Liaw GJ, Chen YY, Yen MH, Chen YH, Mu JJ, Chiang CS. 2008. Antimicrobial susceptibility testing of Bordetella pertussis in Taiwan prompted by a case of pertussis in a paediatric patient. J Med Microbiol 57:1577–1580. doi: 10.1099/jmm.0.2008/002857-0. [DOI] [PubMed] [Google Scholar]
- 13.Fry NK, Duncan J, Vaghji L, George RC, Harrison TG. 2010. Antimicrobial susceptibility testing of historical and recent clinical isolates of Bordetella pertussis in the United Kingdom using the Etest method. Eur J Clin Microbiol Infect Dis 29:1183–1185. doi: 10.1007/s10096-010-0976-1. [DOI] [PubMed] [Google Scholar]
- 14.Dinu S, Guillot S, Dragomirescu CC, Brun D, Lazar S, Vancea G, Ionescu BM, Gherman MF, Bjerkestrand AF, Ungureanu V, Guiso N, Damian M. 2014. Whooping cough in South-East Romania: a 1-year study. Diagn Microbiol Infect Dis 78:302–306. doi: 10.1016/j.diagmicrobio.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 15.Marchand-Austin A, Memari N, Patel SN, Tang P, Deeks SL, Jamieson FB, Crowcroft NS, Farrell DJ. 2014. Surveillance of antimicrobial resistance in contemporary clinical isolates of Bordetella pertussis in Ontario, Canada. Int J Antimicrob Agents 44:82–84. doi: 10.1016/j.ijantimicag.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Shahcheraghi F, Nakhost Lotfi M, Nikbin VS, Shooraj F, Azizian R, Parzadeh M, Allahyar Torkaman MR, Zahraei SM. 2014. The first macrolide-resistant Bordetella pertussis strains isolated from Iranian patients. Jundishapur J Microbiol 7:e10880. doi: 10.5812/jjm.10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J, Wang HX, Yuan L, Gu S, Jiang M, Ding YJ, Guo D, Yao KH, Wang YJ. 2014. Clinical characteristics of whooping cough in neonates and antimicrobial resistance of the pathogenic bacteria. Zhongguo Dang Dai Er Ke Za Zhi 16:975–978. (In Chinese.) [PubMed] [Google Scholar]
- 18.Sommer R, Tautz D. 1989. Minimal homology requirements for PCR primers. Nucleic Acids Res 17:6749. doi: 10.1093/nar/17.16.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassiday PK, Tobin-D'Angelo M, Watson JR, Wu KH, Park MM, Sanden GN. 2008. Co-infection with two different strains of Bordetella pertussis in an infant. J Med Microbiol 57:388–391. doi: 10.1099/jmm.0.47602-0. [DOI] [PubMed] [Google Scholar]



