Abstract
The heterogeneity of members of the Streptococcus anginosus group (SAG) has traditionally hampered their correct identification. Recently, the group was subdivided into 6 taxa whose prevalence among human infections is poorly described. We evaluated the accuracy of the Rapid ID32 Strep test, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), and a PCR multiplex method to identify 212 SAG isolates recovered from human infections to the species and subspecies level by using multilocus sequence analysis (MLSA) as the gold standard. We also determined the antimicrobial susceptibilities of the isolates. Representatives of all SAG taxa were found among our collection. MALDI-TOF MS and the Rapid ID32 Strep test correctly identified 92% and 68% of the isolates to the species level, respectively, but showed poor performance at the subspecies level, and the latter was responsible for major identification errors. The multiplex PCR method results were in complete agreement with the MLSA identifications but failed to distinguish the subspecies Streptococcus constellatus subsp. pharyngis and S. constellatus subsp. viborgensis. A total of 145 MLSA sequence types were present in our collection, indicating that within each taxon a number of different lineages are capable of causing infection. Significant antibiotic resistance was observed only to tetracycline, erythromycin, and clindamycin and was present in most taxa. MALDI-TOF MS is a reliable method for routine SAG species identification, while the need for identification to the subspecies level is not clearly established.
INTRODUCTION
Initially included in the viridans group streptococci, the species now recognized as the Streptococcus anginosus group (SAG) were first described as Streptococcus milleri in 1956 (1), but their taxonomic classification has remained in flux. Currently, the group includes three species, Streptococcus anginosus, Streptococcus constellatus, and Streptococcus intermedius (2–5) with unique genome signature regions (5). The species S. constellatus was subdivided into the subspecies S. constellatus subsp. pharyngis and S. constellatus subsp. constellatus (6). Recently, Jensen et al., based on multilocus sequence analysis (MLSA), divided S. anginosus into two subspecies—S. anginosus subsp. anginosus (the isolates formerly classified as S. anginosus) and S. anginosus subsp. whileyi—and they described the novel subspecies S. constellatus subsp. viborgensis (7).
Most SAG strains share common features, such as small colony size, low growth rate, and a distinctive caramel smell (8, 9). However, SAG also present diversity in their other characteristics. These organisms may be nonhemolytic or display beta- or alpha-hemolysis when grown on blood agar plates (4, 5, 10), and they may present any of the A, C, F, or G Lancefield group antigens or lack Lancefield group antigen altogether (4, 5, 8, 10).
SAG, as with other members of the viridans group streptococci, are part of the human microbiota, colonizing the oral cavity, nasopharynx, and gastrointestinal and genitourinary tracts (4, 11–13). However, these bacteria may also cause infections that can range from mild, such as pharyngitis, to severe, such as bacteremia and abscesses in internal organs, with the three SAG species tending to be associated with different clinical syndromes. Abscesses caused by S. intermedius are more likely to spread hematogenously and be deep seated (4, 14, 15). On the other hand, S. anginosus is less likely to be implicated in abscess formation, but it predominates in blood cultures and is also commonly isolated from urogenital and gastrointestinal sources (4, 11). S. constellatus is often isolated from respiratory tract sources, although it can also cause other infections (4). However, most of the available literature has relied on phenotypic identifications, and none of the studies considered the recent taxonomy.
The changes in taxonomy and the heterogeneous phenotypic reactions exhibited by SAG have hampered their correct identification. Although the first set of phenotypic tests capable of distinguishing the three species was proposed in 1990 (16), rapid phenotypic tests, such as the Rapid ID32 Strep test, are usually found to have a low sensitivity, and it is often suggested that genotypic methods may be necessary to differentiate the taxa (10, 17).
Although the 16S rRNA gene is widely used in the identification of Streptococcus and other genera (2, 7, 18, 19), SAG show mosaic-like structures, confounding their identification and suggesting exchange of DNA between species (20). Other single-gene approaches have been used for SAG species identification (groEL, rpoB, tuf, and sodA) (10, 21–23), but recombination at these loci has also been documented (7, 21, 23), which could result in misidentification when a single-locus approach is used. A PCR based on the amplification of the group-specific penicillin-binding protein 2B gene (pbp2b) and a multiplex assay for identification to the subspecies level have been proposed (24), but the S. constellatus subsp. viborgensis taxon was not considered. Recently, the MLSA scheme proposed for the Streptococcus genus (25) proved to have great discriminatory power for SAG subspecies-level identification (7).
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been increasingly applied in clinical microbiology (26, 27). MALDI-TOF MS has a rapid turnaround time, low sample volume requirements, is cost-effective, and is not influenced by the growth medium (26–30). The size range used is dominated by ribosomal proteins (30, 31), supporting its use as the first-line method for bacterial species identification (26, 32).
In the present study, we used MLSA as a gold standard as we aimed to evaluate the accuracy of the Rapid ID32 Strep test, MALDI-TOF MS, and the previously described PCR scheme (24) in identifying SAG to the species and subspecies levels according to the most recent taxonomy. We analyzed potential associations between the different taxa and patient age, source for sample isolation, and antimicrobial susceptibility profile.
MATERIALS AND METHODS
Bacterial isolates.
Since 2000, the Portuguese Group for the Study of Streptococcal Infections has monitored streptococcal infections in Portugal. Among our activities, our laboratory has occasionally received isolates belonging to the Streptococcus genus from human infections for confirmation of identification. All isolates recovered in our hospital or received from participating laboratories that fulfilled at least one of the following criteria were considered for inclusion in our study: (i) presented colony morphology characteristics compatible with SAG, (ii) were identified by us or the submitting laboratory as belonging to the S. milleri or S. anginosus groups, or (iii) were nonidentified streptococci presenting Lancefield group A, C, G, or F. All blood and pleural fluid isolates (n = 76) and a random sample of 135 isolates recovered from other sources were included in the study. With the aim of increasing the prevalence of the newly described taxa that are thought to be associated with pharyngitis, all other noninvasive isolates recovered from the respiratory tract were also included (n = 23). The final collection included 234 isolates causing human infections, mostly recovered from our hospital but also isolates submitted by 14 other hospital-associated clinical microbiology laboratories. A screening for the pbp2b gene specific for the S. anginosus group (24) was performed with the 234 isolates. A final sample of 212 isolates that amplified the expected fragment of the pbp2b gene was retained for further study, including 64 blood isolates, 11 pleural fluid isolates, and 137 isolates recovered from other sources.
MLSA.
Species and subspecies were assigned based on MLSA, using previously described primers (25). The genes used were map, pfl, ppaC, pyk, rpoB, sodA, and tuf. Since the sodA primer pair occasionally resulted in nonspecific products, it was replaced by the one described by Poyart et al. (22). Sequences of the seven housekeeping genes were edited, aligned, and subjected to phylogenetic analysis using Bionumerics version 7.1 (Applied-Maths, Sint-Martens-Latem, Belgium) and MEGA 6.06 (33). All sequences available in GenBank from the previous study by Jensen et al. (7) and our own sequences were aligned, and the various alleles of each gene were identified by sequential numbers. The different allelic profiles were classified into sequence types (STs) that were given unique numbers. The sequences of each gene as well as the concatenated sequences were subjected to phylogenetic analysis using the minimum evolution (ME) algorithm and a Kimura two-parameter model to compute evolutionary distances. Bootstrap tests were performed with 1,000 replicates for ME trees by using MEGA 6.06 (33). The classifications of the study isolates into taxa were based on their positions in the tree.
The following type strains of the six SAG taxa were used as references: S. anginosus subsp. anginosus ATCC 33397T (DSM20563), S. anginosus subsp. whileyi DSM25818T, S. intermedius ATCC 27335T (DSM20573), S. constellatus subsp. constellatus ATCC 27823T (DSM20575), S. constellatus subsp. pharyngis DSM17475T, and S. constellatus subsp. viborgensis DSM25819T.
Phenotypic characterization and identification.
The isolates were grown on tryptic soy agar (TSA) plates (Oxoid, Hampshire, United Kingdom) supplemented with 5% sheep blood (ProBiológica, Belas, Portugal) and incubated at 37°C under a 5% CO2 atmosphere. After 16 to 24 h of incubation, isolates were classified as beta-hemolytic when complete clearing was seen around the colonies or non-beta-hemolytic (for alpha-hemolytic or nonhemolytic isolates). The Lancefield antigen group was determined by latex agglutination with the streptococcal grouping kit (Oxoid, Hampshire, United Kingdom).
The Rapid ID32 Strep system (bioMérieux, Marcy l'Etoile, France) was used for colonies grown for 48 h under anaerobiosis in Columbia sheep blood agar plates (CBA; Oxoid, Hampshire, United Kingdom) according to the manufacturer's recommendations. For species and subspecies identifications, the API profiles were interpreted using the mini-API computer system (bioMérieux, Marcy l'Etoile, France) with database version 3.0.
Identification by PCR and key substrate degradation capacity.
The multiplex PCR scheme proposed by Takao et al. was used for identification to the species and subspecies levels, with the S. anginosus subsp. whileyi isolates identified based on the amplification pattern described for DNA group 2 strains (24). Since this PCR scheme does not distinguish between S. constellatus subsp. viborgensis and S. constellatus subsp. pharyngis, all isolates identified as “S. constellatus subsp. pharyngis/viborgensis” were tested for their ability to degrade the synthetic fluorogenic substrates 4-methylumbelliferyl-β-d-galactopyranoside (β-Gal) and 4-methylumbelliferyl-N-acetyl-β-d-galactosaminide (β-NAGA) for the detection of β-N-acetylgalactosidase and β-N-acetylgalactosaminidase production, respectively (17). A positive reaction for both enzymes identified the isolate as S. constellatus subsp. pharyngis. If both reactions were negative, the isolates were classified as S. constellatus subsp. viborgensis.
MALDI-TOF MS identifications.
All isolates were analyzed by a direct colony method with MALDI-TOF MS (microflex system; Bruker Corporation, Germany). The analysis was carried out with axenic cultures grown under the same conditions used for the Rapid ID32 Strep test. Spectral analysis was performed with the MALDI Biotyper software (version 3.0), and comparisons with the spectra in the database (version 3.3.1.1) produced the final identifications. The manufacturer's recommended score values were used to interpret the results. Identification scores of <1.700 were considered unreliable and the experiment was repeated, again using a direct colony method. If a similar score was obtained, this was registered as an unreliable identification. For SAG, five strains are included in the reference database: two S. anginosus subsp. anginosus strains (DSM20563T and 0807M10067501 IBS) and the type strains of S. intermedius (DSM20573T), S. constellatus subsp. constellatus (DSM20575T) and S. constellatus subsp. pharyngis (DSM20575T).
Antimicrobial susceptibility.
Susceptibilities to cefotaxime, ceftriaxone, chloramphenicol, clindamycin, erythromycin, levofloxacin, linezolid, penicillin, quinupristin-dalfopristin, tetracycline, and vancomycin were tested by disk diffusion (Oxoid, Hampshire, United Kingdom) according to the CLSI recommendations (34). High-level resistance to gentamicin and streptomycin was tested according to the CLSI recommendations for enterococci (34) but using Mueller-Hinton agar supplemented with 5% sheep's blood and with incubation under a 5% CO2 atmosphere. For all antimicrobials except erythromycin, clindamycin, and tetracycline, any isolate classified as intermediate or resistant was confirmed by determining the MIC using Etest strips (bioMérieux, Marcy l'Etoile, France).
Statistical analysis.
Differences and associations were evaluated by the Fisher exact test with the false discovery rate (FDR) correction for multiple testing (35). A P value of <0.05 was considered significant for all tests. The diversity of classifications was quantitatively evaluated by calculating the Simpson's index of diversity (SID) with 95% confidence intervals (CIs) (36), using the Comparing Partitions website (http://www.comparingpartitions.info).
Nucleotide sequence accession numbers.
The nucleotide sequences were submitted to GenBank and are available under accession no. KT209590 to KT209960.
RESULTS
MLSA analysis and identification.
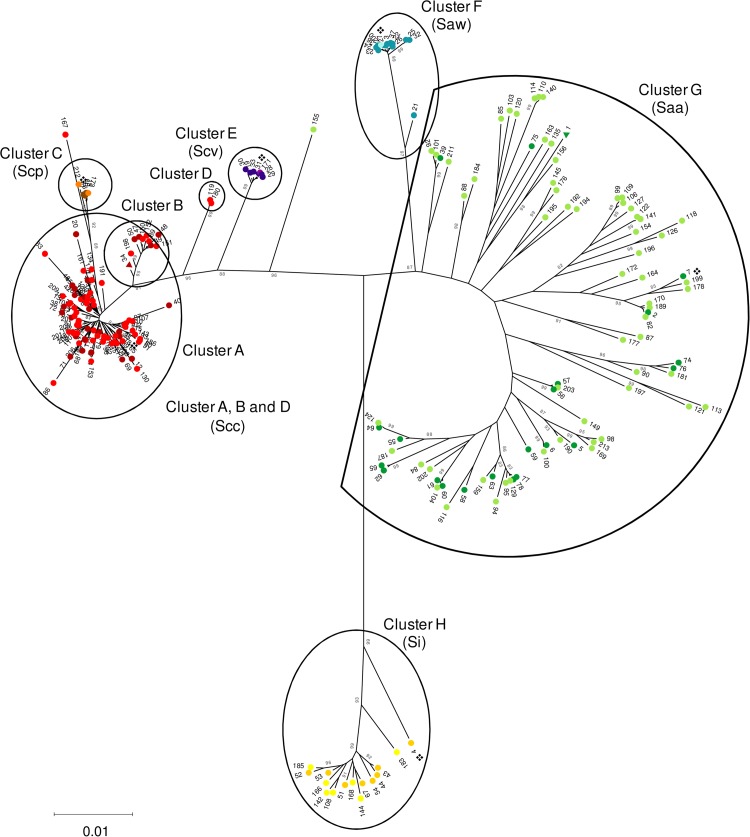
Seventy-eight STs were found among the previously published strains, including the type strains (see Table S1 in the supplemental material). In our collection (n = 212), 145 STs were present (SID and 95% CI, 0.992 ± 0.004), of which 135 were novel STs (ST79 to ST213) (see Table S2 in the supplemental material). The seven genes presented distinct diversities (SID and 95% CI: map, 0.891 ± 0.025; pfl, 0.925 ± 0.015; ppac, 0.970 ± 0.006; pyk, 0.928 ± 0.016, rpoB, 0.946 ± 0.014, sodA, 0.961 ± 0.01, tuf, 0.886 ± 0.021). Phylogenetic trees of individual genes showed little congruence (see Fig. S1a to g in the supplemental material). The ME tree, based on the concatenated sequences of the seven genes for the 213 STs (SID and 95% CI, 0.995 ± 0.003) is shown in Fig. 1. Eight clusters and two outlier isolates (ST167 and ST155) were identified. Clusters A and B were considered representative of S. constellatus subsp. constellatus, since both included solely strains previously identified as S. constellatus subsp. constellatus and no other taxon. Together, they included 72 isolates of the study collection. Cluster C, although not so well defined and branching from within cluster A, grouped together all strains previously identified as S. constellatus subsp. pharyngis, including the type strain, and two study isolates with novel and closely related STs (ST173 and ST212). S. constellatus subsp. viborgensis is represented by cluster E, including six isolates from our collection, five of which represented ST31, the same as the reference strain, and other isolates previously described as S. constellatus subsp. viborgensis (7). The species S. anginosus is represented by clusters F and G. Cluster F represents S. anginosus subsp. whileyi, a very cohesive cluster, in which five study isolates were grouped, four of which represented the same ST as the reference strain. Cluster G includes all sequences previously associated with S. anginosus subsp. anginosus, and we identified 115 isolates of our collection as members of this taxon. Finally, S. intermedius formed the most genetically distant cluster in the tree (cluster H) and included seven study isolates.
FIG 1.
MLSA minimum evolution tree of the concatenated sequences of seven genes defining 213 STs. The Kimura two-parameter method was used to compute evolutionary distances, and branch support was evaluated by bootstrapping with 1,000 replicates (values shown are percentages). Only bootstrap values of >80% are shown. The 213 distinct profiles comprise the different allelic profiles found in the present study and on sequences available in GenBank (7). The eight clusters formed are identified by letters. The isolates were identified according to their position on the tree. For each taxon, two shades of a color are used: the light shade indicates the STs found only among isolates of the present study, and the darker shade indicates STs found first among the isolates characterized previously (7). S. constellatus subsp. constellatus (Scc) is represented by light and dark red (cluster A, B, and D), S. constellatus subsp. pharyngis (Scp) is represented by orange and brown (cluster C), S. constellatus subsp. viborgensis (Scv) is represented by light and dark purple (cluster E), S. anginosus subsp. whileyi (Saw) is represented by light and dark blue (cluster F), S. anginosus subsp. anginosus (Saa) is represented by light and dark green (cluster G), and S. intermedius (Si) is presented by light and dark yellow (cluster H). STs found among isolates present in both collections are represented by triangles of the darker color. STs found in strains assigned to S. anginosus genomosubspecies AJ1 were not distinguished by a particular color (ST1, ST39, and ST75), since the taxon is currently not recognized. The ❖ symbols indicate the type strains.
The position in the tree raised some questions about the identity of cluster D, formed by ST119 (n = 1) and ST180 (n = 2). When each gene was analyzed individually (see Fig. S1a to g in the supplemental material), most of the alleles of these three isolates grouped together with other S. constellatus subsp. constellatus alleles. However, allele 53 of the sodA gene present in these STs seems to be responsible for its detachment from clusters A and B, since sodA53 groups together with the S. anginosus alleles in the ME tree (see Fig. S1f). Thus, we identified isolates that were grouped in cluster D as S. constellatus subsp. constellatus, although these isolates probably reflect the acquisition of a sodA allele from S. anginosus.
Finally, two STs, ST167 and ST155, were so divergent from any cluster that initially their identification was considered uncertain. In the case of ST167, the map and tuf genes were not informative, whereas the pyk and sodA alleles were shared by different subspecies of the S. constellatus species. The alleles of the genes pfl and rpoB indicated that this isolate belongs to S. constellatus subsp. constellatus. This isolate contains the ppac76 allele, which is not present elsewhere. This allele groups together with the S. anginosus subsp. anginosus alleles based on ME and may cause a long-branch attraction effect that results in the grouping of this isolate with the S. constellatus subsp. pharyngis isolates. The allele closest to ppac76 of S. constellatus subsp. constellatus is 90% identical, whereas the closest allele of S. anginosus subsp. anginosus is 98% identical. Overall, we classified this isolate as S. constellatus subsp. constellatus. In contrast, we classified the isolate representing ST155 as S. anginosus subsp. anginosus. As before, the map and tuf genes were not informative. The alleles found in the pfl, rpoB, and sodA genes of this isolate were also found in 10, 14, and 2 isolates, respectively, that were grouped into the S. anginosus subsp. anginosus cluster. Although the allele found for the ppac gene is unique, it groups by ME with other S. anginosus subsp. anginosus isolates. The allele of the pyk gene may have been acquired by recombination, since it is an allele characteristic of the S. constellatus species (found in 14 study isolates). The prevalence of each taxon among the collection studied is presented in Table 1.
TABLE 1.
Distribution of the 212 S. anginosus group clinical isolates studied, by patient age group and source
| Basis of comparison | No. of isolates |
||||||
|---|---|---|---|---|---|---|---|
| S. anginosus subsp. anginosus | S. anginosus subsp. whileyi | S. constellatus subsp. constellatus | S. constellatus subsp. pharyngis | S. constellatus subsp. viborgensis | S. intermedius | Total | |
| Age group (yr) | |||||||
| 0–18 | 11 | 1 | 14 | 1 | 2 | 0 | 29 |
| 19–65 | 66 | 4 | 37 | 1 | 4 | 3 | 115 |
| >65 | 39 | 0 | 25 | 0 | 0 | 4 | 68 |
| Source | |||||||
| Invasive | |||||||
| Blood | 31 | 2 | 25 | 0 | 1 | 5 | 64 |
| Peritoneal fluid | 36 | 0 | 17 | 0 | 1 | 0 | 54 |
| Pleural fluid | 4 | 0 | 6 | 0 | 0 | 1 | 11 |
| SSTIa | 11 | 0 | 7 | 0 | 0 | 1 | 19 |
| Otherb | 3 | 0 | 2 | 0 | 0 | 0 | 5 |
| Total | 85 | 2 | 57 | 0 | 2 | 7 | 153 |
| Noninvasive | |||||||
| Respiratory tract sourcesc | 7 | 2 | 4 | 1 | 3 | 0 | 17 |
| SSTIa | 6 | 0 | 3 | 0 | 0 | 0 | 9 |
| Otherb | 3 | 0 | 4 | 0 | 1 | 0 | 8 |
| Total | 31 | 3 | 19 | 2 | 4 | 0 | 59 |
| Pus (unknown origin) | 15 | 1 | 8 | 1 | 0 | 0 | 25 |
| Total | 116 | 5 | 76 | 2 | 6 | 7 | 212 |
SSTI, skin and soft tissue infections. Sources included pus from phlegmons, Ludwig's anginas, cervical abscesses, internal organ abscesses, surgical wounds, and collected limb abscesses.
Including urine, auricular and ocular exudates, and pus from thoracic abscesses, pelvic abscesses, bone biopsy specimens, and draining abscesses.
Including pharyngeal exudates, sputum, and bronchoalveolar lavage fluid.
Lancefield group, hemolysis, and PCR identification.
The hemolysis and Lancefield group carbohydrate findings for the isolates are given in Table 2. The PCR identification results were in agreement with the MLSA final identifications, including the identification of ST155, ST167, and that of cluster D, discussed above. The six S. constellatus subsp. viborgensis and the two S. constellatus subsp. pharyngis isolates showed identical amplification profiles, as expected (24). However, only the S. constellatus subsp. pharyngis degraded β-Gal and β-NAGA, allowing the correct identification of isolates of both subspecies.
TABLE 2.
Lancefield group and hemolysis group of the 212 S. anginosus group isolates studied
| Taxon and hemolysis group | No. of isolates in Lancefield group: |
Total isolates | ||||
|---|---|---|---|---|---|---|
| A | C | F | G | Absenta | ||
| S. anginosus subsp. anginosus | 11b | 18 | 49b | 16b | 22c | 116 |
| Beta-hemolytic | 1 | 8 | 9 | |||
| Non-beta-hemolyticc | 11 | 18 | 48 | 8 | 22 | 107b |
| S. anginosus subsp. whileyi | 5b | 5 | ||||
| Beta-hemolytic | 5 | 5b | ||||
| S. constellatus subsp. constellatus | 1d | 24 | 51b | 76 | ||
| Beta-hemolytic | 22 | 4 | 26b | |||
| Non-beta-hemolytice | 2 | 48 | 50 | |||
| S. constellatus subsp. pharyngis | 2b | 2 | ||||
| Beta-hemolytic | 2 | 2 | ||||
| S. constellatus subsp. viborgensis | 6b | 6 | ||||
| Beta-hemolytic | 6 | 6b | ||||
| S. intermedius | 7b | 7 | ||||
| Non-beta-hemolyticf | 7 | 7 | ||||
| Total | 11 | 32 | 73 | 16 | 80 | 212 |
The isolate did not react with any of the Lancefield sera tested.
Positive statistically supported association with the characteristic after FDR correction.
Alpha-hemolytic (n = 46) or nonhemolytic (n = 61).
Negative statistically supported association with the characteristic after FDR correction.
Alpha-hemolytic (n = 5) or nonhemolytic (n = 45).
Alpha-hemolytic (n = 1) or nonhemolytic (n = 6).
Identification via MALDI-TOF MS.
Table 3 summarizes the MALDI-TOF MS identifications according to the Bruker score system. Out of 212 isolates, 7 (3.3%) did not reach a score of ≥1.700, and so a reliable identification was not possible, even with repeated testing. All S. constellatus subsp. constellatus isolates (n = 76; 35.8% of total isolates) were identified as S. constellatus subsp. pharyngis, independently of their Bruker score, and were responsible for the high frequency of misidentifications to the subspecies level. S. anginosus subsp. whileyi and S. constellatus subsp. viborgensis were not represented in the database and therefore were identified to the closest taxon present in the database, as they were correctly identified to the species level. Overall, 68.4% of isolates (n = 145) were correctly assigned to the species level (ID score, ≥2.000), a value that increased to 92.5% (n = 196) if identifications with a score of ≥1.700 were considered. All isolates misidentified to the species level were assigned to another SAG species, and one isolate was assigned to a different genus (Table 3).
TABLE 3.
MLSA and MALDI-TOF MS identification results for the 212 isolates studied
| Taxon | No. of isolates with potential identification (Bruker score) to level ofa: |
Total isolates | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Unreliable (<1.700) (n = 7) | Genus (1.700–1.999) (n = 59) |
Species (≥2.000) (n = 146) |
||||||||
| Correct | Incorrect |
Correct | Incorrect |
|||||||
| Subspecies | Species | Genus | Subspecies | Species | Genus | |||||
| S. anginosus subsp. anginosusb | 4 | 17 | 5c | 1d | 88 | 1c | 116 | |||
| S. anginosus subsp. whileyib | 1 | 2e | 2e | 5 | ||||||
| S. constellatus subsp. constellatus | 1 | 26c | 49c | 76 | ||||||
| S. constellatus subsp. pharyngis | 1 | 1 | 2 | |||||||
| S. constellatus subsp. viborgensisf | 1c | 5c | 6 | |||||||
| S. intermedius | 1 | 4 | 2g | 7 | ||||||
| Total | 7 | 22 | 29 | 7 | 1 | 89 | 56 | 1 | 212 | |
IDs are based on the Bruker score system: 0.000 to 1.699, no reliable identification; 1.700 to 1.999, probable genus identification; 2.000 to 2.299, secure genus identification, probable species identification; 2.300 to 3.000, highly probable species identification.
The MALDI-TOF MS system only identifies strains as S. anginosus. Since the two spectra found in the database included the type strain of S. anginosus subsp. anginosus (DSM20563T), we considered a MALDI-TOF MS identification as S. anginosus corresponded to the subspecies S. anginosus subsp. anginosus.
Isolates identified as S. constellatus subsp. pharyngis.
Isolate identified as Clostridium halophilum.
Isolates identified as S. anginosus subsp. anginosus.
No strains of S. constellatus subsp. viborgensis were included in the spectra database; thus, only species-level identification was expected to be achieved.
Isolates identified as S. constellatus subsp. constellatus (n = 1) or S. constellatus subsp. pharyngis (n = 1).
Rapid ID32 Strep identification.
The results with the Rapid ID32 Strep test are shown in Table 4. The analysis of the performance of the Rapid ID32 Strep test in identification to the subspecies level could not be conducted because the system does not consider S. anginosus subspecies nor the newly recognized S. constellatus subsp. viborgensis taxon. Even if, according to the manufacturer, the Rapid ID32 Strep system can identify S. constellatus subsp. pharyngis, all the S. constellatus subsp. pharyngis isolates were incorrectly identified at the subspecies and species levels (Table 4). Only 40.0% and 16.7% of the isolates representing S. anginosus subsp. whileyi and S. constellatus subsp. viborgensis, respectively, were correctly identified to the species level. Approximately 1/10 (10.4%) of the isolates were identified as belonging to a different genus, and a minority (3.8%) were identified as belonging to streptococcal species other than SAG.
TABLE 4.
Rapid ID32 Strep system versus MLSA identification results for the 212 isolates studied
| MLSA ID | Rapid ID32 Strep system ID |
|||||
|---|---|---|---|---|---|---|
| S. anginosus | S. constellatus subsp. constellatus | S. intermedius | Other streptococcal speciesa | Other genusb | Total | |
| S. anginosus subsp. anginosus | 81 | 31 | 1 | 3 | 116 | |
| S. anginosus subsp. whileyic | 2 | 3 | 5 | |||
| S. constellatus subsp. constellatus | 2 | 57 | 5 | 12 | 76 | |
| S. constellatus subsp. pharyngisd | 1 | 1 | 2 | |||
| S. constellatus subsp. viborgensisc | 3 | 1 | 2 | 6 | ||
| S. intermedius | 1 | 3 | 2 | 1 | 7 | |
| Total | 90 | 89 | 3 | 8 | 22 | 212 |
Other Streptococcus species identified were S. oralis (n = 1), S. oralis 2 (n = 1), S. mitis (n = 3), S. gordonii (n = 1), S. suis (n = 1), and S. thermophilus (n = 1).
Other genera identified were Lactococcus lactis subsp. cremoris (n = 19), Erysipelothrix rhusiopathiae (n = 2), and Gemella morbillorum (n = 1).
S. anginosus subsp. whileyi and S. constellatus subsp. viborgensis were only described in 2013 (7), and their profiles were not included in the database.
An S. constellatus subsp. pharyngis profile was included in the database, according to the manufacturer's manual; however, no isolate was assigned that final identification.
Antimicrobial susceptibility.
All isolates were susceptible to cefotaxime, ceftriaxone, linezolid, penicillin, quinupristin-dalfopristin, and vancomycin, and no high-level aminoglycoside resistance was detected. One isolate presented intermediate resistance to chloramphenicol, and another was resistant to levofloxacin. Nonsusceptibility simultaneously to clindamycin and erythromycin was found in 10.4% of isolates, with one isolate presenting the iMLSB phenotype and all others the cMLSB phenotype. Most of the clindamycin- and erythromycin-nonsusceptible isolates (n = 22) were S. anginosus subsp. anginosus (n = 17), corresponding to 14.7% of the taxon. Among both S. constellatus subsp. constellatus and S. intermedius isolates, there were also resistant isolates (5.3% and 14.3%, respectively). Overall, 21.7% (n = 46) of the isolates were nonsusceptible to tetracycline: 31.0% of S. anginosus subsp. anginosus, 7.9% of S. constellatus subsp. constellatus, 50% of S. constellatus subsp. viborgensis, and 14.3% of S. intermedius.
DISCUSSION
In this study, we aimed to evaluate the currently available techniques for the identification of SAG according to its most recent taxonomy. For that purpose, we used MLSA (25) as the gold standard, distinguishing the six currently recognized taxa based on the formation of clusters in an ME tree (7). The tree has three major branches, roughly corresponding to the three species that compose the group. Still, more than one monophyletic cluster was formed within both S. anginosus subsp. anginosus and S. constellatus subsp. constellatus. Two outlier STs were noted in the S. constellatus branch, representing an S. anginosus subsp. anginosus isolate and an S. constellatus subsp. constellatus isolate. The identities of these outliers were established by the analysis of individual genes and were concordant with the results of the PCR and MALDI-TOF MS analyses.
In agreement with previous literature, S. anginosus subsp. anginosus was the most heterogeneous taxon both genotypically, as evaluated by MLSA, and phenotypically (3, 4, 7, 8). The previously reported greater genetic distance of the S. intermedius cluster relative to the other two species was confirmed, as well as the unusual position of the type strain of the species in the cluster, since it has an rpoB allele (rpoB88) more closely related to S. anginosus (Fig. 1; see also Fig. S1e in the supplemental material) (7). All S. anginosus subsp. whileyi, S. constellatus subsp. pharyngis, and S. constellatus subsp. viborgensis isolates formed homogeneous clusters and presented the phenotypic characteristics previously attributed to these subspecies: beta-hemolysis and Lancefield group C antigen. Although S. anginosus subsp. whileyi, S. constellatus subsp. pharyngis, and S. constellatus subsp. viborgensis were found among upper respiratory sources, as indicated in the literature (6, 7), isolates of these subspecies were also found in other sources among our collection. The abundance of S. intermedius among isolates recovered from blood is in agreement with the literature, which indicates that S. intermedius is frequently responsible for deep-seated infections involving hematogenous spread (4, 11, 15). As previously described (4, 11), both S. anginosus subsp. anginosus and S. constellatus subsp. constellatus were found in a wide range of sources.
The Rapid ID32 Strep test should not be used as a routine identification method for SAG. For almost one-third of the isolates, based on Rapid ID32 Strep results the assigned species was incorrect, and 10.4% were assigned to a different genus. SAG is well-known for its intraspecies biochemical variability (3, 4), so misidentifications with enzymatic-based methods are not surprising.
Although some misidentifications were found when using MALDI-TOF MS, most of the discrepancies were minor. Score values of ≥2.000 are frequently considered necessary for reliable species-level identification, but even when considering scores in the range of 1.700 to 1.999 (n = 59), the majority were correct identifications to the species level (86.4%). As suggested previously for the identification of the viridans group streptococci, the application of a lower species-level cutoff score could be equally accurate and enable the identification of more isolates (29, 32, 37, 38). In fact, the proportion of correctly identified isolates to the species level in our study when we considered all scores of ≥1.700 (92.5%) was superior to that described previously for the viridans group streptococci (38) and S. anginosus group (29, 38–41). A previous study reported a similar fraction of correct species-level identifications (93.1%, considering scores of ≥2.000) and found that a high proportion of isolates required full extraction for reliable identification (39). Our data indicated that when considering a lower cutoff value (≥1.700), similar results can be obtained with a faster and cost-saving direct method. In agreement with previous reports (32, 39), MALDI-TOF MS accurately identified S. anginosus and S. constellatus to the species level, but S. intermedius was poorly identified. Although those two studies (32, 39), like ours, had only a few representatives of S. intermedius (n = 6 and n = 9, respectively), this suggests that there is a specific limitation of MALDI-TOF MS when used for the identification of this species.
All S. constellatus subsp. constellatus isolates were wrongly identified as S. constellatus subsp. pharyngis with good scores. Given that the S. constellatus subsp. pharyngis is found on a cluster branching from within S. constellatus subsp. constellatus (Fig. 1), this was not unexpected. In fact, a similar problem is found with the consistent misidentifications of other viridans group streptococci as Streptococcus pneumoniae (32, 41, 42). These observations may be justified by the great similarity of ribosomal proteins, which would limit the technique's resolution, or may reflect deficiencies in the current database (29, 31), since only one spectrum of each SAG taxon (except S. anginosus subsp. anginosus) was included. The spectra are subject to some method-inherent noise (30), with the same isolate rarely producing identical spectra (30), and microbial species generally show a degree of intraspecific variation (28, 30). Occasionally, type strains are not “typical” representatives of a species, as was seen with MLSA (Fig. 1), and may not be appropriate as the sole reference for identifying the subspecies (28). It has been suggested that to allow reliable identifications, each species should be represented by 10 or more strains in order to cover the naturally occurring diversity (26, 28, 41). In the case of SAG, the problem does not seem to be the correct identification of the more genetically diverse group (S. anginosus subsp. anginosus) but the distinction of the closely related taxa, and so it is unclear if including more strains in the database would solve the identification problems we found.
In our study, we evaluated for the first time the accuracy of the multiplex PCR designed by Takao et al. for SAG species-level identification (24), using MLSA results as the gold standard. The PCR method showed excellent concordance for all isolates, and the amplified fragments generated easily distinguishable bands in conventional agarose electrophoresis gels, allowing identification to the species level without sequencing and thus decreasing the time to identification and its associated costs. Also, contrary to all other tested methods, the PCR method enabled robust subspecies identification for S. anginosus and discrimination between S. constellatus subsp. constellatus and S. constellatus subsp. pharyngis/viborgensis. The current method failed to distinguish S. constellatus subsp. viborgensis from S. constellatus subsp. pharyngis, but further study may identify unique markers that could be included and specifically detected by the PCR, allowing their differentiation without further enzymatic tests. However, given the potentially small number of S. constellatus subsp. viborgensis isolates occurring in human infections, this is not a severe limitation. Such PCR procedures could be a promising rapid method for implementation in clinical microbiology laboratories, allowing full SAG identification to the species and subspecies levels without sequencing.
Although antimicrobial resistance is an increasing problem with many bacterial groups (43, 44), SAG remain susceptible to most antibiotic classes, as was observed in the present study. The only significant resistance found was to clindamycin, erythromycin, and tetracycline.
The collection analyzed was extremely diverse, with each subspecies represented by multiple STs. Still, among the subspecies represented by fewer isolates, two STs were dominant: ST31 in S. constellatus subsp. viborgensis (5 of 6 isolates) and ST27 in S. anginosus subsp. whileyi (4 of 5 isolates). In contrast, among the most frequent taxa, S. constellatus subsp. constellatus and S. anginosus subsp. anginosus, the most common STs, accounted for <12% of the isolates. It is not clear if this represents more limited diversity of S. anginosus subsp. whileyi and S. constellatus subsp. viborgensis or a lower sampling of the existing diversity. Nonetheless, it is clear that several genetic lineages within each taxon are capable of causing disease in humans.
Although SAG subspecies can be reliably distinguished by using molecular techniques, such as PCR or MLSA, it is unclear if there is a benefit in their routine identification in clinical microbiology laboratories. For instance, we found no statistically supported association between the subspecies and isolation site or with resistance to antimicrobials (data not shown). Moreover, our study suggested that increased sampling of natural populations uncovers groups that do not clearly cluster into the defined subspecies, an observation that questions the currently recognized subspecies boundaries. The predominance of S. constellatus subsp. constellatus and S. anginosus subsp. anginosus among SAG causing human infections, the uncertainties arising in the genetic boundaries of the different subspecies, and the excellent performance of MALDI-TOF MS in correctly identifying SAG species make MALDI-TOF MS a reliable method for use in routine identification.
Supplementary Material
ACKNOWLEDGMENTS
J.M.-C. has received research grants administered through his university and has received honoraria for serving on the speakers bureaus of Pfizer, Bial, GlaxoSmithKline, and Novartis. M.R. has received honoraria for consulting for GlaxoSmithKline and for serving on the speakers bureau of Pfizer. The other authors declare no conflicts of interest.
No company or financing body had any interference in the decision to publish this report.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01892-15.
REFERENCES
- 1.Guthof O. 1956. Pathogenic strains of Streptococcus viridans; streptococci found in dental abscesses and infiltrates in the region of the oral cavity. Zentralbl Bakteriol Orig 166:553–564. (In German.) [PubMed] [Google Scholar]
- 2.Clarridge JE, Osting C, Jalali M, Osborne J, Waddington M. 1999. Genotypic and phenotypic characterization of “Streptococcus milleri” group isolates from a Veterans Administration hospital population. J Clin Microbiol 37:3681–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whiley RA, Beighton D. 1991. Emended descriptions and recognition of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus anginosus as distinct species. Int J Syst Bacteriol 41:1–5. doi: 10.1099/00207713-41-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Facklam R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev 15:613–630. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson AB, Kent H, Sibley CD, Grinwis ME, Mabon P, Ouellette C, Tyson S, Graham M, Tyler SD, Van Domselaar G, Surette MG, Corbett CR. 2013. Phylogenetic relationship and virulence inference of Streptococcus anginosus group: curated annotation and whole-genome comparative analysis support distinct species designation. BMC Genomics 14:895. doi: 10.1186/1471-2164-14-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whiley RA, Hall LM, Hardie JM, Beighton D. 1999. A study of small-colony, beta-haemolytic, Lancefield group C streptococci within the anginosus group: description of Streptococcus constellatus subsp. pharyngis subsp. nov., associated with the human throat and pharyngitis. Int J Syst Bacteriol 49:1443–1449. doi: 10.1099/00207713-49-4-1443. [DOI] [PubMed] [Google Scholar]
- 7.Jensen A, Hoshino T, Kilian M. 2013. Taxonomy of the anginosus group of the genus Streptococcus and description of Streptococcus anginosus subsp. whileyi subsp. nov. and Streptococcus constellatus subsp. viborgensis subsp. nov. Int J Syst Evol Microbiol 63:2506–2519. doi: 10.1099/ijs.0.043232-0. [DOI] [PubMed] [Google Scholar]
- 8.Doern CD, Burnham C-AD. 2010. It's not easy being green: the viridans group streptococci, with a focus on pediatric clinical manifestations. J Clin Microbiol 48:3829–3835. doi: 10.1128/JCM.01563-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coykendall AL. 1989. Classification and identification of the viridans streptococci. Clin Microbiol Rev 2:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summanen PH, Rowlinson M-C, Wooton J, Finegold SM. 2009. Evaluation of genotypic and phenotypic methods for differentiation of the members of the anginosus group streptococci. Eur J Clin Microbiol Infect Dis 28:1123–1128. doi: 10.1007/s10096-009-0758-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claridge JE, Attorri S, Musher DM, Hebert J, Dunbar S. 2001. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (“Streptococcus milleri group”) are of different clinical importance and are not equally associated with abscess. Clin Infect Dis 32:1511–1515. doi: 10.1086/320163. [DOI] [PubMed] [Google Scholar]
- 12.Ledezma-Rasillo G, Flores-Reyes H, Gonzalez-Amaro AM, Garrocho-Rangel A, del Ruiz-Rodriguez M, Pozos-Guillen SAJ. 2010. Identification of cultivable microorganisms from primary teeth with necrotic pulps. J Clin Pediatr Dent 34:329–333. doi: 10.17796/jcpd.34.4.20124lu111544377. [DOI] [PubMed] [Google Scholar]
- 13.Okayama H, Nagata E, Ito H-O, Oho T, Inoue M. 2005. Experimental abscess formation caused by human dental plaque. Microbiol Immunol 49:399–405. doi: 10.1111/j.1348-0421.2005.tb03742.x. [DOI] [PubMed] [Google Scholar]
- 14.Livingston LV, Perez-Colon E. 2014. Streptococcus intermedius bacteremia and liver abscess following a routine dental cleaning. Case Rep Infect Dis 2014:954046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra AK, Fournier P-E. 2013. The role of Streptococcus intermedius in brain abscess. Eur J Clin Microbiol Infect Dis 32:477–483. doi: 10.1007/s10096-012-1782-8. [DOI] [PubMed] [Google Scholar]
- 16.Whiley RA, Fraser H, Hardie JM, Beighton D. 1990. Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the “Streptococcus milleri group.” J Clin Microbiol 28:1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limia A, Alarcón T, Jiménez ML, López-Brea M. 2000. Comparison of three methods for identification of Streptococcus milleri group isolates to species level. Eur J Clin Microbiol Infect Dis 19:128–131. doi: 10.1007/s100960050444. [DOI] [PubMed] [Google Scholar]
- 18.Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol 45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 19.Bizzini A, Jaton K, Romo D, Bille J, Prod'hom G, Greub G. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J Clin Microbiol 49:693–696. doi: 10.1128/JCM.01463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schouls LM, Schot CS, Jacobs JA. 2003. Horizontal transfer of segments of the 16S rRNA genes between species of the Streptococcus anginosus group. J Bacteriol 185:7241–7246. doi: 10.1128/JB.185.24.7241-7246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glazunova OO, Raoult D, Roux V. 2009. Partial sequence comparison of the rpoB, sodA, groEL and gyrB genes within the genus Streptococcus. Int J Syst Evol Microbiol 59:2317–2322. doi: 10.1099/ijs.0.005488-0. [DOI] [PubMed] [Google Scholar]
- 22.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol 36:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshino T, Fujiwara T, Kilian M. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J Clin Microbiol 43:6073–6085. doi: 10.1128/JCM.43.12.6073-6085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takao A, Nagamune H, Maeda N. 2004. Identification of the anginosus group within the genus Streptococcus using polymerase chain reaction. FEMS Microbiol Lett 233:83–89. doi: 10.1016/j.femsle.2004.01.042. [DOI] [PubMed] [Google Scholar]
- 25.Bishop CJ, Aanensen DM, Jordan GE, Kilian M, Hanage WP, Spratt BG. 2009. Assigning strains to bacterial species via the internet. BMC Biol 7:3. doi: 10.1186/1741-7007-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier P-E, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 27.Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect 16:1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 28.Welker M, Moore ERB. 2011. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol 34:2–11. doi: 10.1016/j.syapm.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Neville SA, Lecordier A, Ziochos H, Chater MJ, Gosbell IB, Maley MW, van Hal SJ. 2011. Utility of matrix-assisted laser desorption ionization–time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J Clin Microbiol 49:2980–2984. doi: 10.1128/JCM.00431-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wieser A, Schneider L, Jung J, Schubert S. 2012. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (minireview). Appl Microbiol Biotechnol 93:965–974. doi: 10.1007/s00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- 31.Emonet S, Shah HN, Cherkaoui A, Schrenzel J. 2010. Application and use of various mass spectrometry methods in clinical microbiology. Clin Microbiol Infect 16:1604–1613. doi: 10.1111/j.1469-0691.2010.03368.x. [DOI] [PubMed] [Google Scholar]
- 32.Kärpänoja P, Harju I, Rantakokko-Jalava K, Haanperä M, Sarkkinen H. 2014. Evaluation of two matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of viridans group streptococci. Eur J Clin Microbiol Infect Dis 33:779–788. doi: 10.1007/s10096-013-2012-8. [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis, version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. 2001. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 36.Carriço JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol 44:2524–2532. doi: 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of Gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 49:2868–2873. doi: 10.1128/JCM.00506-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.López Roa P, Sánchez Carrillo C, Marín M, Romero F, Cercenado E, Bouza E. 2013. Value of matrix-assisted laser desorption ionization-time of flight for routine identification of viridans group streptococci causing bloodstream infections. Clin Microbiol Infect 19:438–444. doi: 10.1111/j.1469-0691.2012.03837.x. [DOI] [PubMed] [Google Scholar]
- 39.Woods K, Beighton D, Klein JL. 2014. Identification of the “Streptococcus anginosus group” by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry. J Med Microbiol 63:1143–1147. doi: 10.1099/jmm.0.076653-0. [DOI] [PubMed] [Google Scholar]
- 40.Friedrichs C, Rodloff AC, Chhatwal GS, Schellenberger W, Eschrich K. 2007. Rapid identification of viridans streptococci by mass spectrometric discrimination. J Clin Microbiol 45:2392–2397. doi: 10.1128/JCM.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Veen SQ, Claas ECJ, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization–time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol 48:900–907. doi: 10.1128/JCM.02071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies AP, Reid M, Hadfield SJ, Johnston S, Mikhail J, Harris LG, Jenkinson HF, Berry N, Lewis AM, El-Bouri K, Mack D. 2012. Identification of clinical isolates of α-hemolytic streptococci by 16S rRNA gene sequencing, matrix-assisted laser desorption ionization–time of flight mass spectrometry using MALDI Biotyper, and conventional phenotypic methods: a comparison. J Clin Microbiol 50:4087–4090. doi: 10.1128/JCM.02387-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chun S, Huh HJ, Lee NY. 2015. Species-specific difference in antimicrobial susceptibility among viridans group streptococci. Ann Lab Med 35:205–211. doi: 10.3343/alm.2015.35.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uh Y, Shin DH, Jang IH, Hwang GY, Lee MK, Yoon KJ, Kim HY. 2004. Antimicrobial susceptibility patterns and macrolide resistance genes of viridans group streptococci from blood cultures in Korea. J Antimicrob Chemother 53:1095–1097. doi: 10.1093/jac/dkh219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.