Abstract
The ability of Escherichia coli O157:H7 to induce cellular damage leading to disease in humans is related to numerous virulence factors, most notably the stx gene, encoding Shiga toxin (Stx) and carried by a bacteriophage. Loss of the Stx-encoding bacteriophage may occur during infection or culturing of the strain. Here, we collected stx-positive and stx-negative variants of E. coli O157:H7/NM (nonmotile) isolates from patients with gastrointestinal complaints. Isolates were characterized by whole-genome sequencing (WGS), and their virulence properties and phylogenetic relationship were determined. Because of the presence of the eae gene but lack of the bfpA gene, the stx-negative isolates were considered atypical enteropathogenic E. coli (aEPEC). However, they had phenotypic characteristics similar to those of the Shiga toxin-producing E. coli (STEC) isolates and belonged to the same sequence type, ST11. Furthermore, EPEC and STEC isolates shared similar virulence genes, the locus of enterocyte effacement region, and plasmids. Core genome phylogenetic analysis using a gene-by-gene typing approach showed that the sorbitol-fermenting (SF) stx-negative isolates clustered together with an SF STEC isolate and that one non-sorbitol-fermenting (NSF) stx-negative isolate clustered together with NSF STEC isolates. Therefore, these stx-negative isolates were thought either to have lost the Stx phage or to be a progenitor of STEC O157:H7/NM. As detection of STEC infections is often based solely on the identification of the presence of stx genes, these may be misdiagnosed in routine laboratories. Therefore, an improved diagnostic approach is required to manage identification, strategies for treatment, and prevention of transmission of these potentially pathogenic strains.
INTRODUCTION
Escherichia coli of serotype O157:H7 was first recognized in 1982 as a human pathogen associated with outbreaks of bloody diarrhea in the United States and is now considered a major cause of foodborne infections (1, 2). The virulence of E. coli O157:H7 depends on the presence of a number of mobile genetic elements (MGE), such as Shiga toxin (Stx)-converting bacteriophages carrying different genes encoding Stx1 and Stx2, the virulence plasmid pO157, the locus of enterocyte effacement (LEE), O islands, an arginine translocation system, and various adhesion factors (3). In Stx-producing E. coli (STEC), Stx is thought to be responsible for the most severe form of the infection causing the life-threatening hemorrhagic colitis (HC) and hemolytic uremic syndrome (HUS) (4). Strains causing these clinical symptoms are also known as enterohemorrhagic E. coli (EHEC) strains (5). The genes encoding Stxs (stx1 and stx2) are found in lysogenic lambdoid bacteriophages (6), which can integrate into the host chromosome via specific insertion sites. Integration sites for stx2-converting phages in STEC O157:H7 include wrbA, argW, sbcB, and yecE, whereas stx1-converting phages integrate in the yehV region (7–11).
STEC O157:H7 strains generally do not ferment sorbitol (non-sorbitol-fermenting [NSF] strains), and this feature is widely used to identify these pathogenic strains. Nevertheless, sorbitol-fermenting (SF) STEC O157:NM (nonmotile) strains, as an emerging and important pathogen in Europe, have been isolated from patients with HUS and diarrhea (12). Both SF and NSF O157:H7/NM strains are thought to have evolved from a common nonpathogenic ancestor of serotype O55:H7 following the modification of the O-antigen gene cluster and the acquisition of a number of virulence-associated MGE via horizontal gene transfers (13). The most current and accepted evolutionary model proposes that E. coli O157:H7 lost the O55 rfb-gnd gene cluster and acquired the Stx2 bacteriophage and the O157 rfb-gnd gene cluster. Subsequently, SF stx2-producing E. coli O157 separated from this lineage. After the diversification of the two lineages, E. coli O157:H7 acquired stx1 via acquisition of the bacteriophage containing the stx1 gene and lost the ability to ferment sorbitol, while the sorbitol-fermenting stx2-producing E. coli O157 lost its motility and evolved into nonmotile E. coli O157:NM (14, 15). Most E. coli O157 isolates produce a large outer membrane protein, intimin (encoded by the eae gene), which is the genetic determinant of the formation of attaching and effacing (A/E) lesions, a central mechanism in the pathogenesis of enteropathogenic E. coli (EPEC) (16). Strains containing eae but not stx are categorized as EPEC (17).
As described in previous studies, stx-negative E. coli O157:H7/NM isolates were obtained from patients with HUS and diarrhea. It was assumed, especially for the HUS cases, that these patients were originally infected with an EHEC strain and that excision of the Stx bacteriophage occurred during the infection. Those stx-negative O157:H7/NM isolates appeared to be closely related to stx-positive O157:H7 isolates as determined by conventional molecular typing methods (18–21). In this study, whole-genome sequencing (WGS) was used to obtain a more detailed molecular characterization of stx-negative E. coli O157:H7/NM isolates and to reveal their genetic relationship with stx-positive O157:H7 isolates obtained from patients with gastrointestinal complaints.
MATERIALS AND METHODS
Selection of isolates for the study.
Fecal samples were collected from patients with gastrointestinal complaints in the regions of Groningen and Rotterdam during the period April 2013 to March 2014 as part of a large multicenter study (STEC-ID-Net, unpublished data). Samples were screened for the presence of stx1, stx2, and escV (used as an alternative marker for the LEE instead of the eae gene) and for O-serogroup determination (O26, O103, O104, O111, O121, O145, and O157) by real-time PCR as described previously (22). This resulted in the collection of 34 E. coli O157 isolates (with or without stx) from 34 different patients. PCR for the detection of the fliCH7 gene was performed as described before (23), and only the isolates positive for fliCH7 were then subjected to WGS.
The publically available genomes of E. coli O157:H7 strains Sakai, EDL933, and SS52, E. coli O157:H45 strain C639_08, E. coli O127:H6 strain E2348/69, and E. coli O55:H7 CB9615 were also included in the comparative analysis. Moreover, as no complete genomes of SF STEC O157:NM and stx-negative O157:H7/NM were available in the NCBI database when the study was performed, we sequenced the genomes of one SF STEC O157:NM (E09/10) strain and one stx-negative O157:NM (E09/224) strain and included them as control strains in all the analyses. The information on the isolates used in this study is presented in Table S1 in the supplemental material.
Phenotypic characterization.
Sorbitol fermentation was determined using CT-SMAC (sorbitol MacConkey agar with cefixime and tellurite) plates, and motility was tested using motility test medium with triphenyltetrazolium chloride (Mediaproducts BV, Groningen, the Netherlands). The production of beta-glucuronidase and urease was checked using MacConkey II agar with 4-methylumbelliferryl-β-d-glucuronide (MUG) (BD Diagnostics, Breda, the Netherlands) and urea-triple sugar iron (TSI) agar (Mediaproducts BV, Groningen, the Netherlands), respectively. The O and H serotypes of the isolates were determined by seroagglutination performed at the National Institute for Public Health and the Environment (RIVM, Bilthoven, the Netherlands).
DNA extraction and WGS.
DNA was extracted using the UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA, USA) according to the manufacturer's protocol. A DNA library was prepared using the Nextera XT kit (Illumina, San Diego, CA, USA) according to the manufacturer's instructions and then run on a MiSeq instrument (Illumina) for generating paired-end 250-bp reads, aiming at a coverage of at least 60-fold.
Data analysis.
De novo assembly was performed using CLC Genomics Workbench v7.0.3 (CLC bio A/S, Aarhus, Denmark) after quality trimming (Qs, ≥28) with optimal word sizes based on the maximum N50 value. Annotation was performed by uploading the assembled genome on the RAST server version 2.0 (24). The sequence type (ST) was identified by uploading the assembled genomes to the multilocus sequence type (MLST) server (version 1.7) (25), and the virulence genes and stx subtypes were determined with virulence finder 1.2 (26). The serogenotype of the isolates was determined using the CGE SeroTypeFinder tool (27).
Comparison of the LEE island, plasmids, and other genes.
To analyze the sequence homology of the LEE pathogenicity island in the isolates, the contigs of each sample were subjected to a BLAST search against the LEE region of E. coli O157:H7 strain 71074 (accession no. GQ338312) as a reference and plotted by BLAST Ring Image Generator (BRIG) (28). To identify plasmids, pO157 (accession no. NC_002128) and pOsak1 (accession no. AB011548) of the E. coli strain Sakai and plasmid pSFO157 (accession no. NC_009602) of the E. coli O157:NM strain 3072/96 were used as references for BLAST analyses. For identifying proposed marker genes for differentiating NSF STEC O157:H7/NM from SF STEC O157:NM, including the complete cdt cluster, the complete sequence of efa1, the tellurite resistance- and adherence-conferring pathogenicity island (TAI), and the urease gene cluster (29), contigs were either subjected to a BLAST search using blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or mapped by CLC Genomics Workbench v7.0.3 using default settings against a reference sequence artificially generated by concatenating sequences of the marker genes.
Phylogenetic analysis.
To determine the phylogenetic relationship of the isolates, a gene-by-gene typing approach was performed using SeqSphere+ v1.0 (Ridom GmbH, Münster, Germany). Briefly, an in-house defined MLST+ (also referred to as core genome [cg] MLST) scheme was developed using the genome of E. coli O157:H7 strain Sakai as reference genome to extract open reading frames (ORFs) from the genome of each isolate by SeqSphere+. Only the ORFs without a premature stop codon and ambiguous nucleotides from contigs of assembled genomes were included. The genes shared by the genomes of all isolates analyzed were defined as the core genome for phylogenetic analysis (30, 31). A neighbor-joining (NJ) tree was constructed based on a distance matrix among the isolates depending on the core genomes of all isolates.
Analyzing phage integration sites.
The most common and well described Stx phage integration sites for STEC O157:H7/NM were analyzed in our stx-negative isolates to reveal if any phage was occupying these sites or that they were available for future integration of a phage, including one that contains stx genes. Five integration sites were studied: yehV (for the Stx1 phage), wrbA and argW (for the Stx2a phage), sbcB (for the Stx2c phage), and yecE (for the Stx2a phage of SF STEC) (11). These five loci were identified in the contigs, the adjacent regions were extracted, and the presence of phage integrases was detected using the blastn algorithm.
Accession numbers.
This whole-genome shotgun project has been deposited in NCBI under BioProject PRJNA285020. The GenBank accession numbers of the isolates analyzed in this study are LDOZ00000000, LFUA00000000, LFUB00000000, LGAZ00000000, LFUH00000000, LGBA00000000, LGBB00000000, LGBC00000000, LGBD00000000, LGBE00000000, LGBF00000000, LGBG00000000, LGBH00000000, LGBQ00000000, LGBI00000000 , LGBJ00000000 , LGBK00000000 , LGBL00000000 , LGBM00000000 , LGBN00000000 , LGBO00000000 , and LGBP00000000 .
RESULTS
Selection of isolates.
Among 34 E. coli O157 isolates initially obtained in this study, 16 were STEC (all were fliCH7 positive) and 18 were categorized as EPEC (as they were stx negative but eae positive). Among the 18 EPEC isolates, four O157:H7, eight O157:H16, two O157:H26, and four O157:H39 isolates were identified. For the subsequent comparative study, 20 isolates positive for fliCH7 (16 STEC O157:H7 and 4 EPEC O157:H7/NM) were used.
Phenotype.
The phenotypic characteristics of the 20 O157:H7 isolates of this study and the two control strains are shown in Table 1. The 16 STEC O157:H7 isolates did not ferment sorbitol (NSF) and were beta-glucuronidase negative, whereas among the four EPEC isolates, one (EPEC 287) was NSF and beta-glucuronidase negative. The remaining three isolates (EPEC 393, EPEC 1572, and EPEC 1669) and the control strain (E09/224) were sorbitol fermenting (SF) and beta-glucuronidase positive, as was the SF STEC control strain E09/10 (see Table S1 in the supplemental material). Among the NSF STEC isolates, six of the 16 isolates were nonmotile, as were all SF isolates. All isolates in this study were urease negative.
TABLE 1.
Phenotypic and molecular characteristics of the isolates
| Pathotype and serotypea | No. of isolates | Beta-glucuronidase activity | Urease production | Motility (no. of isolates) | Stx subtype(s) (no. of isolates) | Intimin typeb | Sequence type by MLST |
|---|---|---|---|---|---|---|---|
| NSF, stx positive, O157:H7/NM | 16 | − | − | + (10) | stx1a+ stx2c (8), stx1a+ stx2a (2), stx2c (6) | Gamma | 11 |
| NSF, stx negative, O157:NM | 1 | − | − | − | NAc | Gamma | 11 |
| SF, stx positive, O157:NM | 1d | + | − | − | stx2a | Gamma | 11 |
| SF, stx negative, O157:NM | 4e | + | − | − | NA | Gamma | 11 |
NSF, non-sorbitol fermenting; SF, sorbitol fermenting; NM, nonmotile.
The type of the eae gene was determined from WGS data using blastn.
NA, not applicable.
This isolate was obtained from Germany and used as a control strain for SF STEC O157:NM isolates.
One of these four isolates was obtained from Germany and used as a control strain for the stx-negative O157:NM isolates.
Molecular typing and presence of virulence genes.
stx subtyping of STEC isolates revealed eight (50%) isolates with stx1a and stx2c subtypes, two (12.5%) with stx1a and stx2a subtypes, and six (37.5%) with the stx2c subtype only. All isolates contained the O-serotyping gene wzxO157 and the flagellar genes fliCH7 and eae (type gamma) and were assigned to ST11 (Table 1). The virulence profiles of the isolates are shown in Table 2. All NSF STEC isolates had identical virulence profiles, with exception of isolate STEC 1109, which contained the cdt-V gene, encoding cytolethal distending toxin. Some of the virulence genes, such as serine protease autotransporter-encoding gene espP, toxin-encoding gene toxB, catalase peroxidase-encoding gene katP, the tellurite resistance- and adherence-conferring island (TAI), and the urease gene cluster, were present only in NSF STEC and in the only NSF EPEC isolate but not in any SF isolates. On the other hand, sfpA, the complete efa1, and the cdt gene cluster were present only in SF isolates. The SF EPEC isolates carried almost all the virulence genes carried by the German SF STEC O157:NM isolate, with the exception of the tccP gene (encoding tir cytoskeletal coupling protein), which was present only in SF EPEC isolates.
TABLE 2.
Distribution of virulence and other genes among stx-positive and stx-negative O157:H7/NM isolates
| Pathotype and serotype (n) | Presence of gene (no. of positive strains) |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adhesin genes |
Fimbrial genes |
Secretion system genes |
Autotransporter gene espP | Toxin genes |
Other genes |
|||||||||||||||||
| eae | tir | efa1a | espB | lpfA | bfpA | sfpA | prfB | espA | espF | espJ | nleA, -B, -C | etpD | tccP | astA | ehxA | cdtb | toxB | katP | TAIc | Urease clusterd | ||
| NSF, STEC O157:H7/NM (16) | + | + | − | + | + | − | − | + | + | + | + | + | + | − | + | + | + | + (1) | + | + | + | + |
| NSF, EPEC O157:NM (1) | + | + | − | + | + | − | − | + | + | + | + | + | + | − | +e | + | + | − | + | + | + | + |
| SF, STEC O157:NMf (1) | + | + | + | + | + | − | + | + | + | − | + | + | + | − | − | + | + | + | − | − | − | − |
| SF, EPEC O157:NM (4) | + | + | + | + | + | − | + | + | + | + (1) | + | + | + | + | − | + | + | (3)g | − | − | − | − |
Complete efa1 gene.
Encoding cytolethal distending toxin A, B, and C subunits.
Tellurite resistance- and adherence-conferring island carrying adhesin gene iha and putative tellurite resistance genes tlrA, tlrB, tlrC, and tlrD.
ure gene cluster containing ureA, ureB, ureC, ureD, ureE, ureF, and ureG.
Only part of the espP gene was present.
This strain was used as a control strain for SF STEC.
cdt was absent in isolate E09/224.
LEE pathogenicity island.
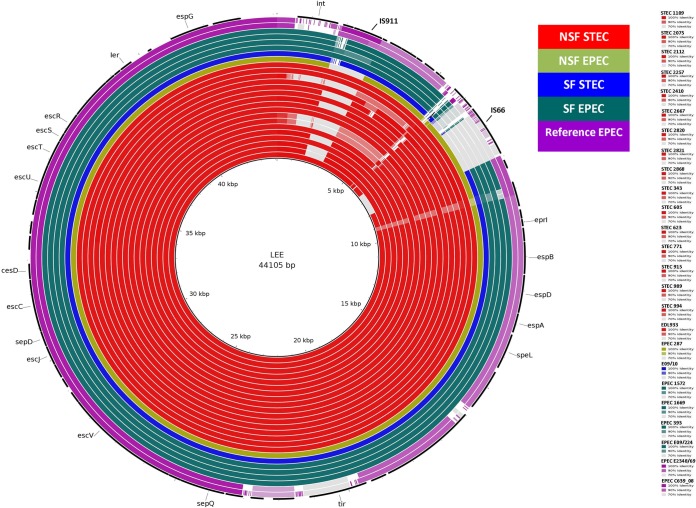
All the STEC isolates contained an LEE region highly similar to the LEE of STEC O157:H7 strain 71074, used as a reference. However, some NSF STEC isolates lacked two genes encoding the mobile element proteins orfA and orfB located in the insertion sequence IS911. Three STEC isolates (STEC 2257, STEC 2820, and STEC 2821) did not possess the intL gene, which is known to encode an integrase of the putative prophage 933L carried in the LEEs of other STEC O157:H7 isolates. The sequences of the LEEs of our NSF and SF EPEC isolates were more similar to those of NSF and SF STEC, respectively, than to the LEE of EPEC reference genomes E2348/69 and C639_08 (Fig. 1).
FIG 1.
Comparison of LEE pathogenicity islands, showing a BLAST comparison of STEC and EPEC isolates, depicted by each ring, against the reference LEE sequence (core black circle). The color of the rings represents sequence identity on a sliding scale; the more gray the ring is, the lower the percent identity. Different colors of the rings represent different groups of isolates. The colors of different groups as well as the order of the rings for each isolate (from inner to outer) with the color gradient for sequence identity are shown at the right.
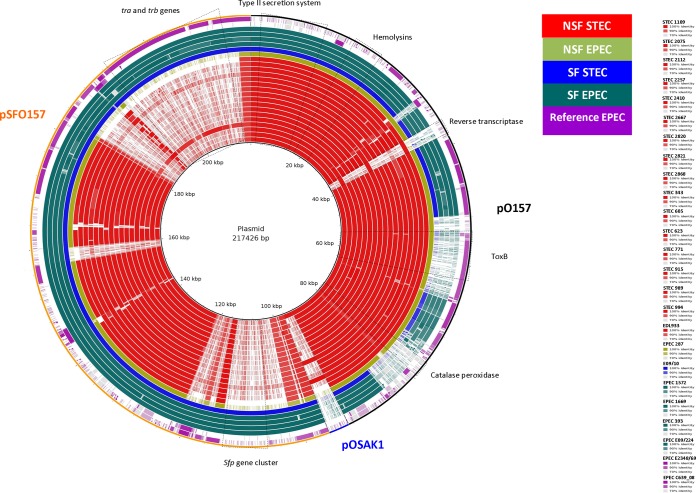
Plasmids.
All the NSF isolates analyzed (including EPEC 287) carried a pO157-like plasmid. No sequence variation was observed in regions carrying putative virulence genes such as, e.g., genes involved in the type II secretion system, hemolysins, toxins, and catalase peroxidase in pO157 of NSF STEC. However, with the exception of the two stx2a-positive isolates (STEC 2112 and STEC 2868), all STEC isolates lacked the pO157p35 gene, encoding a reverse transcriptase. Only three STEC isolates (STEC 605, STEC 989, and STEC 1109) harbored the plasmid pOSAK1. Among the EPEC isolates, the NSF one (EPEC 287) had almost the intact pO157 plasmid, lacking only an intact espP gene. The SF EPEC isolates contained an almost identical copy of plasmid pSFO157 of SF STEC O157:NM (Fig. 2).
FIG 2.
Comparison of plasmids, showing a BLAST comparison of STEC and EPEC isolates, depicted by each ring, against the reference plasmid composed of three plasmids shown in the outermost ring by three different colors (black, blue, and orange represent plasmids pO157, pOSKA1, and pSFO157, respectively). The color of the rings represents sequence identity on a sliding scale; the more gray the ring is, the lower the percent identity. Different colors of the rings represent different groups of isolates. The colors of different groups as well as the order of the rings for each isolate (from inner to outer) with the color gradient for sequence identity are shown at the right.
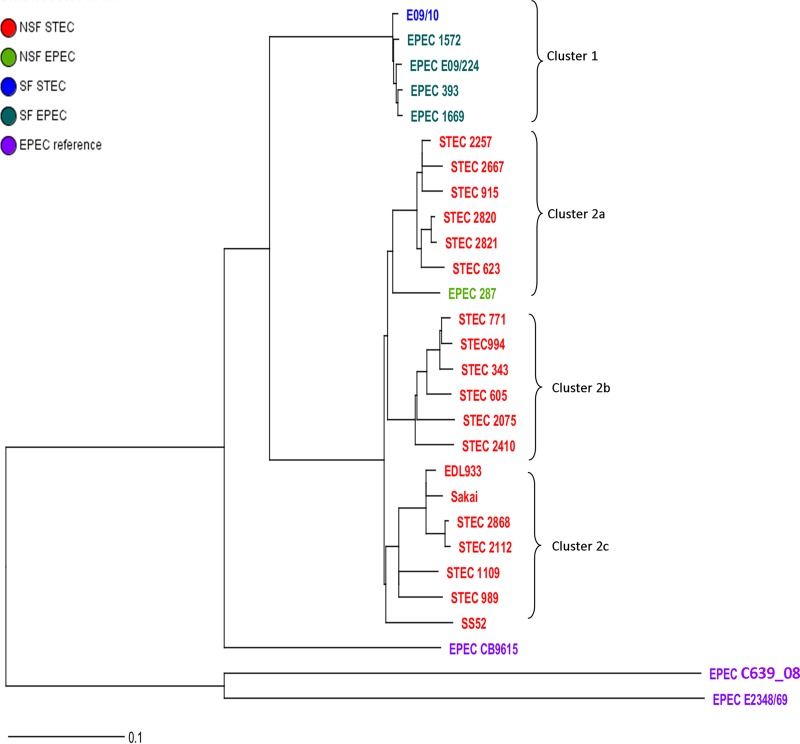
Phylogenetic analysis.
Core genome phylogenetic analysis was performed to evaluate the evolutionary relationship between the stx-positive (STEC) and stx-negative (EPEC) O157:H7/NM isolates. In total, 3,005 ORFs were shared by all isolates analyzed in this study, and these were defined as the core genome for phylogenetic analysis. This analysis separated EPEC C639_08 and EPEC E2348/69 from the O157:H7/NM isolates in this study (Fig. 3). The latter isolates formed two separated clusters: SF isolates (cluster 1) and NSF isolates (cluster 2). Remarkably, in cluster 1, four SF EPEC isolates (EPEC 393, EPEC 1572, EPEC 1669, and E09/224) clustered together with the SF STEC isolate. Cluster 2 (NSF O157:H7 isolates) could be divided into three subclusters: cluster 2a, containing STEC O157:NM (nonmotile) isolates together with one NSF EPEC isolate (EPEC 287); cluster 2b, containing six of the motile STEC O157:H7 isolates; and cluster 2c, containing two stx2a-positive isolates (STEC 2112 and STEC 2868) clustered closely with two previously described STEC outbreak isolates, Sakai and EDL933 (1, 32). The last subcluster also included two other motile isolates (STEC 1109 and STEC 989) and STEC O157:H7 strain SS52 (stx2a and stx2c positive), isolated from super shedder cattle (33). Taken together, the data indicate that the EPEC O157:NM isolates clustered with STEC isolates but not with EPEC isolates (Fig. 3).
FIG 3.
Neighbor-joining (NJ) phylogenetic tree of STEC and EPEC isolates. Different isolate groups are indicated in different colors. The NJ tree was constructed based on a distance matrix among the isolates depending on their core genomes.
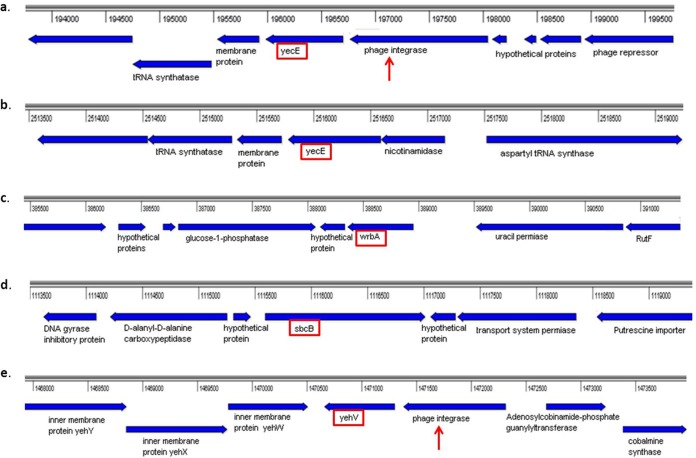
Phage insertion sites.
In the SF EPEC strains (EPEC 393, EPEC 1572, EPEC 1669, and EPEC E09/224), the phage insertion sites analyzed were intact, with the exception of argW, which was occupied in isolates EPEC 393 and EPEC 1572. In EPEC 287, although yehV and argW were occupied by phages, the wrbA and sbcB loci (integration sites for Stx2a and Stx2c phage, respectively) were unoccupied. A comparison of the different integration sites is shown in Fig. 4.
FIG 4.
Phage integration sites. Genes surrounding phage integration sites of the isolates are shown in boxes. A red arrow indicates the presence of a phage integrase adjacent to the integration site. (a) SF STEC isolate E09/10; the yecE region is occupied by phage. (b) SF EPEC isolate EPEC 1572; the yecE region is unoccupied. (c) NSF EPEC isolate EPEC 287; the wrbA region is unoccupied. (d) NSF EPEC isolate EPEC 287; the sbcB region is unoccupied. (e) NSF EPEC isolate EPEC 287; the yehV region occupied by phage.
DISCUSSION
E. coli O157:H7 is one of the major causes of foodborne illness and represents a considerable public health concern worldwide (34). This study aimed at determining the phylogenetic relationships and comparing the virulence factors of stx-negative E. coli O157:H7/NM with those of stx-positive E. coli O157:H7 with the highest resolution and greatest possible detail using a WGS approach. The E. coli O157:H7 isolates used in this study were obtained from patients with diarrhea and other gastrointestinal complaints from two different regions in the Netherlands (Groningen and Rotterdam). Isolates E09/10 and E09/224 were sequenced and used as control strains for SF STEC O157:NM and stx-negative O157:NM isolates, respectively. As the stx-negative isolates were found to be positive for eae but negative for the bfpA gene, they were considered atypical EPEC. Notably, the EPEC isolates belonged to ST11 and contained type gamma intimin, which are not typical features of EPEC but are frequently associated with STEC O157:H7 (35).
All STEC isolates shared a similar virulence pattern. Remarkably, all the typical genes (e.g., ehxA, astA, lpfA, katP, etpD, espP, the pathogenicity island TAI, and the urease gene cluster) of NSF STEC O157:H7 described previously were present in EPEC 287. In contrast, SF EPEC isolates possessed the sfp gene cluster, the cdt gene cluster, and the complete efa1 gene but lacked the genes toxB, katP, espP carried on plasmid pO157, the urease gene cluster, and the pathogenicity island TAI, which are typical features of SF STEC O157:NM (18, 29). Subsequent analyses showed that EPEC 287 harbored a pO157 plasmid almost identical to that of the STEC isolates, with sequence variability mostly in mobile genetic elements. The SF EPEC isolates contained a plasmid similar to the pSFO157 present in the SF STEC O157:NM isolate strain 3072/96. Additionally, our NSF and SF EPEC isolates shared an almost identical LEE pathogenicity island with the STEC isolates containing additional ORFs encoding a putative prophage normally not present in the LEE of EPEC (36). All these results together suggest that the EPEC O157:H7/NM isolates investigated shared a virulence profile similar to that of STEC isolates rather than to the virulence profile of EPEC.
The genetic relationship of the O157:H7/NM isolates of this study was confirmed by the phylogenetic analysis using a gene-by-gene comparison by core genome MLST. The NSF EPEC isolate clustered together with the nonmotile NSF STEC isolates, and the three SF EPEC isolates clustered together with the control strain of SF STEC O157:NM. It has already been described that the nonmotile characteristic of SF STEC O157 is due to a 12-bp deletion in the master regulator of flagellar biosynthesis, the flhC gene (37). We also observed this deletion in the same gene in our nonmotile SF EPEC isolates but not in our nonmotile NSF STEC isolates, further strengthening the hypothesis of their derivation from stx-positive SF O157:NM. Taken together, the results of our analyses provide further evidence that the NSF and SF EPEC isolates were mostly related to the STEC group, but lacking the Stx-converting phages, and might be referred to as EHEC strains that have lost the Shiga toxin (EHEC-LST), as has also been proposed previously (19, 20). Studying in more detail the typical Stx phage insertion sites (yecE for SF EPEC and wrbA and sbcB for NSF EPEC) revealed that they were unoccupied in the stx-negative isolates. Such unoccupied regions could be an indication of loss of the Stx bacteriophage, e.g., in the course of infection or during isolation or subculture (38). Conversely, stx-negative strains could be progenitors of STEC prepared to acquire one or more Stx-converting phages (18, 39). In our case, the stx genes were not lost in the laboratory during isolation or subculture, as they were not detected in the feces of the patients. Therefore, the patients may have been infected with an stx-negative variant, which could explain the mild symptoms they displayed.
In routine diagnostic testing, these stx-negative isolates may be missed, as most microbiological laboratories depend on the molecular screening of STEC by the detection of the stx genes only. Screening for several additional genes (including eae, saa, and sfpA) in routine diagnostics to identify these pathogens has already been proposed (18). Moreover, our findings bring into question whether classifying pathogenic E. coli into STEC and EPEC based on detection of the stx and eae genes, respectively, is reliable enough. Due to integrating and interchanging mobile genetic elements, e.g., the Stx-converting bacteriophages, which could integrate into several E. coli pathogroups, it is sometimes complicated to precisely define the classification of pathogenic E. coli (40, 41). Obviously, screening a number of accessory virulence genes of STEC would be laborious for routine diagnostics. In our opinion, screening of at least the fliCH7 gene, together with the O157-encoding gene, may help to identify EHEC-LST of the most virulent clone of EHEC (O157:H7), as all the fliCH7-positive isolates were genetically related to STEC O157:H7 in this study. Our results are consistent with the finding that the proportion of stx-negative variants among SF O157:NM isolates is generally higher than that among NSF O157:H7 isolates (20, 42). Nevertheless, the number of isolates studied was too low to draw firm conclusions. However, to our best knowledge, this is the first report where the genetic relationship of stx-negative variants of E. coli O157:H7/NM with stx-positive O157:H7/NM has been confirmed using WGS.
In conclusion, stx-negative E. coli O157:H7/NM strains are a cause of gastrointestinal disease in the Netherlands, and because of the presence of the complete set of accessory virulence genes, these isolates should be considered of public health concern similar to that for their stx-positive variants. Additional diagnostic approaches should be implemented to identify these EHEC-LST isolates for surveillance purposes and to allow appropriate treatment measures and for preventing their transmission.
Supplementary Material
ACKNOWLEDGMENTS
This study was partly supported by the Interreg IVa-funded projects EurSafety Heath-net (III-1-02=73) and SafeGuard (III-2-03=025) and by a University Medical Center Groningen Healthy Ageing Pilots grant.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01899-15.
REFERENCES
- 1.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 2.Lim JY, Yoon J, Hovde CJ. 2010. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J Microbiol Biotechnol 20:5–14. [PMC free article] [PubMed] [Google Scholar]
- 3.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A 108:20142–20147. doi: 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J Appl Microbiol 88:729–745. doi: 10.1046/j.1365-2672.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- 5.Levine MM. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis 155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt H. 2001. Shiga-toxin-converting bacteriophages. Res Microbiol 152:687–695. doi: 10.1016/S0923-2508(01)01249-9. [DOI] [PubMed] [Google Scholar]
- 7.De Greve H, Qizhi C, Deboeck F, Hernalsteens JP. 2002. The Shiga-toxin VT2-encoding bacteriophage varphi297 integrates at a distinct position in the Escherichia coli genome. Biochim Biophys Acta 1579:196–202. doi: 10.1016/S0167-4781(02)00539-0. [DOI] [PubMed] [Google Scholar]
- 8.Muniesa M, de Simon M, Prats G, Ferrer D, Pañella H, Jofre J. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect Immun 71:4554–4562. doi: 10.1128/IAI.71.8.4554-4562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielaszewska M, Prager R, Zhang W, Friedrich AW, Mellmann A, Tschäpe H, Karch H. 2006. Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol 72:1900–1909. doi: 10.1128/AEM.72.3.1900-1909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serra-Moreno R, Jofre J, Muniesa M. 2007. Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J Bacteriol 189:6645–6654. doi: 10.1128/JB.00466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shringi S, Schmidt C, Katherine K, Brayton KA, Hancock DD, Besser TE. 2012. Carriage of stx2a differentiates clinical and bovine-biased strains of Escherichia coli O157. PLoS One 7:e51572. doi: 10.1371/journal.pone.0051572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosser T, Dransfield T, Allison L, Hanson M, Holden N, Evans J, Naylor S, La Ragione R, Low JC, Gally DL. 2008. Pathogenic potential of emergent sorbitol-fermenting Escherichia coli O157:NM. Infect Immun 76:5598–5607. doi: 10.1128/IAI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng PC, Monday SR, Lacher DW, Allison L, Siitonen A, Keys C, Eklund M, Nagano H, Karch H, Keen J, Whittam TS. 2007. Genetic diversity among clonal lineages within Escherichia coli O157:H7 stepwise evolutionary model. Emerg Infect Dis 13:1701–1706. doi: 10.3201/eid1311.070381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaikh N, Tarr PI. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J Bacteriol 185:3596–3605. doi: 10.1128/JB.185.12.3596-3605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leopold SR, Magrini V, Holt NJ, Shaikh N, Mardis ER, Cagno J, Ogura Y, Iguchi A, Hayashi T, Mellmann A, Karch H, Besser TE, Sawyer SA, Whittam TS, Tarr PI. 2009. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc Natl Acad Sci U S A 106:8713–8718. doi: 10.1073/pnas.0812949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco M, Blanco JE, Dahbi G, Mora A, Alonso MP, Varela G, Gadea MP, Schelotto F, González EA, Blanco J. 2006. Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: identification of two novel intimin variants (muB and xiR/beta2B). J Med Microbiol 55:1165–1174. doi: 10.1099/jmm.0.46518-0. [DOI] [PubMed] [Google Scholar]
- 17.Bentancor A, Vilte DA, Rumi MV, Carbonari CC, Chinen I, Larzábal M, Cataldi A, Mercado EC. 2010. Characterization of non-Shiga-toxin-producing Escherichia coli O157 strains isolated from dogs. Rev Argent Microbiol 42:46–48. doi: 10.1590/S0325-75412010000100010. [DOI] [PubMed] [Google Scholar]
- 18.Bielaszewska M, Köck R, Friedrich AW, von Eiff C, Zimmerhackl LB, Karch H, Mellmann A. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One 2:e1024. doi: 10.1371/journal.pone.0001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bielaszewska M, Middendorf B, Köck R, Friedrich AW, Fruth A, Karch H, Schmidt MA, Mellmann A. 2008. Shiga toxin-negative attaching and effacing Escherichia coli: distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin Infect Dis 47:208–217. doi: 10.1086/589245. [DOI] [PubMed] [Google Scholar]
- 20.Friedrich AW, Zhang W, Bielaszewska M, Mellmann A, Köck R, Fruth A, Tschäpe H, Karch H. 2007. Prevalence, virulence profiles, and clinical significance of Shiga toxin-negative variants of enterohemorrhagic Escherichia coli O157 infection in humans. Clin Infect Dis 45:39–45. doi: 10.1086/518573. [DOI] [PubMed] [Google Scholar]
- 21.Themphachana M, Nakaguchi Y, Nishibuchi M, Seto K, Rattanachuay P, Singkhamanan K, Sukhumungoon P. 2014. First report in Thailand of a stx-negative Escherichia coli O157 strain from a patient with diarrhea. Southeast Asian J Trop Med Public Health 45:881–889. [PubMed] [Google Scholar]
- 22.de Boer RF, Ferdous M, Ott A, Scheper HR, Wisselink GJ, Heck ME, Rossen JW, Kooistra-Smid AM. 2015. Assessing the public health risk of Shiga toxin-producing Escherichia coli by use of a rapid diagnostic screening algorithm. J Clin Microbiol 53:1588–1598. doi: 10.1128/JCM.03590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perelle S, Dilasser F, Grout J, Fach P. 2004. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Mol Cell Probes 18:185–192. doi: 10.1016/j.mcp.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedrich AW, Köck R, Bielaszewska M, Zhang W, Karch H, Mathys W. 2005. Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J Clin Microbiol 43:546–550. doi: 10.1128/JCM.43.2.546-550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 33.Katani R, Cote R, Raygoza Garay JA, Li L, Arthur TM, DebRoy C, Mwangi MM, Kapur V. 2015. Complete genome sequence of SS52, a strain of Escherichia coli O157:H7 recovered from supershedder cattle. Genome Announc 3:e01569-14. doi: 10.1128/genomeA.01569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olaimat AN, Holley RA. 2012. Factors influencing the microbial safety of fresh produce: a review. Food Microbiol 32:1–19. doi: 10.1016/j.fm.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 35.Tennant SM, Tauschek M, Azzopardi K, Bigham A, Bennett-Wood V, Hartland EL, Qi W, Whittam TS, Robins-Browne RM. 2009. Characterisation of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiol 9:117. doi: 10.1186/1471-2180-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perna NT, Mayhew GF, Pósfai G, Elliott S, Donnenberg MS, Kaper JB, Blattner FR. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun 66:3810–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monday SR, Minnich SA, Feng PC. 2004. A 12-base-pair deletion in the flagellar master control gene flhC causes nonmotility of the pathogenic German sorbitol-fermenting Escherichia coli O157:H− strains. J Bacteriol 186:2319–2327. doi: 10.1128/JB.186.8.2319-2327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng P, Dey M, Abe A, Takeda T. 2001. Isogenic strain of Escherichia coli O157:H7 that has lost both Shiga toxin 1 and 2 genes. Clin Diagn Lab Immunol 8:711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karch H, Bielaszewska M. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H(−) strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J Clin Microbiol 39:2043–2049. doi: 10.1128/JCM.39.6.2043-2049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 41.Tozzoli R, Grande L, Michelacci V, Ranieri P, Maugliani A, Caprioli A, Morabito S. 2014. Shiga toxin-converting phages and the emergence of new pathogenic Escherichia coli: a world in motion. Front Cell Infect Microbiol 4:80. doi: 10.3389/fcimb.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellmann A, Bielaszewska M, Zimmerhackl LB, Prager R, Harmsen D, Tschäpe H, Karch H. 2005. Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin Infect Dis 41:785–792. doi: 10.1086/432722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.