Abstract
Members of the Mycobacterium abscessus group (MAG) cause lung, soft tissue, and disseminated infections. The oral macrolides clarithromycin and azithromycin are commonly used for treatment. MAG can display clarithromycin resistance through the inducible erm(41) gene or via acquired mutations in the rrl (23S rRNA) gene. Strains harboring a truncation or a T28C substitution in erm(41) lose the inducible resistance trait. Phenotypic detection of clarithromycin resistance requires extended incubation (14 days), highlighting the need for faster methods to detect resistance. Two real-time PCR-based assays were developed to assess inducible and acquired clarithromycin resistance and tested on a total of 90 clinical and reference strains. A SYBR green assay was designed to distinguish between a full-length and truncated erm(41) gene by temperature shift in melting curve analysis. Single nucleotide polymorphism (SNP) allele discrimination assays were developed to distinguish T or C at position 28 of erm(41) and 23S rRNA rrl gene mutations at position 2058 and/or 2059. Truncated and full-size erm(41) genes were detected in 21/90 and 69/90 strains, respectively, with 64/69 displaying T at nucleotide position 28 and 5/69 containing C at that position. Fifteen isolates showed rrl mutations conferring clarithromycin resistance, including A2058G (11 isolates), A2058C (3 isolates), and A2059G (1 isolate). Targeted sequencing and phenotypic assessment of resistance concurred with molecular assay results. Interestingly, we also noted cooccurring strains harboring an active erm(41), inactive erm(41), and/or acquired mutational resistance, as well as slowly growing MAG strains and also strains displaying an inducible resistance phenotype within 5 days, long before the recommended 14-day extended incubation.
INTRODUCTION
Rapidly growing mycobacteria (RGM) are important emerging human pathogens. Three closely related taxa of RGM with a controversial species/subspecies status—i.e., Mycobacterium abscessus subsp. abscessus (here M. abscessus), M. abscessus subsp. massiliense (M. massiliense), and M. abscessus subsp. bolletii (M. bolletii)—comprise the so-called “M. abscessus group” (MAG) (1–3). MAG is commonly associated with chronic lung infections in susceptible hosts, such as cystic fibrosis (CF) (4), as well as wound infection and postsurgical site infections (5, 6). Macrolides such as clarithromycin and azithromycin are frequently the only oral antibiotics that are active against MAG and are commonly used to treat pulmonary infections (7, 8). Members of the MAG differ in their susceptibility to clarithromycin. M. abscessus and M. bolletii, commonly display inducible macrolide resistance conferred by the ribosomal methyl transferase erm(41) gene (9). In contrast, most M. massiliense strains do not show inducible resistance due to a large, 274-bp deletion in the ribosomal methyl transferase erm(41) gene that renders it nonfunctional (9, 10), and clinical studies have shown a better response to clarithromycin in M. massiliense compared to M. abscessus (8, 10). Besides this major truncation, erm(41) can lose functionality by a T-to-C substitution at position 28 (T28C) yielding a Trp-to-Arg amino acid change (9, 11). The second mechanism of clarithromycin resistance is acquired through mutations in the drug-binding pocket of the 23S rRNA rrl gene at nucleotide positions 2058 and 2059 (12–15, 27). Phenotypic detection of clarithromycin resistance requires incubation of MAG with clarithromycin for up to 14 days. While MAG strains displaying acquired resistance show high MICs to clarithromycin after 3 to 5 days, those with an inducible active erm(41) gene typically show low MICs at 3 to 5 days and require longer incubation times (up to 14 days) for induction of resistance (16).
Targeted sequencing is routinely used in the clinical laboratory to determine the intraspecies genetic diversity in mycobacterial isolates and discriminate among closely related taxa. Multiple genes, including rpoB, hsp65, secA, and others (17–22), as well as PCR-based assays (10, 23) have been evaluated as tools to discriminate between the closely related subspecies of the MAG. Since a truncated erm(41) gene was described as a hallmark of M. massiliense, size differences in PCR-amplified erm(41) PCR products were proposed as a simple method to differentiate M. massiliense from M. abscessus and M. bolletii (10). However, this method can misclassify some strains, such as two recently described M. massiliense strains harboring a full-length and functional erm(41) gene (23).
Since standard susceptibility testing of clarithromycin requires up to 14 days, there is a need for faster methods to detect clarithromycin resistance. The present study describes the development and evaluation of two novel rapid real-time PCR assays for assessment of clarithromycin resistance on 87 clinical and 3 reference MAG strains. The first assay discriminates between full-length and truncated erm(41) based on differences in the melting temperature (Tm) of amplified products. The second assay identifies mutations associated with clarithromycin susceptibility and resistance using fluorescent locked nucleic acid (LNA) probes, including single nucleotide polymorphisms (SNPs) in the 23S rRNA rrl A2058G/C and A2058G positions (Escherichia coli rrl numbering) and T/C at the position 28 of the erm(41) gene. Our assays decreased the turnaround time of detection of resistance from 14 days to ∼3 h, and results were concordant with targeted sequencing and phenotypic testing. This assay is feasible to use in laboratories set up to perform molecular tests and can help overcome technical challenges of determining MICs in slowly growing strains.
(This work was presented in part at the 115th General Meeting of the American Society for Microbiology, New Orleans, LA, 30 May to 2 June 2015 [poster 2003].)
MATERIALS AND METHODS
DNA isolation and identification of clinical isolates.
Eighty-seven clinical isolates belonging to the M. abscessus group were obtained between 2005 and 2015 from sputum, bronchoalveolar lavage fluid, blood cultures, skin, or lymph node from patients with bronchiectasis, cystic fibrosis, and interleukin-21 receptor gene primary immunodeficiency. In addition, 3 reference strains were used, for a total of 90 isolates: M. abscessus ATCC 19977T, M. massiliense CCUG 48898T, and M. bolletii CCUG 50184T. The bacterial strains were stored at −80°C in Tween albumin broth (Remel, Thermo Scientific, Lenexa, KS). Prior to use, the organisms were subcultured onto Middlebrook 7H11 agar (Remel, Lenexa, KS). DNA was extracted from a 10-μl loopful of each mycobacterial colony with the Ultra Clean microbial DNA isolation kit (MoBio Laboratories, Solana Beach, CA), according to the manufacturer's instructions. Most clinical isolates before 2011 were previously identified by multilocus genomic sequencing with secA1, rpoB, and hsp65 (22). More recent isolates were identified as MAG by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) followed by subspecies-level identification either by multilocus genomic sequencing (22) or a recently developed PCR method (23). All primers (Table 1) and probes (Table 2) in this study were purchased from IDT (Integrated DNA Technologies, Coralville, IA).
TABLE 1.
Primers used in the full and truncated erm(41) gene discrimination assay, single nucleotide polymorphism, and sequencing of the erm(41) and 23S rRNA rrl genes of the M. abscessus group
| Primer | Sequence | Purpose | Gene ID no. | Product size (bp) |
|---|---|---|---|---|
| erm225 Forward | 5′-GGCACATCTGGTTGCCGC-3′ | SYBR green erm(41) real-time assay | MAB_2297 M. abscessus | 321 or 47 |
| erm546Rev_RC Reverse | 5′-CGGCAATGTGATTCCGGCC-3′ | |||
| MycoSNP_Fex Forward | 5′-GAGCATGGGCATATTCATGATGG-3′ | erm(41) position 28 SNP real-time assay | MAB_2297 M. abscessus | 125 |
| MycoSNP_Rex Reverse | 5′-TGAGCGAACACCGGATTCG-3′ | |||
| 23S_SNPF Forward | 5′-GCGAAATTGCACTACGAGTAAAG-3′ | 23S rRNA position 2058/2059 SNP real-time assay | MAB_5052 M. abscessus | 110 |
| 23S_SNPR Reverse | 5′-CCTATCCTACACAAACCGAACC-3′ | |||
| ermF Forward | 5′-GACCGGGGCCTTCTTCGTGAT-3′ | erm(41) sequencing | MAB_2297 M. abscessus | 672 or 396 |
| ermR1 Reverse | 5′-GACTTCCCCGCACCGATTCC-3′ | |||
| 23S_18 Forward | 5′-AGTCGGGACCTAAGGCGAG-3′ | 23S rRNA sequencing | MAB_5052 M. abscessus | 1,525 |
| 23S_21 Reverse | 5′-TTCCCGCTTAGATGCTTTCAG-3′ | |||
| 23SrRNAF_207 Forward | 5′-AGCGAAATTCCTTGTCGGGT-3′ | 23S rRNA sequencing | MAB_5052 M. abscessus | 207 |
| 23SrRNAR_207 Reverse | 5′-CTGCTTCACAGTCTCCCACC-3′ |
TABLE 2.
SNP detection probes of the 23S rRNA rrl gene 2058A-to-G or -C and 2059A-to-G mutation and the erm(41) gene 28T-to-C mutation
| Probe | Gene | Nucleotide position(s) | Base change | LNA probe sequencea | Tm (°C) |
|---|---|---|---|---|---|
| WT_MAB | 23S rRNA | 2058–2059 | AA to AA | 5′-HEX-ACG+A+A+A+AGA+C+CC-IABkFQ-3′ | 65.1 |
| AA_2058G | 23S rRNA | 2058 | AA to GA | 5′-Cy5-ACG+A+G+A+AGA+CCC-IABkFQ-3′ | 68.2 |
| AA_2058C | 23S rRNA | 2058 | AA to CA | 5′-6FAM-ACG+A+C+A+AGA+CCC-IABkFQ-3′ | 66.8 |
| AA_2059G | 23S rRNA | 2059 | AA to AG | 5′-6FAM-ACG+A+A+G+AGA+CCC-IABkFQ-3′ | 68.2 |
| C Allele | erm(41) | 28 | T to C | 5′-6FAM-CCA+G+C+GGGGC/IABkFQ-3′ | 67.4 |
| T Allele | erm(41) | 28 | T to T | 5′-HEX-CCA+G+T+GGGGC-IABkFQ-3′ | 69.3 |
HEX, hexachlorofluorescein; 6FAM, 6-carboxyfluorescein; IABkFQ, Iowa Black fluorescent quencher.
Real-time SYBR green assay to discriminate between full-length and truncated erm(41) genes.
Primers erm225 and erm546Rev_RC (Table 1) were designed using the M. abscessus ATCC 19977T genome erm(41) gene (MAB_2297), spanning the 274-bp region, which is truncated (nonfunctional) in most M. massiliense strains. The SYBR green assay was carried out in a 20-μl reaction mixture, which consisted of 1 μl of the forward and reverse primers (10 μM stock) at a final concentration of 500 nm each and 2 to 3 μl of genomic DNA (30 to 50 ng), nuclease-free water (5 to 6 μl), and 10 μl of the 2× Fast SYBR green master mix (Thermo Fisher Scientific, Life Technologies, Grand Island, NY). The cycling conditions were as follows: a pre-PCR step of 95°C for 20 s followed by 40 cycles of 95°C for 10 s, 60°C for 30 s and 72°C for 30 s. This was followed by a melt curve step of 2 cycles of 95°C for 15 s and 60°C for 1 min in a 96-well PCR plate and processing on a 7500 Fast thermocycler (Thermo Fisher Scientific, Applied Biosystems, Grand Island, NY). The M. abscessus and M. massiliense type strains were used as positive controls for the full-length and truncated erm(41) genes, respectively. A negative control containing nuclease-free water instead of DNA template was included in all experiments. The reaction was set up in triplicate in a 96-well PCR plate and processed on a 7500 Fast thermocycler (Thermo Fisher Scientific, Applied Biosystems, Grand Island, NY). In silico PCR was done using NCBI BLASTn (http://blast.ncbi.nlm.nih.gov/), and the publicly available M. abscessus ATCC 19977T and M. massilense CCUG 48898T genomes yielded the expected PCR product sizes for each subspecies.
Real-time 28C/T SNP detection in the inducible clarithromycin resistance erm(41) gene.
Primers (MycoSNP_Fex and MycoSNP_Rex (Table 1) were designed using the M. abscessus ATCC 19977T genome spanning a 124-bp region of the erm(41) gene containing the position 28 C/T SNP. Probes were reconstituted in Tris-EDTA (TE) buffer at a stock concentration of 100 μM and stored at −70°C before use. The probes were then diluted to a working concentration of 10 μM, with a final concentration in the reaction mixture of 250 nM for each probe. Allele-specific fluorescent-labeled LNA probes with a C or T nucleotide at position 28 of the erm(41) gene with LNA bases located on the SNP base and the flanking nucleotides were designed (probes T Allele and C Allele in Table 2). The SNP assay was carried out in a 20-μl reaction mixture, which consisted of 1 μl of the forward and reverse primers at a concentration of 20 μM (1,000 nM final concentration), 0.5 μl of each of the Allele T and Allele C LNA probes at a concentration of 10 μm (250 nM final concentration), 2 to 3 μl of genomic DNA (30 to 50 ng), nuclease-free water (5 to 6 μl), and 10 μl of the TaqMan GTXexpress master mix (Thermo Fisher Scientific, Life Technologies, Grand Island, NY). The reaction was set up in a 96-well PCR plate and processed on a 7500 Fast thermocycler (Applied Biosystems, Life Technologies, Grand Island, NY). The cycling conditions for the assays were a pre-PCR step at 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s, followed by a post-PCR step at 72°C for 2 min. The extent of binding and the resulting fluorescence signal during the PCR at each time point are measured as Rn values for each probe (where Rn is the normalized reporter value). The fluorescence intensity for each probe is shown in allelic discrimination plots; the probe showing a high-intensity signal represents the allele on the DNA template. Allelic discrimination plots were generated using the Applied Biosystems 7500 Fast real-time PCR system software (Thermo Fisher Scientific, Applied Biosystems, Grand Island, NY).
Real-time single nucleotide polymorphism detection for acquired clarithromycin resistance on the rrl (23S rRNA) gene.
Primers 23S_SNPF and 23S_SNPR (Table 1) were designed using the M. abscessus ATCC genome 23S rRNA rrl gene (MAB_r5052). Allele-specific fluorescent-labeled LNA probes with the wild-type allele AA at nucleotides 2058 to 2059 and G or C at the nucleotide 2058 position or G at 2059 were designed (Table 2) with LNA bases located on the SNP base and the flanking bases. The SNP assay was carried out with the pairs WT_MAB and AA_2058G to detect the A2058G SNP, WT_MAB and AA_2058C to detect the A2058C SNP, and WT_MAB and AA_2059G to detect the A2059G SNP. The PCR, cycling conditions, and analysis of results were as described for the real-time 28C/T SNP detection assay.
Sequencing of the erm(41) and rrl genes to confirm deletions and SNPs.
PCR was performed for the erm(41) and rrl genes on 87 clinical isolates and three reference strains using Qiagen Taq polymerase (Qiagen, Carlsbad, CA) in a 25-μl total reaction volume consisting of 12.5 μl master mix comprised of buffer, deoxynucleoside triphosphates (dNTPs), Taq polymerase, and 2 to 3 μl (∼30 to 50 ng) of extracted DNA. Primers ermF and ermR1 (Table 1) (10) were used for amplification and sequencing of the erm(41) gene. Primers 23S_18 and 23S_21 were used for partial amplification of a region of the rrl gene spanning nucleotides 2058 and 2059, and sequencing was performed using the PCR primers and the internal primers 23SrRNAF_207 and 23SrRNAR_207. PCR products from the erm(41) and rrl genes were purified for sequencing using Amicon Ultra-0.5 ml 100K centrifugal filters (Millipore, Ltd., Carrigtwohill, Ireland). Sequencing of PCR amplicons was performed on the ABI Prism 3100 genetic analyzer (Applied Biosystems, Carlsbad, CA), and data were assembled using Lasergene SeqMan Pro technology (DNAStar, Inc., Madison, WI). Sequences were compared against the GenBank database using NCBI BLASTn (24; http://blast.ncbi.nlm.nih.gov/).
Clarithromycin susceptibility testing.
Clarithromycin MIC values were determined in Mueller-Hinton medium by the broth microdilution method, using Sensititre RAPMYCO plates for strains isolated after July 2010 (Trek Diagnostic Systems, Thermo Fisher Scientific, Oakwood Village, OH). Earlier isolates were tested with frozen panels from PML Microbiologicals (Wilsonville, OR). Plates were evaluated at 3 to 5 days and were further incubated for 14 days at 30°C for a final reading to ensure detection of inducible resistance, according to the CLSI M24-A2 document (25) requiring extended incubation. The interpretative breakpoints used were those recommended by the CLSI (25): ≤2 μg/ml, susceptible; 4 μg/ml, intermediate; and ≥8 μg/ml, resistant (see Table S1 in the supplemental material).
RESULTS
A total of 90 clinical and reference isolates of M. abscessus subspecies were used to develop and validate two real-time PCR-based assays aimed at the detection of clarithromycin resistance, which include differentiation between full-length and truncated inducible clarithromycin erm(41) and SNP detection in erm(41) and 23S rRNA rrl genes.
Discrimination between full-length and truncated erm(41) genes by real-time SYBR green assay.
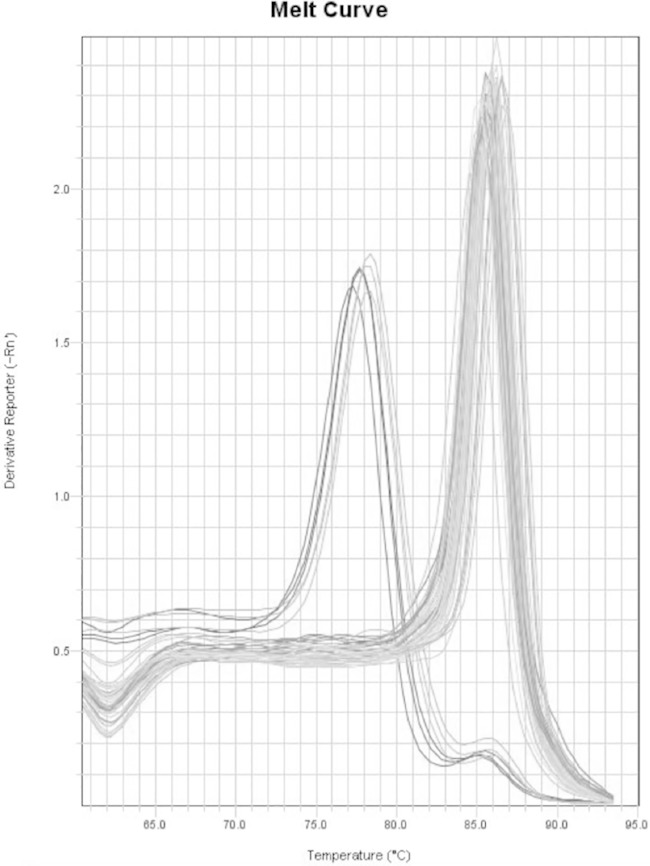
This assay takes advantage of the size difference between a full-length erm(41) gene (characteristic of M. abscessus and M. bolletii) versus a truncated erm(41) gene (seen in most M. massiliense strains) (9, 11). In this assay, primers were designed spanning the 274-bp deletion region of the erm(41) gene, generating a 321-bp product for the full-length erm(41) gene or a 47-bp product with a truncated product as observed by in silico PCR on M. abscessus ATCC 19977T and M. massiliense CCUG 48898T genomes. This size difference results in an ∼7°C change in Tm in real-time PCR melting curve analysis (Fig. 1). Most of our isolates (69/90 [77%]), including M. abscessus and M. bolletii type strains, showed a Tm of 85.8°C (±0.9°C), corresponding to a full-length erm(41) gene, while (21/90 [23%]) of the isolates (including the M. massiliense type strain) showed a Tm of 78.4°C (±0.4°C), consistent with a truncated erm(41) gene. The water negative control had a melt curve temperature of 73.9°C, about 5°C lower than the Tm of the truncated erm(41) gene melt curve. The standard deviations of Tm values of replicates were 0.09 to 0.43. Sequencing of the erm(41) gene performed on all isolates confirmed the results of the real-time PCR assay (see Table S1 in the supplemental material). Most of the 69 isolates with the full-length erm(41) gene were M. abscessus. Four isolates (CI5-9, CI7-8 CI20-27, and CI20-38) were identified as M. bolletii, and three isolates (CI20-39, CI20-40, and CI80-82) with full-length erm(41) were identified as M. massiliense.
FIG 1.
Melting curve peaks for the full-length erm(41) gene at 85.8°C (median) for 69/90 (77%) of isolates and truncated erm(41) gene at 78.4°C (median) for 21/90 (23%) isolates. The assay was run in triplicates. Isolates with a median Tm of 85.8°C include CI12-1, CI12-8, CI12-9, CI12-13, CI12-14, CI12-15, CI12-16, CI12-19, CI12-20, CI4-1, CI6-7, CI6-5, CI6-4, and CI6-3, M. abscessus ATCC 19977T, and M. bolletii 501898T. The isolates with a median Tm of 78.4°C are CI12-10, CI12-2, and M. massiliense 48898T.
Real-time assay for T/C SNP at position 28 of the erm(41) gene.
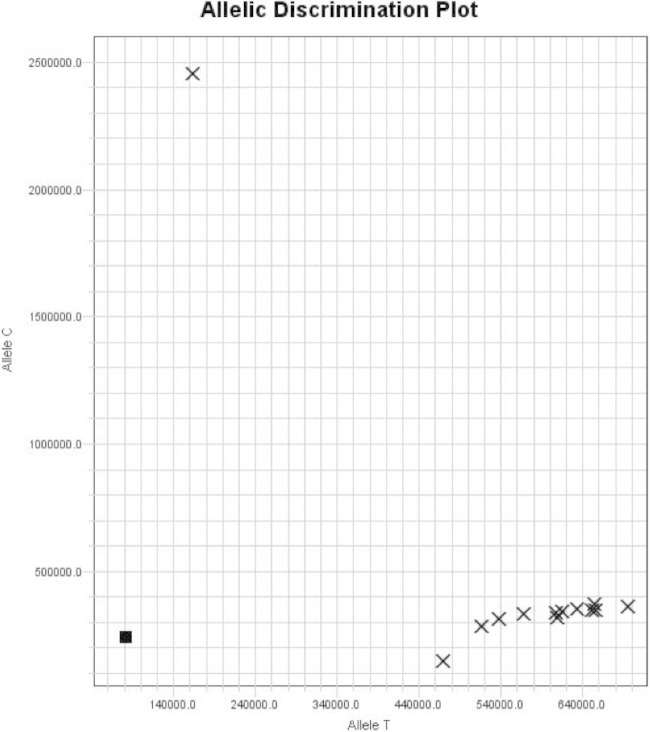
Another mechanism by which the inducible clarithromycin erm(41) gene can lose functionality (besides a major truncation) is through a T-to-C substitution at position 28 (T28C) (9, 11). The erm(41) SNP allele discrimination assay was designed to differentiate between both nucleotides at position 28. Out of a total of 90 isolates in this study, the majority had a T at position 28 (85/90 [95%]), and 5/90 (5%) had a C (see Table S1 in the supplemental material). Sixty-four of 69 isolates with full-length erm(41) contained a T at position 28 characteristic of active erm(41) genes, while 5/69 contained a C (strains CI6-2, CI6-6, CI7-9, CI12-1, and CI20-43—all M. abscessus). An allelic discrimination plot of T and C alleles at position 28 is shown in Fig. 2. Isolates with T are seen in the far right quadrant along the x axis (CI6-3, CI20-19, CI20-25, CI20-30, CI20-31, CI20-35, CI20-36, CI20-37, CI20-38, CI20-42, and CI20-44), and an isolate with C is located in the upper left quadrant on the y axis. The negative control with water instead of genomic DNA is located in the left lower quadrant of the graph close to zero.
FIG 2.
SNP allelic discrimination plot for SNP at position 28 in the erm(41) gene. This is a representative figure of allelic discrimination between probes Allele C and Allele T with isolate CI20-43 with C28 shown in the upper quadrant of the y axis. The isolates with T28 are located in the far right quadrant along the x axis: CI6-3, CI20-19, CI20-25, CI20-30, CI20-31, CI20-35, CI20-36, CI20-37, CI20-38, CI20-42, CI20-44, and M. abscessus ATCC 19977T . The negative control with water instead of genomic DNA is located in the left lower quadrant of the graph close to zero.
SNP assignment by this assay was confirmed by partial amplification and sequencing of erm(41). Susceptibility testing of most of the isolates with a T at position 28 of erm(41) yielded initial MIC values between 0.5 and 2.0 μg/ml (susceptible) at 3 to 5 days and MIC values of ≥8 μg/ml (resistant) at 14 days (see Table S1 in the supplemental material). Similar results were obtained with M. bolletii isolates (CI7-8, CI20-38, and CCUG 50184T) and M. massiliense full-length erm(41) isolates CI20-40 and CI81-82). In contrast, all 5 isolates with C at position 28 of erm(41) (CI6-2, CI6-6, CI7-9, CI12-1, and CI20-43) remained susceptible to clarithromycin even at 14 days incubation, with MIC values of 0.25 to 2.0 μg/ml.
All 24 M. massiliense isolates in the study had a T at position 28, of which 21 harbored a truncated erm(41). Of the 21 strains with truncated erm(41), 13/21 (62%) had no acquired resistance and displayed clarithromycin MIC values of 0.12 to 0.5 μg/ml at 3 to 5 days after the initial reading and remained susceptible to clarithromycin at 14 days.
Eight MAG isolates harboring a full erm(41) gene with T at position 28 and no SNPs associated with acquired resistance showed clarithromycin MICs above the susceptible range at the initial reading. These isolates were categorized into two groups: (i) isolates with a MIC of 4 (intermediate) to clarithromycin at the initial reading, including 3 M. abscessus strains CI20-2, CI20-15, and CI20-24, and (ii) isolates with a MIC of ≥8 (resistant) to clarithromycin at 3 to 5 days. This group includes 5 isolates: M. abscessus CI20-32, CI20-33, and CI20-44, M. bolletii isolate CI20-27, and M. massiliense isolate CI20-39.
Another noteworthy finding is the presence of very-slow-growing MAG strains with a rough-colony morphology requiring 6 to 8 days of incubation for the first clarithromycin MIC reading, due to insufficient growth at 3 to 5 days. The slow-growing isolates in this group are M. abscessus CI20-2, CI20-6, CI20-8, CI20-10, CI20-11, and CI20-12. Interestingly, isolates CI20-2, CI20-6, CI20-8, and CI20-10 belong to 4 patients from which earlier isolates (7 to 11 years prior) with a standard growth pattern were available (CI20-1, CI20-5, CI20-7, and CI20-9, respectively) and were included in this study. Isolates CI20-11 and CI20-12 are an early strain and later strain from a 5th patient (5 years apart), both showing slow growth. All of these isolates showed inducible resistance to clarithromycin by day 14. Repeat antibiotic susceptibility tests on some of the slow-growing isolates showed faint growth in the positive-control well but no growth in any of the clarithromycin wells within the 5-day initial reading period. This highlights the technical difficulty in antibiotic susceptibility testing of slowly growing isolates.
Real-time assays for 23S rRNA SNPs associated with acquired clarithromycin resistance.
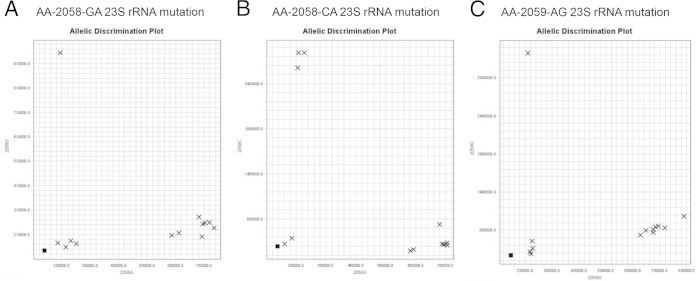
Allele determination for common mutations conferring acquired clarithromycin resistance in the 23S rRNA rrl gene was performed using probes targeting an A-to-G or -C change at position 2058 and an A-to-G change at position 2059. This assay was done in a pairwise manner in 3 reactions comprising WT_MAB with mutant probe AA_2058G, WT_MAB with mutant probe AA_2058C, and WT_MAB with mutant probe AA_2059G, respectively. We identified 15/90 (17%) isolates harboring resistance mutations in the 23S rRNA gene, including 11/90 (12%) isolates with the A2058G mutation comprising 8 M. massiliense isolates with a truncated erm(41) gene (CI6-14, CI6-15, CI7-3, CI1-1, CI20-42, CI15-1, CI20-26, and CI20-34) and 3 M. abscessus isolates with a full-length erm(41) gene (CI5-7, CI20-16, and CI20-18). Figure 3A is a representative allelic discrimination plot of an A or G SNP at position 2058, with isolate CI20-42 containing the 2058G mutation in the upper quadrant of the y axis, isolates with A at position 2058 (wild-type allele) in the far right quadrant of the x axis (CI20-19, CI20-30, CI20-31, CI20-35, CI20-38, CI20-43, and CI20-44, M. abscessus ATCC 19977T), and the negative control (water) or isolates with a different mutation (2058C or 2059G, strains CI6-3, CI20-25, CI20-36, and CI20-37) on the x-y coordinate close to zero. Three isolates in this study (CI6-3, CI20-25, and CI20-37) harbored the A2058C mutation in the 23S rRNA gene (Fig. 3B), and one isolate (CI20-36) contained the A2059G mutation (Fig. 3C). Again, strains harboring an SNP different from the wild type or the one assayed, A2058C (Fig. 3B) or A2059G (Fig. 3C), respectively, clustered next to the negative control, close to zero. SNP assignments by these assays were confirmed by partial amplification and sequencing of the 23S rRNA gene.
FIG 3.
(A) SNP allelic discrimination plot for SNP A2058G at position 2058 in the rrl (23S rRNA) gene. This is a representative figure of allelic discrimination between probes 2059A (WT) and 2058G (mutation). The upper quadrant of the y axis includes isolate CI20-42 with the A2058G mutation. Strains CI20-19, CI20-30, CI20-31, CI20-35, CI20-38, CI20-43, CI20-44, and M. abscessus ATCC 19977T are A2058A and cluster in the right lower quadrant. Isolates CI6-3, CI20-25, CI20-36, and CI20-37, which have A2058C or A2059G, are in the lower quadrant with the negative-control water. (B) SNP allelic discrimination plot for SNP A2058C at position 2058 in the rrl (23S rRNA) gene. This is a representative figure of allelic discrimination between probes 2058A (WT) and 2058C (mutation). Strains CI6-3, 20-25, and 20-37 with the A2058C mutation are clustered in the upper quadrant of the y axis. Similar to panel A, isolates with the A2058A mutation are clustered in the right lower quadrant. CI20-42 and CI20-36 have A2058G or A2059G and appear in the left lower quadrant with the negative-control water. (C) SNP allelic discrimination plot for SNP A2059G at position 2059 in the rrl (23S rRNA) gene. This is a representative figure of allelic discrimination between probes 2059A (WT) and 2059G (mutation). The upper quadrant of the y axis includes isolate CI20-36 with the A2059G mutation. Similar to Fig. 3A, A2059A isolates cluster in the right lower quadrant. Isolates CI6-3, CI20-25, CI20-37, and CI20-42, which have A2058G or A2058C, are in the left lower quadrant with the negative-control water.
All 15 isolates, M. abscessus or M. massiliense with rrl mutations conferring clarithromycin resistance, showed phenotypic resistance to clarithromycin at 3 to 5 days of incubation, with MIC values of ≥8 μg/ml (see Table S1 in the supplemental material). There was 100% concordance between SNP assay results, sequencing of the 23S rRNA gene, and phenotypic detection of resistance by the broth microdilution method.
DISCUSSION
Macrolides such as clarithromycin and azithromycin are key antimicrobials in the treatment of pulmonary infections by members of the M. abscessus group (MAG) (7, 8, 26). Clarithromycin resistance can be acquired via point mutations in the rrl gene encoding the peptidyl transferase domain of the 23S rRNA (27) via A2058G/C and/or A2059G transition or can be induced by the drug via a functional full-length inducible clarithromycin resistance erm(41) gene with a T at position 28 (9). In this study, we developed a highly reproducible real-time PCR-based assay to differentiate between full-length and truncated erm(41) genes and SNP detection assays for mutations associated with clarithromycin resistance on the 23S rRNA and erm(41) genes using 87 clinical isolates of MAG obtained between 2005 and 2015 and 3 reference strains. All isolates were identified to the subspecies level by genomic sequencing and/or a PCR-based assay (23).
Our SYBR green real-time PCR-based assay distinguished between the full-length (77%) and truncated (23%) erm(41) genes with a sensitivity of 100% compared to sequencing of the erm(41) gene. The assay is highly reproducible and does not require setting reactions with replicates based on the large, 7°C, difference in Tm between the two products and the low standard deviation observed when tested in triplicate. Similarly, assignment of the T or C SNP at position 28 of erm(41) by our real-time SNP assay showed 100% concordance to erm(41) gene sequencing. In this study, the majority (93% [64/69]) of the isolates with full-length erm(41) had a T at position 28, and 7% (5/69) had a C. Thus, the rate of inactive C28 erm(41) in our study (7%) is lower than the recently reported rate of 18% of M. abscessus with an inactive C28 erm(41) sequevar (28).
Current CLSI guideline M24-A2 (25) recommends reading the clarithromycin MIC at 3 days (or 4 or 5 days if there is insufficient growth) and a final reading at 14 days. All isolates in this study harboring full-length erm(41) with the T28 nucleotide (and no acquired resistance) showed inducible resistance to clarithromycin within 14 days. While most isolates showed MIC values of ≤2 μg/ml (sensitive) in the first reading at 3 to 5 days, some strains showed a MIC of >2 (i.e., CI20-32, CI20-33, CI20-44, CI20-27, and CI20-39). One hypothesis is the possibility of an earlier induction of the erm(41) gene upon incubation with the drug for some strains. Another explanation lies in the technical challenges of this test. In a recent article, Brown-Elliott et al. (28) reported that some C28 isolates with a MIC of 8 upon extended incubation showed a MIC of ≤2 μg/ml upon repeat testing. In the same study, the authors proposed to change clarithromycin susceptibility breakpoints from ≤2 μg/ml to ≤4 μg/ml. Using those newly proposed breakpoints, our 3 isolates with a MIC of 4 at 3 to 5 days would be considered susceptible at initial reading. In any case, more important than determination of the exact days of incubation at which inducible resistance appears is to make sure inducible resistance is phenotypically detected by day 14, when the final reading of the clarithromycin MIC occurs.
A more challenging issue is the slow-growing MAG strains often seen in multiply treated chronically infected patients for which incubation for 3 to 5 days in Mueller-Hinton broth yields insufficient growth for recording MICs and for which, according to CLSI M24-A2 (25), testing should be repeated. Examples of these isolates are CI20-2, CI20-6, CI20-8, and CI20-10. In the absence of guidelines for phenotypic testing of slowly growing isolates of MAG, sequencing of the erm(41) gene as suggested by Brown-Elliott et al. (28) could be particularly useful for predicting clarithromycin susceptibility patterns for these MAG strains.
In the work-up of cultures for identification and susceptibility testing, it is important to note any evidence of mixed populations on the culture plate reflecting a heterogeneous population of MAG in the patient sample. This is best illustrated by M. abscessus strains CI20-43 and CI20-44 and M. abscessus strains CI20-15 and CI20-16, which were coisolated on the same cultures from sputum samples. CI20-43 is a smooth-colony-morphology strain that harbors a C28-inactive erm(41) gene and remains sensitive to clarithromycin upon extended incubation, while the rough morphotype CI20-44 contains a T28-active erm(41) gene conferring inducible resistance to the clarithromycin MIC. Isolate CI20-15 is a rough-colony-morphology isolate with an active erm(41) gene and no mutational resistance, while CI20-16 is a smooth-colony-morphology isolate with acquired mutational resistance to clarithromycin (A2058G mutation in the 23S rRNA gene).
In this study, we have identified 15 (out of 90 MAG) isolates with mutations on the 23S rRNA gene comprising 7/61 (11%) M. abscessus and 8/24 (33%) M. massiliense. The most common mutation was A2058G, seen in all M. massiliense isolates and 3 M. abscessus isolates with acquired resistance, followed by A2058C (three M. abscessus isolates) and A2059G (one M. abscessus isolate). The high frequency of mutational resistance among M. massiliense isolates should not be surprising, as this is the only known mechanism of resistance available for organisms harboring a truncated erm(41) gene. This study included early (CI15-4 and CI20-41) and late (CI15-1 and CI20-42) M. massiliense isolates (4 to 7 years apart) from the 2 cystic fibrosis (CF) patients. Early isolates were susceptible to clarithromycin, with MIC values of 0.25 μg/ml after 14 days of incubation, as expected for strains harboring truncated erm(41) genes; however, late isolates from each patient showed MIC values of >16 μg/ml due to the acquisition of the A2058G mutation on the 23S rRNA gene. Acquisition of mutational resistance to clarithromycin on patients treated with macrolides has been documented in serial isolates of M. abscessus (29) and M. massiliense (2). Our study confirms this finding with these 2 CF patients who received prolonged treatment with azithromycin and/or clarithromycin.
There are few studies describing molecular assays for clarithromycin resistance that are amenable to implementation in a clinical laboratory. In a recent study, Lee et al. (30) described an amplification refractory mutation system and PCR (ARMS-PCR) and a real-time PCR melting peak analysis method for detection of erm(41) T28 and C28 mutations in M. abscessus clinical isolates. The ARMS-PCR method (cheaper but more time-consuming than real-time PCR) detected all 17 C28 isolates and all 140 T28 isolates, in agreement with sequence analysis, which was used as a reference standard. Their real-time PCR melting peak method also detected all 17 C28 isolates; however, the authors only reported the results from 50 of the 140 T28 isolates. Curry et al. developed a real-time assay consisting of two duplex PCRs for identification of both erm(41) and rrl clarithromycin mutations using SYBR green chemistry and melt curve analysis (C. Curry, R. Luo, and N. Banaei, poster 118, presented at the 114th General Meeting of the American Society for Microbiology, Boston, MA, 17 to 20 May 2014). Phenotypic testing (either Etest or broth MIC) was used as the reference standard to define clarithromycin resistance. They reported a sensitivity of 88% (23/26 isolates) for detection of resistance due to a functional erm(41) gene and 100% sensitivity for detection of isolates with a nonfunctional or truncated erm(41) gene in 37/37 isolates. The assay was 100% sensitive in detecting rrl mutations in 3/3 isolates.
In our study, the SYBR green and position 28 T/C SNP real-time PCR-based assays were 100% sensitive in differentiating between functional and nonfunctional erm(41) compared to erm(41) gene sequencing and correctly predicted the presence and absence of inducible clarithromycin resistance by phenotypic testing. These assays were particularly helpful for predicting resistance in slowly growing strains of MAG. Our real-time SNP assay for acquired mutational clarithromycin resistance in the 23S rRNA gene was 100% sensitive compared to 23S rRNA gene sequencing and also predicted the phenotypic resistance observed in all 15 strains. Of note, both the RAPMYCO and PML Microbiologicals susceptibility testing systems test a limited range of clarithromycin MIC values (up to 8 or 16 μg/ml) far below the MICs (>128 μg/ml) reported in MAG with 23S rRNA mutations (10, 11, 27).
Based on this study, our proposed testing algorithm includes an initial assessment of the full or truncated erm(41) gene using the SYBR green assay followed by SNP detection in isolates that have an intact erm(41) gene for the presence of a C or T base at position 28. This is followed by detection of SNPs of acquired clarithromycin resistance in the 23S rRNA gene. The most common mutation in this cohort was the A2058G substitution on the 23S rRNA gene and should be tested first. Moreover, the A2058G SNP assay by itself will also distinguish non-wild-type isolates with other mutations at position 2058 and/or 2059, as they will be clustering with the negative control in SNP allelic discrimination plots. In conclusion, our real-time melting curve analysis and SNP detection assays for inducible and acquired clarithromycin resistance are easy to implement in the laboratory and provide clinically useful results within ∼3 h, a significant improvement in turnaround time compared to the 14-day incubation required by phenotypic detection of resistance by broth microdilution.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christina Henderson and the technologists of the Mycology and Mycobacteriology Laboratory, Microbiology Service, at the National Institutes of Health for their help. We also thank Barbara Brown-Elliott, Mycobacteria/Nocardia Laboratory, the University of Texas Health Center at Tyler, TX, for providing some of the strains used in this study.
This research was supported in part by the Intramural Research Programs of the Clinical Center, NIAID, and NHLBI, NIH.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01714-15.
REFERENCES
- 1.Leao SC, Tortoli E, Euzeby JP, Garcia MJ. 2011. Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus. Int J Syst Evol Microbiol 61:2311–2313. doi: 10.1099/ijs.0.023770-0. [DOI] [PubMed] [Google Scholar]
- 2.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tettelin H, Davidson RM, Agrawal S, Aitken ML, Shallom S, Hasan NA, Strong M, de Moura VC, De Groote MA, Duarte RS, Hine E, Parankush S, Su Q, Daugherty SC, Fraser CM, Brown-Elliott BA, Wallace RJ Jr, Holland SM, Sampaio EP, Olivier KN, Jackson M, Zelazny AM. 2014. High-level relatedness among Mycobacterium abscessus subsp. massiliense strains from widely separated outbreaks. Emerg Infect Dis 20:364–371. doi: 10.3201/eid2003.131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivier KN, Weber DJ, Lee JH, Handler A, Tudor G, Molina PL, Tomashefski J, Knowles MR. 2003. Nontuberculous mycobacteria. II. Nested-cohort study of impact on cystic fibrosis lung disease. Am J Respir Crit Care Med 167:835–840. [DOI] [PubMed] [Google Scholar]
- 5.Leao SC, Viana-Niero C, Matsumoto CK, Lima KV, Lopes ML, Palaci M, Hadad DJ, Vinhas S, Duarte RS, Lourenco MC, Kipnis A, das Neves ZC, Gabardo BM, Ribeiro MO, Baethgen L, de Assis DB, Madalosso G, Chimara E, Dalcolmo MP. 2010. Epidemic of surgical-site infections by a single clone of rapidly growing mycobacteria in Brazil. Future Microbiol 5:971–980. doi: 10.2217/fmb.10.49. [DOI] [PubMed] [Google Scholar]
- 6.Benwill JL, Wallace RJ Jr. 2014. Mycobacterium abscessus: challenges in diagnosis and treatment. Curr Opin Infect Dis 27:506–510. doi: 10.1097/QCO.0000000000000104. [DOI] [PubMed] [Google Scholar]
- 7.Brown BA, Wallace RJ Jr, Onyi GO De Rosas V, Wallace RJ III. 1992. Activities of four macrolides including clarithromycin against Mycobacterium fortuitum, Mycobacterium chelonae, and M. chelonae-like organisms Antimicrob Agents Chemother 36:180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 9.Nash KA, Brown-Elliott BA, Wallace RJ Jr. 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother 53:1367–1376. doi: 10.1128/AAC.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HY, Kim BJ, Kook Y, Yun YJ, Shin JH, Kim BJ, Kook YH. 2010. Mycobacterium massiliense is differentiated from Mycobacterium abscessus and Mycobacterium bolletii by erythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol Immunol 54:347–353. doi: 10.1111/j.1348-0421.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 11.Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, Cambau E. 2011. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother 55:775–781. doi: 10.1128/AAC.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander PPT, Meier A. 1997. The role of ribosomal RNAs in macrolide resistance. Mol Microbiol 26:469–480. doi: 10.1046/j.1365-2958.1997.5811946.x. [DOI] [PubMed] [Google Scholar]
- 13.Pfister P, Jenni S, Poehlsgaard J, Thomas A, Douthwaite S, Ban N, Bottger EC. 2004. The structural basis of macrolide-ribosome binding assessed using mutagenesis of 23S rRNA positions 2058 and 2059. J Mol Biol 342:1569–1581. doi: 10.1016/j.jmb.2004.07.095. [DOI] [PubMed] [Google Scholar]
- 14.Pfister P, Corti N, Hobbie S, Bruell C, Zarivach R, Yonath A, Bottger EC. 2005. 23S rRNA base pair 2057-2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A→G. Proc Natl Acad Sci U S A 102:5180–5185. doi: 10.1073/pnas.0501598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akshay S, Bertea M, Hobbie SN, Oettinghaus B, Shcherbakov D, Bottger EC, Akbergenov R. 2011. Phylogenetic sequence variations in bacterial rRNA affect species-specific susceptibility to drugs targeting protein synthesis. Antimicrob Agents Chemother 55:4096–4102. doi: 10.1128/AAC.01398-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurer FP, Castelberg C, Quiblier C, Bottger EC, Somoskovi A. 2014. Erm(41)-dependent inducible resistance to azithromycin and clarithromycin in clinical isolates of Mycobacterium abscessus. J Antimicrob Chemother 69:1559–1563. doi: 10.1093/jac/dku007. [DOI] [PubMed] [Google Scholar]
- 17.Kim BJ, Yi SY, Shim TS, Do SY, Yu HK, Park YG, Kook YH, Kim BJ. 2012. Discovery of a novel hsp65 genotype within Mycobacterium massiliense associated with the rough colony morphology. PLoS One 7:e38420. doi: 10.1371/journal.pone.0038420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blauwendraat C, Dixon GL, Hartley JC, Foweraker J, Harris KA. 2012. The use of a two-gene sequencing approach to accurately distinguish between the species within the Mycobacterium abscessus complex and Mycobacterium chelonae. Eur J Clin Microbiol Infect Dis 31:1847–1853. doi: 10.1007/s10096-011-1510-9. [DOI] [PubMed] [Google Scholar]
- 19.Zelazny AM, Calhoun LB, Li L, Shea YR, Fischer SH. 2005. Identification of Mycobacterium species by secA1 sequences. J Clin Microbiol 43:1051–1058. doi: 10.1128/JCM.43.3.1051-1058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sassi M, Ben Kahla I, Drancourt M. 2013. Mycobacterium abscessus multispacer sequence typing. BMC Microbiol 13:3. doi: 10.1186/1471-2180-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macheras E, Roux AL, Bastian S, Leao SC, Palaci M, Sivadon-Tardy V, Gutierrez C, Richter E, Rusch-Gerdes S, Pfyffer G, Bodmer T, Cambau E, Gaillard JL, Heym B. 2011. Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J Clin Microbiol 49:491–499. doi: 10.1128/JCM.01274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ Jr, Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J Clin Microbiol 47:1985–1995. doi: 10.1128/JCM.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shallom SJ, Gardina PJ, Myers TG, Sebastian Y, Conville P, Calhoun LB, Tettelin H, Olivier KN, Uzel G, Sampaio EP, Holland SM, Zelazny AM. 2013. New rapid scheme for distinguishing the subspecies of the Mycobacterium abscessus group and identifying Mycobacterium massiliense isolates with inducible clarithromycin resistance. J Clin Microbiol 51:2943–2949. doi: 10.1128/JCM.01132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams KJ, Bryant WA, Jenkins VA, Barton GR, Witney AA, Pinney JW, Robertson BD. 2013. Deciphering the response of Mycobacterium smegmatis to nitrogen stress using bipartite active modules. BMC Genomics 14:436. doi: 10.1186/1471-2164-14-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CLSI. 2011. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, 2nd ed, p 14 CLSI document M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 26.Mushatt DM, Witzig R. 1995. Successful treatment of Mycobacterium abscessus infections with multidrug regimens containing clarithromycin. Clin Infect Dis 20:1441–1442. doi: 10.1093/clinids/20.5.1441. [DOI] [PubMed] [Google Scholar]
- 27.Wallace RJ Jr, Meier A, Brown BA, Zhang Y, Sander P, Onyi GO, Bottger EC. 1996. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother 40:1676–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown-Elliott BA, Vasireddy S, Vasireddy R, Iakhiaeva E, Howard ST, Nash K, Parodi N, Strong A, Gee M, Smith T, Wallace RJ Jr. 2015. Utility of sequencing the erm(41) gene in isolates of Mycobacterium abscessus subsp. abscessus with low and intermediate clarithromycin MICs. J Clin Microbiol 53:1211–1215. doi: 10.1128/JCM.02950-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurer FP, Ruegger V, Ritter C, Bloemberg GV, Bottger EC. 2012. Acquisition of clarithromycin resistance mutations in the 23S rRNA gene of Mycobacterium abscessus in the presence of inducible erm(41). J Antimicrob Chemother 67:2606–2611. doi: 10.1093/jac/dks279. [DOI] [PubMed] [Google Scholar]
- 30.Lee SH, Yoo HK, Kim SH, Koh WJ, Kim CK, Park YK, Kim HJ. 2014. Detection and assessment of clarithromycin inducible resistant strains among Korean Mycobacterium abscessus clinical strains: PCR methods. J Clin Lab Anal 28:409–414. doi: 10.1002/jcla.21702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.