Abstract
Transcription elongation programs are vital for the precise regulation of several biological processes. One key regulator of such programs is the P-TEFb kinase, which phosphorylates RNA polymerase II (Pol II) once released from the inhibitory 7SK small nuclear ribonucleoprotein (snRNP) complex. Although mechanisms of P-TEFb release from the snRNP are becoming clearer, how P-TEFb remains in the 7SK-unbound state to sustain transcription elongation programs remains unknown. Here we report that the PPM1G phosphatase (inducibly recruited by nuclear factor κB [NF-κB] to target promoters) directly binds 7SK RNA and the kinase inhibitor Hexim1 once P-TEFb has been released from the 7SK snRNP. This dual binding activity of PPM1G blocks P-TEFb reassembly onto the snRNP to sustain NF-κB-mediated Pol II transcription in response to DNA damage. Notably, the PPM1G-7SK RNA interaction is direct, kinetically follows the recruitment of PPM1G to promoters to activate NF-κB transcription, and is reversible, since the complex disassembles before resolution of the program. Strikingly, we found that the ataxia telangiectasia mutated (ATM) kinase regulates the interaction between PPM1G and the 7SK snRNP through site-specific PPM1G phosphorylation. The precise and temporally regulated interaction of a cellular enzyme and a noncoding RNA provides a new paradigm for simultaneously controlling the activation and maintenance of inducible transcription elongation programs.
INTRODUCTION
Precise transcriptional regulation in response to intrinsic and extrinsic stimuli enables normal biological processes and cell fate responses (1). Gene transcription by RNA polymerase II (Pol II) is controlled at multiple steps, including initiation and elongation (2–4). Specifically, the transition from transcription initiation to elongation in response to environmental cues is regulated by posttranslational modifications, primarily phosphorylation on the C-terminal domain (CTD) of Pol II (5–8). One of the major CTD kinases is the positive transcription elongation factor (P-TEFb), which is composed of cyclin-dependent kinase 9 (Cdk9) and a regulatory cyclin subunit (T1, T2, or K) (9–11). Cdk9 phosphorylates the Pol II CTD, relieving transcriptional pausing at promoter-proximal regions of many genes and thereby promoting gene activation (12–14). P-TEFb is required for a wide array of transcriptional programs and is thus recruited to gene promoters by a number of pathway-specific transcriptional regulators, including nuclear factor κB (NF-κB), the bromodomain-containing protein BRD4, p53, Myc, and HIV Tat (14–19).
Nuclear levels of active P-TEFb are tightly regulated by its reversible assembly into the 7SK small nuclear ribonucleoprotein (snRNP) complex, which is composed of the 7SK small nuclear RNA, hexamethylene bisacetamide-inducible proteins (Hexim1/2), La-related protein (Larp7), and the 7SK methyl phosphate capping enzyme (MePCE) (11, 20–22). In this complex, the P-TEFb kinase is held catalytically inactive through interactions with the kinase inhibitor Hexim1, which directly binds to 7SK RNA to tether P-TEFb to the snRNP. Therefore, the 7SK snRNP serves to directly control the transcription cycle by sequestering primed P-TEFb, which becomes active when released from the snRNP via dephosphorylation of residue T186 on the activating T-loop (21, 23, 24).
Despite the initial discovery that the 7SK snRNP complex roams the nucleoplasm, we and others have found that it is also recruited to promoter-proximal regions along with paused Pol II, probably to direct the rapid induction of transcriptional programs in response to activating stimuli (25–29). Therefore, P-TEFb release from the promoter-bound 7SK snRNP complex is a prerequisite for transcription factors to capture the kinase to activate transcription. Along this line, we have recently discovered that the nuclear, metal-dependent PPM1G phosphatase is a key regulator of P-TEFb dissociation from the 7SK snRNP. PPM1G is recruited to NF-κB target gene promoters to enzymatically disassemble the promoter-bound 7SK snRNP in response to an inflammatory stimulus to activate gene transcription (26). PPM1G directly interacts with the 7SK snRNP to dephosphorylate T186 at the T-loop of Cdk9 to release P-TEFb from the snRNP. This temporarily inactivated Cdk9 is then rapidly phosphorylated either by the adjacent Cdk7 (part of the preinitiation complex) or through intrinsic autophosphorylation, thereby allowing rapid gene activation by Pol II (26, 30, 31). Furthermore, we have previously reported that PPM1G associates with 7SK RNA in vitro and in vivo (26). However, the mechanism through which PPM1G recognizes 7SK RNA and the functional relevance of this molecular interaction in the transcriptional cycle remain to be elucidated.
In order to investigate the role of PPM1G in transcriptional activation, we biochemically defined minimal protein domains and RNA elements required for the assembly of the PPM1G-7SK protein-RNA complex. Here we present evidence that PPM1G specifically recognizes 7SK RNA via the C-terminal domain and that assembly of a PPM1G-Hexim1 complex on 7SK RNA prevents P-TEFb reassociation onto the RNA and the formation of the 7SK snRNP complex. Additionally, given that PPM1G has been functionally linked with the DNA damage response (DDR) and is a transcriptional coactivator of NF-κB (26, 32, 33), we investigated the role of the PPM1G-7SK RNA interaction in the context of NF-κB transcription in response to genotoxic stress. We found that in vivo, PPM1G is rapidly recruited to NF-κB target genes to activate transcription during the DDR. PPM1G then remains bound to Hexim1-7SK RNA off chromatin (7SK-PPM1G snRNP) to sustain transcription elongation by preventing the reassembly of P-TEFb back onto the inhibitory 7SK snRNP complex. In agreement with this model, PPM1G binds to the C-terminal domain of Hexim1, which is also required for P-TEFb recruitment. Assembly of the 7SK-PPM1G snRNP complex is transient, kinetically follows the recruitment of PPM1G to 7SK-regulated NF-κB target genes, and precedes RNA synthesis. Consistent with the transcriptional role of PPM1G in the response to genotoxic stress, we found that a major DNA damage regulator, the ATM kinase, participates in the site-specific phosphorylation of PPM1G to facilitate its recruitment to the 7SK snRNP complex and to facilitate NF-κB transcription. Our findings provide evidence that PPM1G couples the disassembly of the promoter-bound 7SK snRNP complex with the formation of the 7SK-PPM1G snRNP to initiate and maintain NF-κB-mediated Pol II transcription in response to DNA damage. The precise and temporally regulated interplay of PPM1G and the 7SK snRNP provides a new paradigm for simultaneously controlling the activation and maintenance of inducible transcription elongation programs with a noncoding RNA.
MATERIALS AND METHODS
Cell culture and plasmids.
HeLa and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C with 5% CO2 (all plasmids used in this study are listed in Table 1 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html). Plasmids for heterologous expression of full-length (FL) PPM1G and PPM1G lacking the Lys-rich region (ΔLys) were cloned into the pET30a vector (Merck Millipore) between the NdeI and NotI restriction sites and fused to an N-terminal 6×His tag. The acidic (Ac), C-terminal (Ct), and the acidic and C-terminal (Ac-Ct) domains were cloned similarly but express a C-terminal 6×His tag. To generate plasmids for in vitro transcription, the genes encoding human 7SK RNA, individual stems, or mutants were cloned into pUC19 between the EcoRI and XbaI sites. The T7 promoter followed by a GG dinucleotide was incorporated into the forward primer used for PCR to yield high levels of RNA synthesis. The individual stem-loops of 7SK and stem I mutants were cloned similarly. The 7SK stem I loop and bulge mutants (ΔLoop and ΔBulge) were cloned by using overlap extension, a DNA polymerase I large (Klenow) fragment (New England BioLabs), and PCR extension, as described previously (34). The stem I proximal and distal bulge mutants were cloned by using synthesized oligonucleotides containing the desired mutations and overhangs for the desired restriction sites (Sigma). The 5′ ends of these oligonucleotides were phosphorylated with T4 polynucleotide kinase (New England BioLabs) prior to their ligation into the pUC19 vector (all oligonucleotides used in this study for cloning to generate PPM1G expression vectors, 7SK RNA, and mutants are listed in Table 2 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html). FLAG-tagged Hexim1 and domains were cloned into the pCMV2 backbone as previously reported, including FL Hexim1 and Hexim1 with an N-terminal deletion (ΔN) (35), a C-terminal deletion (ΔC) and a coiled-coil 2 deletion (ΔCR2) (36), and a deletion of the basic region (BR) (ΔBR or mB2 mutant, as previously defined by Barboric et al. [37]).
siRNA-mediated RNAi assay.
HeLa cells were plated at a density of ∼0.5 × 106 cells per well in a 6-well plate and transfected with the PPM1G or nontarget control small interfering RNAs (siRNAs) (Qiagen) (see Table 3 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html) at a concentration of 30 pmol by using Lipofectamine RNAi Max (Life Technologies) according to the manufacturer's guidelines (for the siRNA sequence, see reference 26). Cells were transfected with siRNAs for 48 h, and knockdown (KD) efficiency was calculated by quantitative real-time PCR (qRT-PCR) (with primers listed in Table 4 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html) and Western blot analysis (with antibodies listed in Table 5 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html).
Inducible RNAi.
Lentiviral particles were made by transfecting HEK 293T cells with three plasmids: the short hairpin RNA (shRNA) transfer vector pLKO (Sigma), pMD2.G (vesicular stomatitis virus G protein [VSV-G]), and psPAX2 (Gag-Pol). Two days posttransfection, supernatants containing lentivirus were harvested, and their titer was determined by an enzyme-linked immunosorbent assay (ELISA), and stored at −80°C. HeLa cells (2 × 105) were spinoculated (inoculated by centrifugation) at 2,900 rpm for 2 h with lentiviral particles (50 to 200 μl; ∼1 × 107 transducing units [TU] as determined by a p24 ELISA) in the presence of 8 μg/ml Polybrene. Efficiently transduced cells were selected with 1 μg/ml puromycin for 5 days. At this point, cells were treated with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 days to induce KD. RNA interference (RNAi) validation efficiency was determined by qRT-PCR and Western blotting using the corresponding primer pair and primary antibody, respectively. The shRNAs used for RNAi were pLKO-puro-IPTG-3xLacO-NT shRNA (catalog number SHC332; Sigma) and pLKO-puro-IPTG-3xLacO-PPM1G shRNA (catalog number TRCN0000001213; Sigma).
qRT-PCR assay.
Total RNA was isolated by using TRIzol (Life Technologies) and phenol-chloroform-isoamyl alcohol (PCA) (25:24:1) extraction, and first-strand cDNA was synthesized by using Moloney murine leukemia virus (M-MuLV) reverse transcriptase (New England BioLabs). qRT-PCR was performed by using Sybr green master mix on an ABI7500 instrument (Applied Biosystems). Threshold cycle (CT) values and fold changes were calculated as previously described (38) (qRT-PCR primer sequences are listed in Table 4 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html).
Affinity purification (AP), coimmunoprecipitation, and Western blot analysis.
Expression plasmids containing PPM1G or domains were transformed into Rosetta(DE3) cells (EMD) and grown in cultures overnight. LB cultures (500 ml) were inoculated with a 1/50 dilution of a culture grown overnight and grown at 37°C until an optical density at 600 nm (OD600) of ∼0.6 was reached. Cultures were induced for protein expression with 1 mM IPTG and grown for 3 h at 30°C. To purify the recombinant proteins, cells were lysed by sonication in lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 20 mM imidazole, 2 mM MnCl, 5 mM dithiothreitol [DTT], 5% glycerol) containing protease inhibitors (Roche). Triton X-100 was added to a final concentration of 1% after sonication, and the mixture was nutated at 4°C for 15 min. The protein lysate was clarified by centrifugation at 10,000 × g for 15 min, and the clarified lysate was loaded onto a Ni-Sepharose column (Qiagen). Beads were prewashed three times in high-salt buffer (HSB) (20 mM Tris-HCl [pH 7.4], 500 mM NaCl, 20 mM imidazole, 5 mM DTT, 0.1% Triton X-100, and 5% glycerol) and then washed in low-salt buffer (LSB) (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 20 mM imidazole, 5 mM DTT, 0.1% Triton X-100, and 5% glycerol). Proteins were eluted in LSB supplemented with 150 mM imidazole. The purity of the eluted fractions was tested by using SDS-PAGE and silver staining (Thermo Fisher). Protein fractions were stored at −80°C. For Western blot analysis, immunoprecipitated samples were electrophoresed on 10% Tris-glycine-SDS gels, transferred onto 0.45-μm nitrocellulose membranes (Bio-Rad), blocked in Tris-buffered saline (TBS) containing 0.2% Tween 20 and 5% nonfat dry milk for 2 h, and incubated with primary antibodies at 4°C from 1 h to overnight. Primary antibodies used were described previously (26), except for γH2AX, ATM, and PPM1G pS183 antibodies (see Table 5 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html). Secondary antibodies coupled to horseradish peroxidase (HRP), including donkey anti-rabbit IgG–HRP (catalog number sc-2313) and goat anti-mouse IgG–HRP (catalog sc-2005; Santa Cruz Biotechnologies), were incubated at 1:10,000 dilutions for 1 h, and blots were developed by using chemiluminescence.
Fluorescent antibodies used for Fig. 6, including donkey anti-rabbit antibody–IRDye 800CW (product number 926-32213; Li-Cor) and donkey anti-mouse antibody–IRDye 680LT (product number 926-68022; Li-Cor), were incubated at 1:10,000 dilutions for 1 h, and bands were visualized by using the Li-Cor Odyssey instrument.
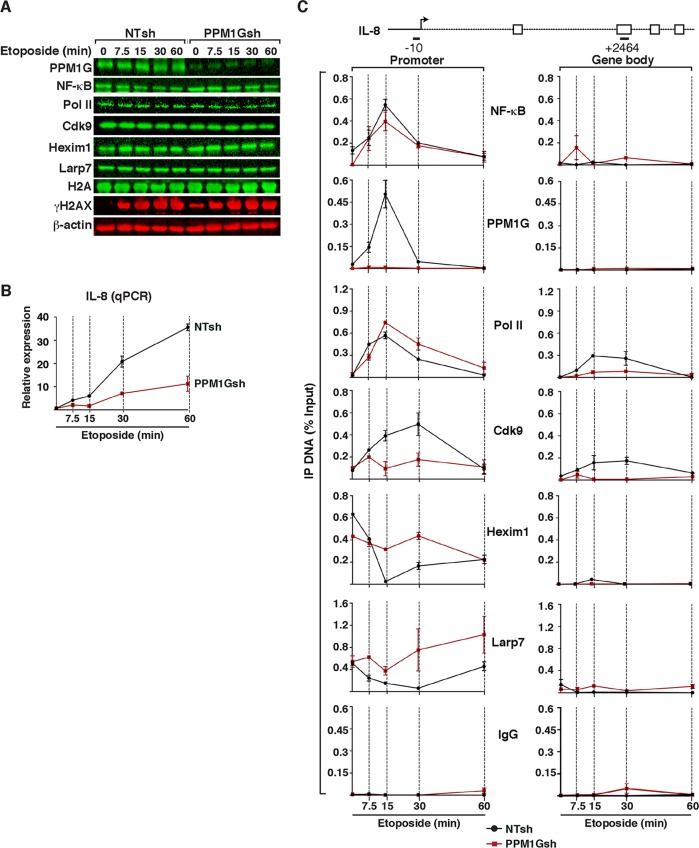
FIG 6.
PPM1G functions downstream of NF-κB and Pol II recruitment to gene promoters to induce 7SK snRNP disassembly and Pol II elongation in response to DNA damage. (A) HeLa cells were transduced with lentiviruses (pLKO) inducibly expressing nontarget (NTsh) or PPM1G (PPM1Gsh) shRNAs, induced with IPTG for 3 days, and treated with etoposide. PPM1G KD efficiency, induction of DNA damage (γH2AX), and stability of transcriptional components were evaluated by Western blotting over time. (B) PPM1G KD antagonizes IL-8 gene expression in response to etoposide. The cell lines from panel A were used for total RNA extraction, and IL-8 gene expression in response to etoposide was quantified by qRT-PCR (means ± standard errors of the means; n = 3). The data were normalized to levels of the β-actin transcript. (C) ChIP assay of cell lines from panel A showing levels of NF-κB, PPM1G, Pol II, and 7SK snRNP components (Cdk9, Hexim1, and Larp7) at the IL-8 promoter-proximal region (−10 amplicon) and gene body (+2464 amplicon) in response to a time course of etoposide treatment (means ± standard errors of the means; n = 3). Normal serum (IgG) was used as a negative control to demonstrate the specificity of enrichment with the antibodies used.
RNA binding gel shift assay.
Wild-type (WT) 7SK RNA and mutants were in vitro transcribed using T7 RNA polymerase from plasmid templates linearized with XbaI. For radiolabeling, the RNAs were transcribed with [α-32P]UTP as described previously (26). RNAs were electrophoresed on a native gel (1× Tris-glycine, pH 8.3), eluted from gels by using RNA elution buffer (0.6 M sodium acetate [pH 6.0], 1 mM EDTA, 0.01% SDS), ethanol precipitated twice, resuspended in sterile deionized water, and stored at −80°C. For gel shift assays, the purified RNA was denatured at 95°C for 10 min on a heat block and renatured by slowly cooling back to room temperature. Protein and radiolabeled RNA were incubated on ice for 20 min in a reaction mixture containing binding buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM DTT, 5% glycerol, and 50 μg/ml Escherichia coli tRNA). Protein-RNA complexes were resolved on 5% and 8% native PAGE gels for full-length 7SK RNA and individual stems, respectively. Gels were electrophoresed at 120 V at 4°C, dried, and autoradiographed.
PPM1G-7SK RNA binding curves.
Binding reactions for gel shift assays were set up as described above. The bands representing unbound RNA and protein-RNA complexes were quantified by using ImageJ. Binding curves and apparent binding constant (Kdapp) values were calculated by using GraphPad Prism software.
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed as previously described (25). HeLa cells were treated with 50 μM etoposide (Fisher) or dimethyl sulfoxide (DMSO) (control) for the indicated times. Protein lysates were prepared and used for ChIP at the interleukin-8 (IL-8) and A20 loci by using the indicated antibodies (see Table 5 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html) and amplicons for qPCR (see Table 6 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html).
Cross-linking RNA immunoprecipitation (X-RIP) assay.
HeLa cells treated with 50 μM etoposide (Fisher) or DMSO (vehicle control) were cross-linked with formaldehyde to stably trap RNA-protein complexes formed in vivo (39). Briefly, HeLa cells were cross-linked in 1% formaldehyde for 10 min at room temperature, quenched with 0.125 M glycine for 5 min, and washed twice with 1× phosphate-buffered saline (PBS). Cell pellets were resuspended in lysis buffer (50 mM Tris-HCl [pH 7.4], 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS, 2 mM MgCl2, and 150 mM NaCl), and the whole-cell lysate was passed through a QiaShreder spin column (Qiagen). The extracts were incubated for 3 h at 4°C with protein G Dynabeads prebound with the PPM1G and normal IgG mouse antibodies. Bound samples were washed four times with RNA immunoprecipitation (RIP) assay buffer for 5 min each and then resuspended in a solution containing 50 mM Tris-HCl (pH 6.3), 5 mM EDTA, 1% SDS, and 10 mM DTT. Samples were reversed cross-linked by heating them at 70°C for 30 min. RNAs were purified by using PCA extraction, and first-strand cDNA synthesis was performed as described above. 7SK RNA was detected by qRT-PCR using gene-specific primers (see Table 4 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html) and Sybr green master mix on an ABI7500 instrument (Applied Biosystems).
UV cross-linking of RNA-protein complexes in vivo.
HeLa cells (grown on a 10-cm2 plate) were irradiated with 125 mJ/cm2 of UV light by using a Spectrolinker XL-1500 UV cross-linker, harvested, lysed by using denaturing lysis buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.0], 5% glycerol, 1.5 mM MgCl2, 1 mM DTT, 1% NP-40, 0.25% sodium deoxycholate, and 0.1% SDS), and sonicated by using the Bioruptor water bath sonicator (Diagenode) with 8 cycles (30 s on and 30 s off). PPM1G was affinity purified by using Strep-tactin Superflow resin (IBA Life Sciences) and washed by using denaturing washing buffer (250 mM NaCl, 20 mM Tris-HCl [pH 7.0], 5% glycerol, 1.5 mM MgCl2, 1 mM DTT, 0.05% NP-40, 0.1% sodium deoxycholate, 0.1% SDS). Coimmunoprecipitating RNAs were extracted by using proteinase K digestion (0.5 mg/ml proteinase K, 20 mM Tris-HCl [pH 7.0], 5 mM EDTA, 0.5% SDS) (39), followed by PCA extraction and ethanol precipitation. The immunoprecipitated material was analyzed by qRT-PCR (primers are listed in Table 4 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html).
In vitro phosphatase assay using pNPP as the substrate.
para-Nitrophenyl phosphate (pNPP; New England BioLabs) was used in enzymatic reactions to determine the catalytic activities of PPM1G and mutants. Phosphatase assays were carried out with a reaction buffer containing 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM MnCl2, and 2.5 mM DTT, at 37°C for 30 min, with 400 ng purified recombinant phosphatase and various pNPP concentrations (0, 1, 2.5, 5, 10, 15, and 20 mM) in a total volume of 100 μl. Enzyme-catalyzed dephosphorylation was halted by the addition of 10 μl of 5 N NaOH, and the absorbance of p-nitrophenol (pNP) was measured at 405 nm. Initial linear rates were determined by using a molar extinction coefficient (ε) of 18,800 M−1 cm−1 for the product pNP at 405 nm at pH 8.0. The acquired data were fitted by using the Lineweaver-Burk equation with GraphPad Prism, where 1/V = (Km + [S])/(Vmax × [S]) or 1/V = [(Km/Vmax) × (1/[S])] + 1/Vmax.
ATM kinase assay.
HEK 293T cells (2 × 106) were lysed in 0.5 ml of fresh lysis buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 0.2% Tween 20) containing protease inhibitors (Roche), 20 mM NaF, and 2 mM NaVO4 by rotation at 4°C for 30 min. The lysate was centrifuged at 12,000 × g for 15 min at 4°C, and the supernatant was transferred to a new tube containing 1 μl of anti-ATM IgG (catalog no.MAT3 4G10/8) (Sigma) and incubated for 2 h at 4°C. Next, 25 μl of preequilibrated protein G Dynabeads (Life Technologies) was added, and the mixture was incubated for 2 h at 4°C. The ATM-bound beads were collected on a magnetic stand and washed with 2× lysis buffer followed by 2× kinase buffer. Beads were aliquoted for the different reaction mixtures and incubated with 5 μl of the substrate (50 pmol WT PPM1G or the S183A mutant) in 20 μl of kinase buffer (10 mM HEPES [pH 7.4], 50 mM NaCl, 10 mM MgCl2, 10 mM MnCl2, and 1 mM DTT) and 1 μl of 0.5 mM ATP (Roche). Kinase reactions were performed at 30°C for 30 min and stopped with the addition of 12 μl 3× Laemmli sample buffer, and reaction mixtures were boiled for 5 min. Reaction mixtures were directly electrophoresed on polyacrylamide gels, and Western blot analyses were performed with the indicated antibodies.
RESULTS
PPM1G binds 7SK RNA by using the C-terminal Lys-rich domain.
The PPM1G phosphatase directly participates in the disassembly of promoter-bound 7SK snRNP complexes to release the P-TEFb kinase, uncoupling it from the kinase inhibitor Hexim1 and thereby activating transcription elongation in response to inflammatory stimuli (26) (Fig. 1A). During this process, PPM1G also appears to bind the 7SK snRNP lacking P-TEFb, henceforth referred to as “7SK-PPM1G snRNP” to differentiate it from the P-TEFb-bound 7SK snRNP complex containing all subunits (26). Based on this evidence, we hypothesized that PPM1G directly contacts 7SK RNA once P-TEFb has been released from the 7SK snRNP to prevent its reassociation, thereby maintaining a local pool of 7SK-unbound, catalytically active P-TEFb to sustain the transcription elongation program. In this study, we investigated the molecular basis of this protein-RNA interaction and the mechanism underlying signal-dependent transcriptional regulation by PPM1G.
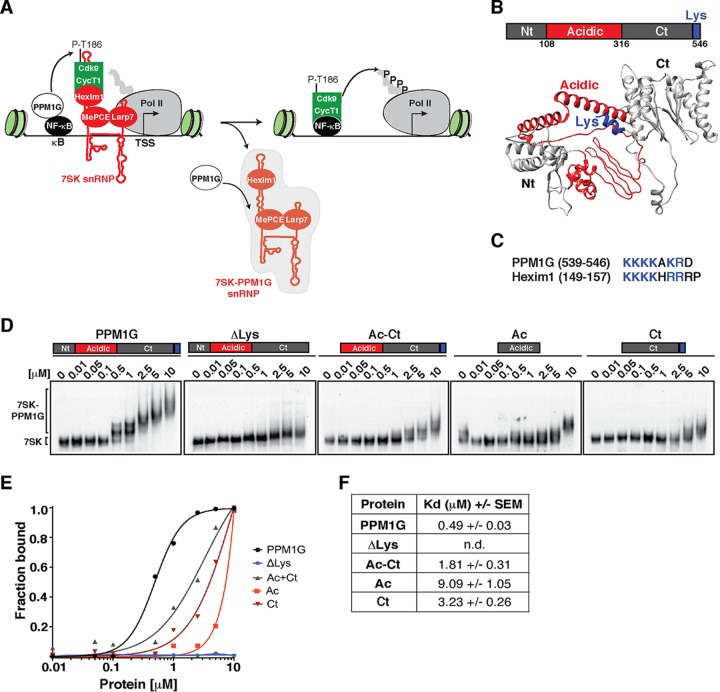
FIG 1.
The PPM1G phosphatase binds 7SK RNA using its C-terminal Lys-rich domain. (A) Proposed model for the role of PPM1G in the NF-κB transcriptional program. PPM1G is recruited by NF-κB to target genes in response to environmental stimuli, where it dephosphorylates the T-loop of Cdk9 (P-T186), thereby promoting its uncoupling from Hexim1 and its release from the inhibitory, promoter-bound 7SK snRNP complex. In this process, PPM1G subsequently binds 7SK RNA, possibly forming a 7SK-PPM1G snRNP complex lacking P-TEFb. (B) Domain organization of human PPM1G and representation of each domain in the predicted three-dimensional structure. Nt, N-terminal domain; Ac, acidic region; Ct, C-terminal domain; Lys, Lys-rich region. (C) Alignment of PPM1G and Hexim1 Lys-rich regions with their positions indicated in parentheses. (D) Gel shift assays with 7SK RNA and increasing amounts of full-length PPM1G or domains. (E) Binding curves from the gel shifts shown in panel D. (F) Calculations of the apparent dissociation constants (Kdapp) from data in panel E. Kdapp values represent the averages of data from three independent experiments with the standard errors of the means (n = 3). n.d., not determined, because the data set does not enable accurate calculation of the Kdapp values.
PPM1G is composed of three discrete modules: the N-terminal (Nt) and C-terminal (Ct) phosphatase domains with an intervening acidic region (Ac) that is thought to modulate substrate specificity (40) (Fig. 1B). The Ct domain bears a lysine (Lys)-rich region that resembles the RNA-binding motif of Hexim1 (Fig. 1C). To determine how PPM1G recognizes 7SK RNA, we performed an in vitro binding assay where full-length PPM1G or discrete domains were incubated with purified 7SK RNA (gel shift assays) (Fig. 1D). Strikingly, we observed that PPM1G binds 7SK RNA with an apparent dissociation constant (Kdapp) of ∼0.49 μM and that deletion of the Lys-rich region (PPM1GΔLys) sharply reduces the binding affinity in vitro (Kdapp of >10 μM) (Fig. 1D to F). If the Lys-rich region was the minimal domain involved in 7SK binding, we would expect a Ct domain to bind with an affinity similar to that of full-length PPM1G. However, unexpectedly, we found that the Ct domain alone binds with a low affinity (Kdapp of ∼3.23 μM) compared to that of full-length PPM1G, implying that other domains in PPM1G may contribute to binding affinity or that the Ct domain requires other regions of the protein for proper folding. In accordance with this, the Ac-Ct construct binds 7SK RNA with an intermediate affinity (Kdapp of ∼1.81 μM) compared to those of full-length PPM1G and the Ct domain (Fig. 1D to F). Given these results, we reasoned that the Lys-rich region directs the in vitro assembly of PPM1G on 7SK RNA but that other protein domains are required for the observed high-affinity binding and perhaps oligomerization of PPM1G on the RNA. In fact, we observed that PPM1G appears to oligomerize on the RNA in a Lys-rich-region-dependent manner (Fig. 1D). Furthermore, we found that the PPM1G-7SK RNA interaction is specific because PPM1G does not bind an antisense 7SK or U6 snRNA in vitro (see Fig. S1 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html), in agreement with previously reported observations (26). Collectively, these results demonstrate that full-length PPM1G is required for high-affinity 7SK RNA binding and that deletion of the Lys-rich region in the C-terminal domain, resembling the RNA-binding motif of the kinase inhibitor Hexim1, virtually abolishes 7SK RNA recognition.
PPM1G preferentially recognizes 7SK RNA stem I.
7SK RNA is composed of four stem-loops (stems I, II, III, and IV) containing specific binding sites for the integral 7SK snRNP components (P-TEFb, Hexim1, Larp7, and MePCE) (41–44) (Fig. 2A). Stem I (5′-terminal hairpin) carries two independent Hexim1-binding sties in the proximal and distal regions, with the latter one directing P-TEFb recruitment to 7SK RNA (42, 44–46). In addition, P-TEFb binding also requires stem IV (3′-terminal hairpin) (42). To more precisely pinpoint how PPM1G recognizes 7SK RNA and to determine how this fits into the model of PPM1G blocking P-TEFb reassembly into 7SK RNA, we mapped RNA elements that contribute to binding affinity using gel shifts assays (Fig. 2B). We found that PPM1G binds stem I with a higher affinity (Kdapp of ∼3.22 μM) than those for other stems (Kdapp of >10 μM) (Fig. 2B to D), suggesting that stem I serves as the primary binding site, with possibly other stems contributing to binding affinity and structural stability. In support of this model, deletion of stem I from 7SK caused an ∼5-fold reduction in PPM1G binding affinity, but it did not completely block the PPM1G-7SK RNA interaction (data not shown). Furthermore, because the affinity of PPM1G for full-length 7SK RNA was greater than that for stem I (Kdapp values of ∼0.49 μM and ∼3.22 μM, respectively) and because deletion of stem I reduces, but does not eliminate, PPM1G binding, other low-affinity binding sites in stems III and IV contribute to binding affinity, supporting the model that PPM1G homo-oligomerizes on the RNA (Fig. 1).
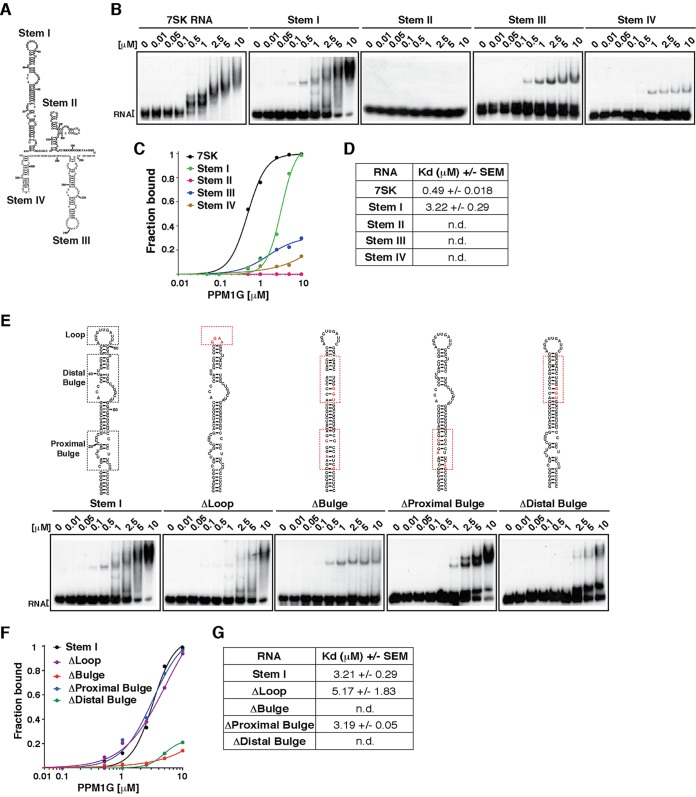
FIG 2.
PPM1G binds the 7SK RNA stem I distal bulge. (A) Primary sequence and predicted secondary structure of human 7SK RNA with the corresponding stem-loops (Stem). (B) Gel shift assays with full-length PPM1G and 7SK RNA or each of the individual stems. (C) Binding curves from the gel shifts shown in panel B. (D) Calculations of the Kdapp values from data shown in panel C. Kdapp values represent the averages of data from three independent experiments and the standard errors of the means (n = 3). (E) Gel shift assays with PPM1G and WT 7SK stem I or mutants (ΔLoop, ΔBulge, ΔProximal Bulge, and ΔDistal Bulge). The schemes above the gel shifts show the positions of the different RNA elements and the corresponding mutations. (F) Binding curves from the gel shifts shown in panel E. (G) Calculations of the Kdapp from data shown in panel F (means ± standard errors of the means; n = 3). n.d., not determined, because the data set does not enable accurate calculation of the Kdapp values.
To more precisely define elements in the high-affinity binding site (stem I), we created internal deletions within the stem-loop structure: ΔLoop, where the loop was replaced with a tetraloop (GGAA), and ΔBulge, where all bulges were either deleted or replaced with sequences to establish perfect base pairing between RNA strands (Fig. 2E). While replacement of stem I with a tetraloop (ΔLoop) caused a minimal reduction in binding affinity (Kdapp of ∼5.17 μM, compared to ∼3.21 μM for stem I), deletion of all bulges (ΔBulge) virtually abolished PPM1G binding (Fig. 2E to G). Given that stem I is composed of two sets of bulges (three bulges in the proximal region close to the 5′ end and three in the distal region closer to the loop), we further mapped structural elements specifically recognized by PPM1G. Mutations and deletions were introduced to eliminate the proximal (ΔProximal Bulge) or distal (ΔDistal Bulge) bulges. Interestingly, the results showed that while deletion of the proximal bulge did not alter PPM1G's ability to bind the RNA (Kdapp of ∼3.19 μM), disruption of the distal bulge virtually eliminated the PPM1G-7SK RNA interaction (Kdapp of >10 μM) (Fig. 2E to G).
Collectively, these results indicate that PPM1G preferentially recognizes the stem I distal bulge, which is in close proximity to the site where Hexim1 binds 7SK RNA to recruit P-TEFb in order to assemble the 7SK snRNP complex (22, 42, 44).
PPM1G binds 7SK RNA and Hexim1 to block P-TEFb recruitment and 7SK snRNP complex formation.
Given that PPM1G and Hexim1 have similar Lys-rich RNA-binding motifs and that they both bind to 7SK RNA stem I, we speculated that PPM1G could recognize 7SK and block P-TEFb reassembly onto the 7SK snRNP. To test this possibility, we first affinity purified Strep-tag II (Strep; IBA Lifesciences)-tagged PPM1G and tandem-affinity-purified (TAP) Hexim1–P-TEFb (P-TEFb–Strep and Hexim1-Flag) to homogeneity, as judged based on silver staining (Fig. 3A). We incubated Hexim1–P-TEFb with 7SK RNA in the absence or presence of increasing amounts of PPM1G (from 0.05 to 5 μM) and resolved the protein-RNA complexes in native gels (gel shift assays). Interestingly, we observed that while Hexim1–P-TEFb binds 7SK RNA, PPM1G blocks the association of P-TEFb with Hexim1 and 7SK RNA in a dose-dependent manner (Fig. 3B).
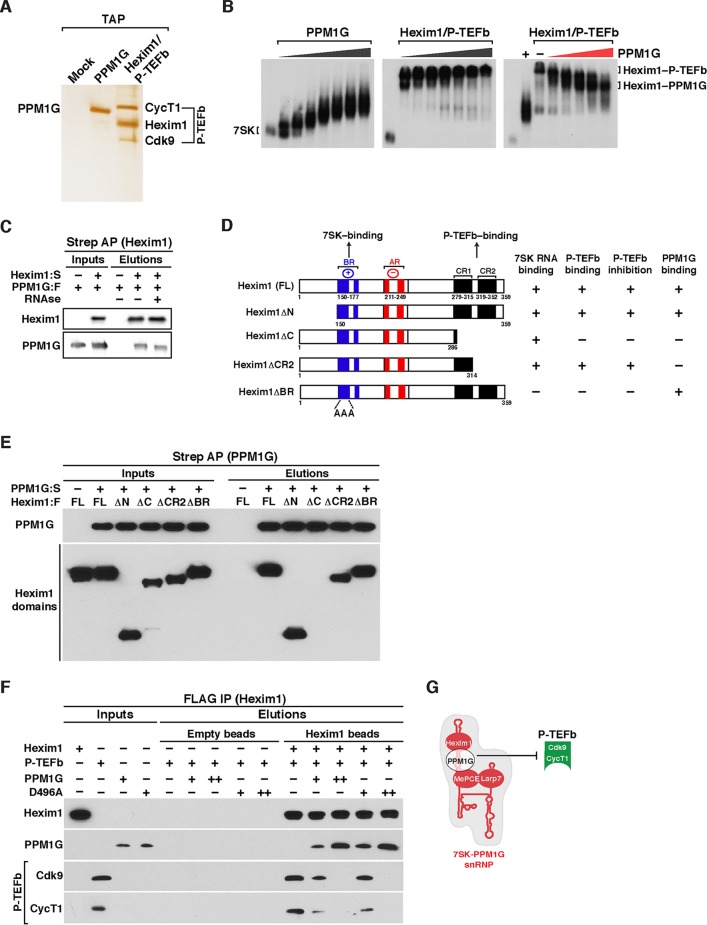
FIG 3.
PPM1G interacts with Hexim1 and assembles onto 7SK RNA to block P-TEFb binding and 7SK snRNP formation. (A) Affinity purification of Strep-tagged PPM1G and Hexim1–P-TEFb (FLAG-Hexim1, Strep-CycT1, and untagged Cdk9). Proteins were electrophoresed on an SDS-PAGE gel and silver stained. (B) Gel shift assays with full-length 7SK RNA and increasing amounts of PPM1G (left), the Hexim1/P-TEFb complex (middle), and 7SK RNA-bound Hexim1-P-TEFb with increasing amounts of PPM1G (right). (C) Hexim1 binds PPM1G. FLAG-tagged PPM1G (PPM1G:F) was transfected with (+) or without (−) Strep-tagged Hexim1 (Hexim1:S) into HEK 293T cells. Protein lysates treated with (+) or not treated with (−) RNase were used for Strep-Tactin affinity purifications (Strep AP) of Hexim1 and analyzed by Western blotting with the indicated antibodies. (D) Scheme of full-length (FL) Hexim1 and domains alongside their respective biochemical properties in relation to 7SK RNA, P-TEFb, and PPM1G binding (derived from the data shown in panel E). BR denotes the basic region, and AR denotes the acidic region in Hexim1. (E) PPM1G contacts the Hexim1 CR1 domain in vitro. Strep-tagged PPM1G (PPM1G:S) and FLAG-tagged Hexim1 (FL or domains) were purified from HEK 293T cells (inputs). Proteins were used for an in vitro Strep-binding assay where Strep-tagged PPM1G coupled to beads (or empty beads [−]) was incubated with FL Hexim1 or domains in the absence of 7SK RNA, and elutions were analyzed by Western blotting. (F) PPM1G associates with Hexim1 to block P-TEFb binding in vitro. The indicated proteins or protein complexes were bound to beads and incubated with P-TEFb in the presence of 7SK RNA (because it is needed for the Hexim1–P-TEFb interaction) and analyzed by Western blotting. (G) Proposed model where PPM1G associates with Hexim1 on 7SK RNA stem I (referred to as 7SK-PPM1G snRNP) to block Hexim1-mediated P-TEFb recruitment and formation of the 7SK snRNP complex.
However, we noticed that upon the addition of PPM1G and disassembly of P-TEFb from Hexim1, the resulting protein-RNA complex (Fig. 3B, right) migrated more slowly than did the PPM1G-7SK RNA complex (left). Therefore, we speculated that PPM1G might interact with Hexim1 (Hexim1-PPM1G) on 7SK RNA to prevent P-TEFb association and 7SK snRNP formation. To test this possibility, we transfected HEK 293T cells with FLAG-tagged PPM1G with or without Strep-tagged Hexim1 and performed Strep affinity purifications (APs) in the presence or absence of RNase to degrade any possible bridging 7SK RNA interaction. Remarkably, we observed that PPM1G interacts with Hexim1 even in the presence of RNase (Fig. 3C), suggesting that the Hexim1-PPM1G protein-protein interaction is direct and that both proteins can assemble a protein-RNA complex with 7SK RNA (Fig. 3B).
To further define Hexim1 domains involved in PPM1G binding, we performed a domain mapping analysis in vitro (Fig. 3D). The Hexim1 domains used were Hexim1ΔN (containing a deletion of the N-terminal domain including residues 1 to 149), Hexim1ΔC (containing a deletion of the C-terminal domain including residues 287 to 359, which has deletions of both coiled-coil region 1 [CR1] and CR2), Hexim1ΔCR2 (containing a shorter deletion of the C-terminal domain [only CR2] including residues 315 to 359), and Hexim1ΔBR (containing a mutation of the basic region [BR], which impairs 7SK RNA binding) (Fig. 3D). Full-length Strep-tagged PPM1G (PPM1G:S) or empty beads were incubated with the FLAG-tagged full-length (FL) Hexim1 protein or domains expressed and purified from HEK 293 cells in the presence of RNase, and after Strep-Tactin AP, interacting domains were screened by Western blot analysis. We observed that in addition to Hexim1, ΔN, ΔCR2, and ΔBR still interact with Hexim1. However, deletion of both CR1 and CR2 (ΔC construct) did not interact with Hexim1, thereby indicating that the Hexim1 CR1 domain (residues 286 to 314) is responsible for PPM1G binding in the absence of 7SK RNA (Fig. 3E), consistent with the coimmunoprecipitation data shown in Fig. 3C.
Interestingly, it is known that the Hexim1 CR1 domain (Fig. 3D) mediates its interaction with P-TEFb to assemble the 7SK snRNP and inhibit transcription (36). This indicates that the assembly of PPM1G with Hexim1 (through its CR1 domain) may be crucial in preventing the Hexim1–P-TEFb protein-protein interaction. To test this possibility and also to determine if the catalytic activity of PPM1G is required for this effect, we immobilized Strep-tagged Hexim1, Hexim1 bound to FLAG-tagged WT PPM1G or to a catalytically inactive mutant (D496A), or unbound controls (empty beads) and incubated them with purified Strep-tagged P-TEFb in an in vitro binding assay in the presence of 7SK RNA (because of the RNA dependence of the Hexim1–P-TEFb interaction). Interestingly, we found that Hexim1 binds P-TEFb, but conversely, preincubation of Hexim1 with PPM1G or the D496A mutant can block (in a dose-dependent manner) P-TEFb recognition (Fig. 3F), thus providing evidence that the PPM1G interaction with the Hexim1 CR1 domain prevents P-TEFb recruitment in vitro, despite the presence of 7SK RNA. Given that the catalytic activity of PPM1G is required for the release of P-TEFb from the 7SK snRNP complex but is not needed to prevent P-TEFb association with Hexim1, PPM1G may block the P-TEFb-binding site on Hexim1 through steric hindrance.
Together, the data indicate that PPM1G assembles a complex with 7SK RNA and Hexim1 (7SK-PPM1G snRNP), thus blocking the P-TEFb/Hexim1/7SK RNA interaction and 7SK snRNP formation in vitro (Fig. 3G). Below, we elucidate the significance of this molecular interaction in the context of signal-dependent inducible transcriptional programs in vivo.
PPM1G binds 7SK RNA transiently in response to DNA damage.
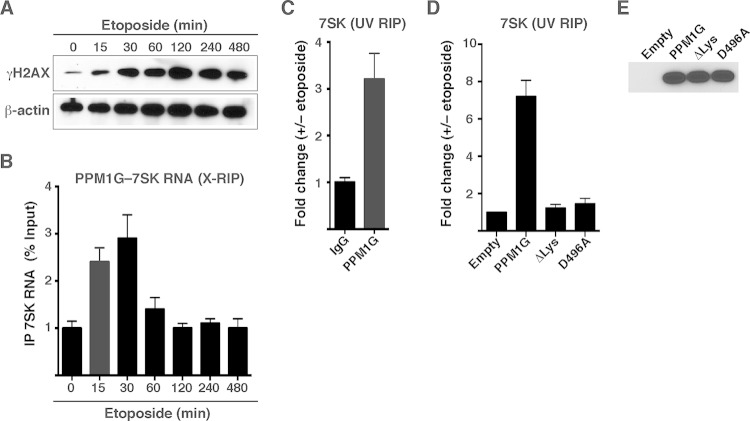
If the PPM1G-7SK protein-RNA interaction was functional in the context of transcription, we would expect to observe changes in this interaction in response to signal-dependent transcription activation. Given that PPM1G is involved in the DDR pathway (32, 33), and NF-κB mediates transcriptional activation in response to DNA damage (47, 48), we further investigated the role of the PPM1G-7SK RNA interaction in the context of transcriptional programs activated in response to genotoxic stress (Fig. 4). To study this, we established an assay that recapitulates DNA damage in tissue cultures using etoposide, a well-known DNA-damaging agent that directly interferes with topoisomerase I DNA unwinding and thus triggers a rapid generation of double-strand breaks (DSBs) (49–51). We used etoposide concentrations that mimic peak plasma levels of circulating chemotherapy agents in cancer patients (∼50 μM) (52) and monitored its DNA-damaging effect by measuring phosphorylation levels of the histone variant H2AX at residue Ser139 (known as γH2AX) (50). In this system, HeLa cells treated with etoposide showed a rapid increase (as early as 15 min posttreatment) and persistence in levels of γH2AX (Fig. 4A), consistent with data from previous reports (47).
FIG 4.
PPM1G interacts directly and transiently with 7SK RNA in response to DNA damage. (A) Etoposide induces rapid DNA damage in HeLa cells. Induction of the DNA damage response (as shown by the accumulation of the DNA damage marker γH2AX) in HeLa cells treated with etoposide over a time course is shown. (B) RIP assay (X-RIP) showing the kinetics of the PPM1G-7SK snRNP interaction in response to etoposide treatment. HeLa cells were treated with etoposide as described above for panel A and cross-linked with formaldehyde, and endogenous PPM1G was immunoprecipitated to monitor the interaction between PPM1G and 7SK RNA by qRT-PCR (means ± standard errors of the means; n = 3). (C) UV RIP showing direct interactions between PPM1G and 7SK RNA in response to etoposide treatment. HeLa cells were treated with etoposide (+) or DMSO (−), endogenous PPM1G was immunoprecipitated after UV treatment, and levels of PPM1G-bound 7SK RNA were quantified by qRT-PCR (means ± standard errors of the means; n = 3). (D) The Lys-rich region of catalytically active PPM1G directs the interaction with 7SK RNA in response to etoposide. WT, ΔLys, and catalytically dead D496A mutant PPM1G proteins or an empty vector was transfected into HeLa cells, and cells were subsequently treated with etoposide and UV cross-linked. Levels of coimmunoprecipitating 7SK RNA were quantified by qRT-PCR (means ± standard errors of the means; n = 3). (E) Western blot analyses of lysates of HeLa cells transfected with an empty vector, Strep-tagged PPM1G, and the ΔLys and catalytically dead (D496A) mutants, as described above for panel D.
Using this approach, we first asked whether the PPM1G-7SK RNA interaction is modulated in cells in response to etoposide. To monitor the protein-RNA interaction in cells, we used an RNA immunoprecipitation (X-RIP) assay by fixing etoposide-treated cells with formaldehyde to trap in vivo interactions and prevent the reassortment of protein-RNA complexes in vitro (39, 53). Interestingly, we observed that etoposide causes a rapid (15 to 30 min) increase (∼2.5- to 3-fold) in bulk levels of PPM1G-7SK RNA interactions (Fig. 4B) that temporally correlate with the onset of γH2AX phosphorylation (Fig. 4A). Unlike γH2AX, the interaction between PPM1G and 7SK RNA was rapidly reversed after 60 min of etoposide treatment (Fig. 4B), demonstrating that PPM1G is inducibly recruited to 7SK RNA after DNA damage and that the interaction is reversible.
Despite the observed interaction between PPM1G and 7SK RNA in cells using X-RIP, this assay does not provide evidence of direct protein-RNA interactions. To test whether PPM1G binds 7SK RNA directly, we performed UV cross-linking RIP (UV-RIP) (39) followed by RNA quantitation using qRT-PCR. With this method, which specifically cross-links proteins to nucleic acids in vivo, we observed that endogenous PPM1G coimmunoprecipitates with 7SK RNA in response to a short (30-min) etoposide treatment (Fig. 4C), demonstrating a direct interaction between PPM1G and 7SK RNA in response to the stimulus. To further provide evidence that the Lys-rich region in PPM1G is responsible for directly contacting 7SK RNA, we transfected HEK 293T cells with Strep-tagged PPM1G, ΔLys, or an empty plasmid and measured the levels of coimmunoprecipitated 7SK RNA by qRT-PCR. Remarkably, we observed that deletion of the Lys-rich domain (PPM1GΔLys) abolishes 7SK RNA binding in response to etoposide treatment (Fig. 4D) without affecting protein steady-state levels (Fig. 4E). These data demonstrate that the polybasic motif is necessary and sufficient for 7SK RNA binding in cells in response to DNA damage, in agreement with our in vitro interaction data (Fig. 1).
To define whether the catalytic activity of PPM1G is required for binding 7SK RNA in response to etoposide, we transfected cells with a catalytically dead phosphatase mutant (D496A) and observed that this nonfunctional mutant is unable to bind RNA in response to stimuli in cells (Fig. 4D), even though it is correctly expressed (Fig. 4E). However, in vitro, the binding affinity of the recombinant D496A mutant was similar to that of WT PPM1G (see Fig. S2 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html). Together, the data suggest that in response to a stimulus, both molecular events (PPM1G-mediated release of P-TEFb from 7SK snRNP and 7SK RNA binding post-kinase release) are temporally coupled.
PPM1G regulates NF-κB-mediated 7SK RNA-dependent Pol II transcription in response to DNA damage.
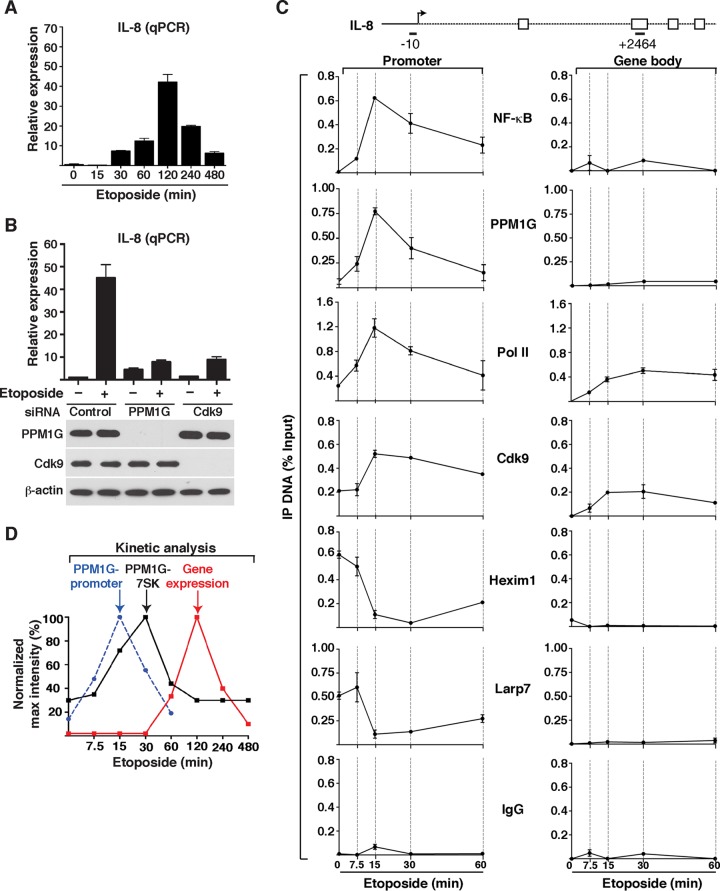
Given that DNA damage activates the NF-κB signaling pathway and that PPM1G is a transcriptional coactivator of NF-κB, we investigated the role of the PPM1G-7SK RNA interaction in the context of transcription activation in response to DNA damage. Upon the use of induction stimuli, NF-κB translocates from the cytoplasm to the nucleus to activate proinflammatory genes in a P-TEFb-dependent manner (15, 26). Consistent with data from those reports, several NF-κB target genes, such as the IL-8 and tumor necrosis factor alpha (TNF-α) genes, are rapidly activated in HeLa cells in response to etoposide without interfering with the expression of nonresponsive, or housekeeping, genes (Fig. 5A and data not shown). Importantly, in the presence of a paninhibitor that blocks IKK (inhibitor of κB kinase), an upstream activating kinase of the NF-κB signaling pathway, IL-8 is not activated in response to etoposide, demonstrating that NF-κB is indeed the master transcriptional regulator (see Fig. S3 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html). Synthesis of IL-8 RNA increases gradually upon etoposide treatment, with a maximum at 120 min posttreatment. Therefore, we used this time point to examine the effect of PPM1G knockdown (KD) on IL-8 gene expression. Expectedly, PPM1G knockdown decreased IL-8 gene expression in response to etoposide by ∼5-fold (Fig. 5B). If PPM1G functions through the 7SK snRNP to activate IL-8 gene transcription, we would expect that the KD of P-TEFb affects IL-8 induction in response to etoposide. We found that RNAi-mediated KD of Cdk9, using a previously validated siRNA (26), abolished (like PPM1G KD) IL-8 induction in response to etoposide but without affecting basal levels (Fig. 5B). Together, these data indicate that IL-8 expression in response to DNA damage is regulated by both PPM1G and P-TEFb.
FIG 5.
PPM1G controls transcription of 7SK snRNP-regulated NF-κB target genes in response to DNA damage. (A) Transcription of NF-κB target genes (IL-8 gene) in response to etoposide. HeLa cells were treated with etoposide over a time course, and RNA was isolated for quantitation of gene expression by qRT-PCR. The expression level of IL-8 was normalized to that of β-actin (means ± standard errors of the means; n = 3). (B) KD of PPM1G and Cdk9 abolishes transcription of the IL-8 gene in response to etoposide. HeLa cells were transfected with nontarget control, PPM1G, or Cdk9 siRNAs for 48 h to knock down the indicated proteins, followed by treatment with etoposide (+) or DMSO (−) for 120 min. Total RNA was extracted, and IL-8 gene expression was quantified by qRT-PCR (means ± standard errors of the means; n = 3). The bottom panel shows Western blotting for validation of the RNAi-mediated KD. (C) ChIP assay showing levels of NF-κB, PPM1G, Pol II, and 7SK snRNP components (Cdk9, Hexim1, and Larp7) at the IL-8 promoter and gene body in response to a time course of etoposide treatment (means ± standard errors of the means; n = 3). Normal serum (IgG) was used as a negative control to demonstrate the specificity of enrichment with the antibodies used. (D) Kinetic plots for IL-8 gene expression (as determined by qRT-PCR as described above for panel A), PPM1G recruitment to the IL-8 promoter (as determined by ChIP as described above for panel C), and PPM1G-7SK RNA interactions (as determined by an X-RIP assay as described for Fig. 4B) in HeLa cells in response to etoposide. Note that the time scale (x axis) is on a log2 scale for better data visualization.
To test whether PPM1G and NF-κB (RelA/p65 subunit) are corecruited to the IL-8 promoter in response to etoposide, we performed ChIP assays on HeLa cells treated with etoposide for various times (7.5, 15, 30, and 60 min) preceding peak RNA synthesis (120 min) (Fig. 5A). Similar to the increased NF-κB occupancy detected at the IL-8 promoter upon etoposide treatment, there was a rapid and transient recruitment of PPM1G with a peak at 15 min (Fig. 5C), which precedes the kinetics of PPM1G-7SK RNA interaction (peak at 30 min) (Fig. 4B) and IL-8 gene expression (peak at 120 min) (Fig. 5A). The profile of PPM1G recruitment to the IL-8 locus in response to DNA damage is specific for the promoter because we were unable to detect significant enrichment over background levels at two promoter-distal regions located either upstream of the transcription start site (TSS) (position −2891) or within the IL-8 gene (position +2464) (see Fig. S4 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html).
Like NF-κB and PPM1G, Pol II and Cdk9 were strongly recruited to the IL-8 promoter, with their levels peaking at 15 min after-etoposide treatment and gradually resetting to those under basal conditions. Expectedly, Pol II levels at the gene body in the absence of stimuli were undetectable. However, both Pol II and Cdk9 (which travels with Pol II) levels were increased in the gene body in response to etoposide (Fig. 5C), in agreement with increased transcription elongation in response to stimuli. PPM1G and NF-κB were not detected at the gene body, thus revealing that they are recruited to the promoter only in response to stimuli and do not travel with elongating Pol II (Fig. 5C).
Furthermore, the occupancy of the inhibitory 7SK snRNP components (Hexim1 and Larp7) at the promoter decreased significantly with etoposide treatment (∼2- to 8-fold depending on the time point evaluated) and was temporally correlated with PPM1G recruitment (Fig. 5C). The snRNP complex was detected at the promoter but not at the gene body, consistent with previously reported data (25–27). More importantly, the peak of 7SK snRNP eviction from the promoter was observed after PPM1G recruitment (15 to 30 min) but before the maximum increase in the formation of the 7SK-PPM1G snRNP complex, thus providing evidence that these molecular events are intimately linked, both kinetically and functionally.
Of note, we found that low levels of P-TEFb and the 7SK snRNP (Hexim1 and Larp7) were detected at the promoter before stimulation, probably to rapidly promote the first round of transcription elongation in response to etoposide. However, increased levels of Cdk9 were detected at the promoter in response to the stimuli but without an evident increase in the 7SK snRNP level. The fact that more Cdk9 was detected during activation is consistent with the model where additional P-TEFb is recruited to the promoter from the nucleoplasmic pool to stimulate multiple rounds of elongation. Because the 7SK snRNP complex is enzymatically disassembled at the promoter, only Cdk9 that is loaded onto Pol II, but not the inhibitory snRNP components (Hexim1 and Larp7), is readily detected (Fig. 5C).
In summary, the kinetics of PPM1G recruitment to the IL-8 gene promoter in response to etoposide precedes the increase in bulk levels of the PPM1G-7SK RNA complex and peak gene expression (Fig. 5D). Interestingly, both PPM1G-promoter and PPM1G-7SK RNA interaction steps precede RNA synthesis, are transient and reversible, and show a similar unimodal “binding/unbinding” distribution (Fig. 5D). These results are consistent with a model in which PPM1G couples the disassembly of the promoter-bound 7SK snRNP with 7SK RNA binding to initiate and sustain the NF-κB transcriptional program in response to DNA damage.
PPM1G functions downstream of NF-κB and Pol II recruitment to gene promoters to induce 7SK snRNP disassembly and Pol II elongation in response to DNA damage.
Given that PPM1G appears to be important in the induction of P-TEFb-dependent NF-κB target genes, such as the IL-8 gene, in response to DNA damage (Fig. 5), we examined the functional consequences of the loss of PPM1G on the activation kinetics of these genes (Fig. 6). Because excessive siRNA-mediated knockdown of PPM1G in HeLa cells causes abnormal growth, which impeded us from obtaining a large number of cells, we switched to an inducible-RNAi approach (see Materials and Methods). In this system, HeLa cells were transduced with self-inactivating lentiviruses expressing IPTG-inducible nontarget shRNA (NTsh) or PPM1G shRNA (PPM1Gsh). After 72 h of IPTG induction of shRNAs, cells were treated with etoposide for different times (0, 7.5, 15, 30, and 60 min) to analyze effects at the level of gene expression and the dynamics of NF-κB, PPM1G, 7SK snRNP, and Pol II recruitment to target genes.
Despite the efficient PPM1G KD, we did not observe profound changes in bulk protein levels for the transcription factor NF-κB and cofactors required for target gene activation, including Pol II, Cdk9, Hexim1, and Larp7 (Fig. 6A). Importantly, the etoposide treatment functioned as expected based on the appearance of the DNA damage marker γH2AX. Different from the NTsh cell line, the PPM1Gsh line showed a slight increase in the level of γH2AX in the absence of etoposide treatment (time zero), owing to PPM1G's role in endogenous DNA repair (33; R. P. McNamara and I. D'Orso, unpublished data). Despite these slight increases in endogenous DNA damage as well as basal IL-8 RNA levels (∼3.2-fold [data not shown]), which were similar to those in the siRNA-mediated RNAi experiment (Fig. 5B), PPM1G KD largely reduces IL-8 expression over time (∼4-fold) in response to etoposide (Fig. 6B).
Having established the inducible RNAi system to define the importance of PPM1G in the induction of NF-κB target genes in response to DNA damage, we asked whether the loss of PPM1G affects 7SK snRNP disassembly at the promoter and Pol II elongation, thus conferring reduced induction in response to stimuli (Fig. 6B). To examine this, we performed detailed analysis of the kinetics of NF-κB and cofactor occupancy at the IL-8 locus in both the NTsh and PPM1Gsh cell lines. Expectedly, PPM1G was rapidly recruited (15 min) to the IL-8 promoter but not to the gene body in response to etoposide in the NTsh, but not in the PPM1Gsh, cell line. These data are consistent with the ChIP data for HeLa cells in the absence of RNAi (Fig. 5C). Importantly, while the dynamics of NF-κB and Pol II recruitment to the promoter remained mostly unaffected, Cdk9 levels were increased (∼3- to 5-fold at between 15 and 30 min post-etoposide treatment) at the IL-8 promoter in response to DNA damage only in the NTsh cell line, indicating that 7SK snRNP disassembly promotes further Cdk9 recruitment for multiple rounds of elongation. Consistently, the snRNP (Hexim1 and Larp7) was normally disassembled at the promoter of the NTsh cell line in response to etoposide, but levels remained more constant in the PPM1Gsh cell line, indicating that in the absence of PPM1G, the 7SK snRNP is not properly ejected, and levels of Cdk9 (promoter-bound 7SK snRNP) remain low. Even when Pol II recruitment to the promoter remained very similar, Pol II did not efficiently transition into the elongation phase because the level of Pol II occupancy in the gene body in PPM1Gsh cells was ∼3-fold lower than that in NTsh cells (Fig. 6C). In addition, the ChIP data demonstrate that NF-κB, PPM1G, and 7SK snRNP binding is restricted to the IL-8 promoter but not to the gene body. As a ChIP control, normal IgG showed no enrichment in the promoter or gene body, thus revealing the specificity of the antibodies used.
Collectively, the data indicate that the loss of PPM1G interferes with Pol II transition into the elongation phase because of reduced 7SK snRNP disassembly and Cdk9 activity at the promoter. However, importantly, the loss of PPM1G does not affect NF-κB and Pol II recruitment to the promoter in response to the stimuli.
PPM1G is required for NF-κB-mediated Pol II transcription elongation at other target genes.
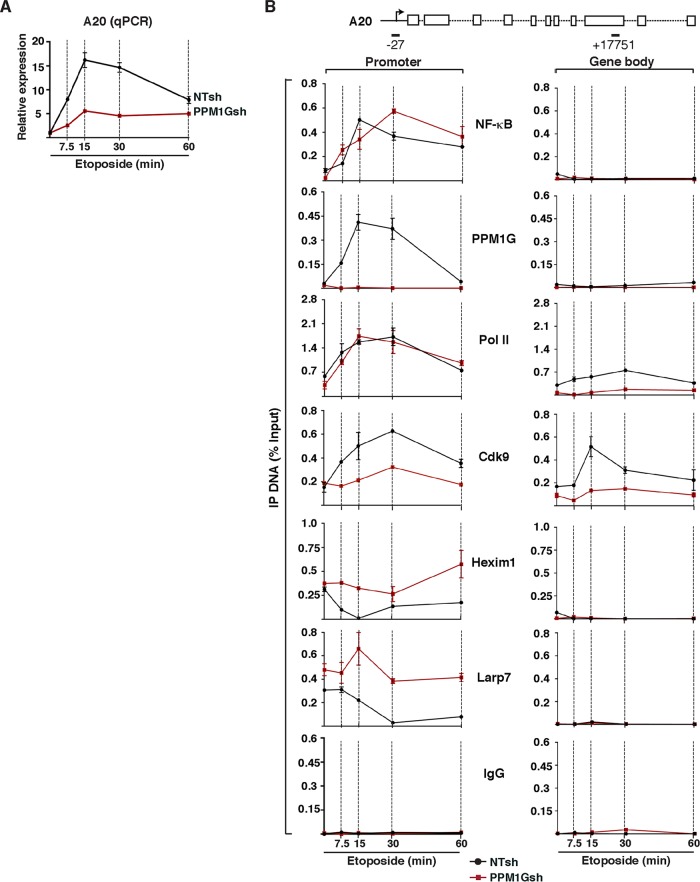
Given that we found a critical role for PPM1G in the NF-κB-mediated induction of IL-8 in response to etoposide, we asked whether other target genes are also regulated by a similar mechanism. We selected a few target genes, including the A20/TNFAIP3 gene (which encodes a deubiquitinase required to shut off the NF-κB signaling pathway) and analyzed their activation in the inducible RNAi system discussed above (NTsh and PPM1Gsh). Remarkably, we found that A20 is rapidly induced by etoposide (peaking at 15 min) in the NTsh cell line but that its expression is virtually impaired in the PPM1Gsh cell line (∼3- to 4-fold decrease compared with the control) (Fig. 7A). Of note, A20 was more rapidly induced than IL-8, with a peak at 15 to 30 min post-etoposide treatment.
FIG 7.
PPM1G is required for NF-κB-mediated Pol II transcription elongation at the A20 gene in response to etoposide. (A) PPM1G KD abolishes A20 gene expression in response to etoposide. The cell lines from Fig. 6A (HeLa NTsh and PPM1Gsh) were used for total RNA extraction, and A20 gene expression in response to etoposide was quantified by qRT-PCR (means ± standard errors of the means; n = 3). The data were normalized to levels of the β-actin transcript. (B) ChIP assay of cell lines from Fig. 6A showing levels of NF-κB, PPM1G, Pol II, and 7SK snRNP components (Cdk9, Hexim1, and Larp7) at the A20 promoter-proximal region (−27 amplicon) and gene body (+17751 amplicon) in response to a time course of etoposide treatment (means ± standard errors of the means; n = 3). Normal serum (IgG) was used as a negative control to demonstrate the specificity of enrichment with the antibodies used.
Because PPM1G appears to be involved in the NF-κB-mediated induction of A20 in response to etoposide, we performed ChIP assays to examine NF-κB, PPM1G, 7SK snRNP, and Pol II recruitment dynamics at two sites within the A20 locus (promoter and gene body). Similar to the results with the IL-8 gene, the loss of PPM1G did not have much of an effect on the kinetics or levels of NF-κB and Pol II recruitment to the promoter-proximal region, but Pol II levels in the gene body were increased (∼2- to 3-fold) in response to etoposide only in the NTsh cell line, indicating that PPM1G is required for the Pol II transition to the elongation phase. Expectedly, PPM1G was recruited to the A20 promoter region only in the NTsh, but not in the PPM1Gsh, cell line (Fig. 7B). Moreover, while the loss of PPM1G did not affect the levels of Cdk9 and inhibitory 7SK snRNP components (Hexim1 and Larp7) recruited to the A20 promoter in the absence of stimuli, the ejection of the snRNP components from the promoter in response to etoposide was mostly impaired, consistent with the loss of further Cdk9 recruitment (probably from the nucleoplasmic pool) to the promoter. In agreement, Cdk9 levels detected in the gene body in response to etoposide dropped between ∼1.5- and 5-fold (depending on the time of treatment) in the PPM1Gsh cell line. Similar to the ChIP data for the IL-8 locus, normal IgG serum showed a very low level of enrichment at the A20 loci, thus revealing the specificity of all the antibodies used (Fig. 7B).
Together, we have identified two NF-κB target genes that are controlled by PPM1G at the elongation step, thus suggesting that this regulatory mechanism might be more common than expected. Undoubtedly, further genome-wide experiments are required to determine how widespread this regulatory mechanism is and to identify all target genes within the NF-κB transcriptional program.
PPM1G-7SK RNA interaction and PPM1G-dependent NF-κB transcription in response to DNA damage are regulated by the ATM kinase.
PPM1G has been shown to play a major role in the DDR pathway, where ataxia telangiectasia mutated (ATM) kinase-dependent phosphorylation of PPM1G is required for its activation (33, 54). ATM activation and phosphorylation of its substrates are two of the primary responses to DNA damage. Through ATM signaling, the cell activates multiple pathways to facilitate DSB repair, activation of cell cycle checkpoints, and induction of NF-κB-activated prosurvival genes (48, 55–57). We therefore asked whether NF-κB-mediated IL-8 gene activation in response to etoposide treatment is controlled by the ATM kinase. To assess this, we treated HeLa cells with a well-characterized ATM inhibitor (ATMi) (KU55933) or vehicle (DMSO) for 120 min, followed by treatment with etoposide or DMSO for different times (Fig. 8A). To validate that ATMi indeed inhibited ATM activity, we monitored the γH2AX signal in response to short-term etoposide treatment. Indeed, pharmacological inhibition of ATM virtually abolished γH2AX after etoposide treatment (Fig. 8B), consistent with previously reported data (47). Using this system, we monitored IL-8 gene expression in the presence of etoposide or DMSO in cells pretreated or not pretreated with ATMi by qRT-PCR (Fig. 8A). Expectedly, treatment with ATMi drastically abrogated transcription from the IL-8 promoter in response to DNA damage (Fig. 8C; see also Fig. S5 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html), thus providing evidence that NF-κB-mediated IL-8 induction upon genotoxic stress requires the ATM kinase.
FIG 8.
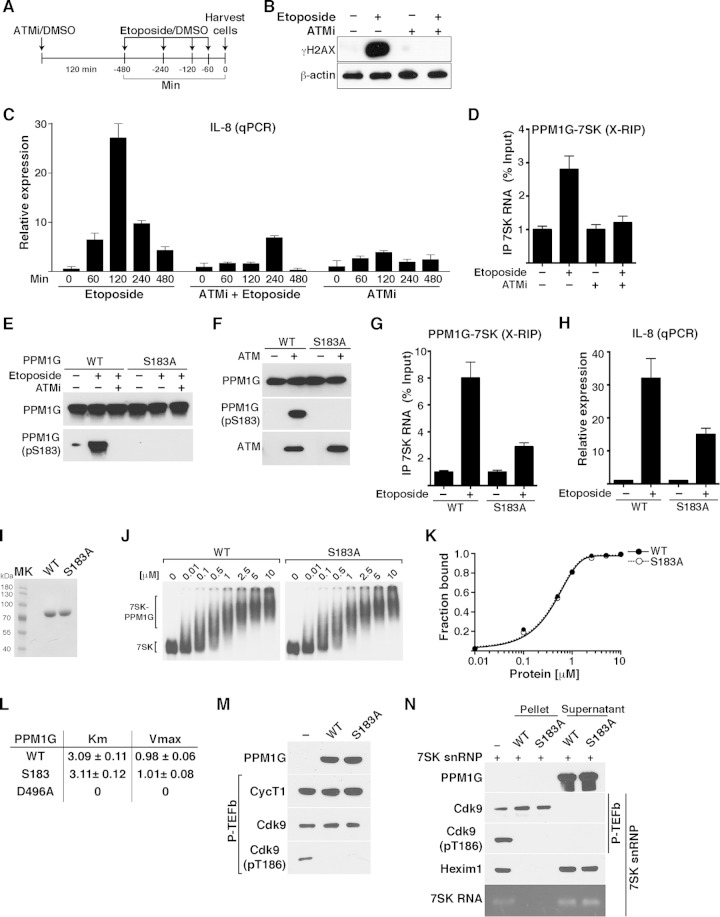
PPM1G-7SK RNA interaction and activation of NF-κB transcription in response to DNA damage are ATM kinase dependent. (A) Scheme showing the protocol used to pretreat HeLa cells with ATMi (or DMSO as a control) for 120 min, followed by a time course of etoposide treatment or DMSO. (B) Validation of ATM inhibition with an ATMi by Western blotting with γH2AX antibody. HeLa cells were pretreated for 120 min with an ATMi or DMSO as a control, followed by a 120-min etoposide or DMSO treatment. Cell pellets were used for Western blotting with the indicated antibodies (β-actin was used as a loading control). (C) ATM inhibition blocks NF-κB transcription (IL-8 gene) from HeLa cells in response to etoposide. Cells were pretreated with an ATMi or vehicle (DMSO) for 120 min and sequentially treated with etoposide or vehicle (DMSO) for the time points indicated. Total RNA was then extracted, and the IL-8 gene expression level, normalized to the β-actin level, was calculated by qRT-PCR (means ± standard errors of the means; n = 3). (D) ATM inhibition abolishes the PPM1G-7SK protein-RNA interaction in response to etoposide. HeLa cells were pretreated (+) or not pretreated (−) with an ATMi (120 min), followed by a 30-min treatment with etoposide or DMSO (−). Cells were cross-linked with formaldehyde, and endogenous PPM1G was immunoprecipitated to quantify the levels of coprecipitated 7SK RNA in response to etoposide by qRT-PCR (means ± standard errors of the means; n = 3). (E) PPM1G is phosphorylated at Ser183 (S183) in response to etoposide, and an ATMi blocks site-specific PPM1G phosphorylation (pS183) upon etoposide treatment. HeLa cells were transfected with Strep-tagged WT PPM1G or the S183A mutant and treated as described above for panel D. Proteins were affinity purified and used for Western blot assays with the indicated antibodies. (F) PPM1G is phosphorylated by the ATM kinase at S183. Strep-affinity-purified WT PPM1G or the S183A mutant was incubated with (+) or without (−) ATM kinase, and Western blot analyses (total PPM1G and the pS183 form) were performed. (G) Mutation of the Ser183 residue (S183A) in PPM1G reduces the PPM1G-7SK RNA interaction in response to etoposide. HeLa cells were transfected with Strep-tagged WT PPM1G or the S183A mutant, treated with etoposide (+) or DMSO (−) for 30 min, and cross-linked with formaldehyde, and a RIP assay (X-RIP) was performed to quantitate the association between PPM1G and 7SK RNA by using qRT-PCR (means ± standard errors of the means; n = 3). (H) Mutation of Ser183 in PPM1G (S183A) reduces activation of IL-8 gene expression in response to etoposide. HeLa cells were transfected with siRNA-resistant (HS_PPM1G_6 [see Table S3 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html]) Strep-tagged WT PPM1G or the S183A mutant and retransfected 24 h later with a PPM1G siRNA to knock down endogenous PPM1G. Forty-eight hours later (at which time the PPM1G KD level was ∼80%), cells were treated with etoposide (+) or DMSO (−) for 120 min, RNA was extracted, and IL-8 gene expression was quantified by qRT-PCR and normalized to the β-actin level (means ± standard errors of the means; n = 3). (I) Strep-tagged WT PPM1G and S183A mutant proteins were affinity purified and visualized by Coomassie staining. MK, protein molecular size marker. (J) Gel shift assays with 7SK RNA and increasing amounts of WT PPM1G or the S183A mutant. (K) Binding curves for the gel shifts shown in panel J. (L) Determination of the kinetic parameters Km and Vmax for WT PPM1G, the S183A mutant, and the catalytically dead mutant (D496A) on the phosphatase substrate pNPP. (M) Purified P-TEFb was incubated with WT PPM1G or the S183A mutant under dephosphorylation conditions, and Cdk9 T-loop phosphorylation at Thr186 (pT186) was monitored by Western blotting. (N) The S183A mutation in PPM1G does not affect the enzymatic release of P-TEFb from the 7SK snRNP complex. 7SK-bound P-TEFb complexes were incubated with WT PPM1G or the S183A mutant under dephosphorylation conditions. Subsequent purification of P-TEFb using FLAG beads was done to monitor released (supernatant) and retained (pellet) components by Western blotting.
The idea that ATM is required for the phosphorylation of PPM1G at specific SQ/TQ motifs (54, 58) prompted us to test if ATM regulates the PPM1G-7SK RNA interaction. To test this hypothesis, we performed an X-RIP assay on cells treated with etoposide in the presence and absence of an ATMi. Interestingly, we found that upon etoposide treatment, PPM1G's inducible recruitment to 7SK RNA is lost in the presence of ATMi (Fig. 8D). This supports the model that the ATM kinase plays an important role in regulating the interaction between PPM1G and 7SK RNA (as part of the inhibitory snRNP) in response to DNA damage.
Previous studies reported that etoposide triggers PPM1G phosphorylation at residue S183, which is part of a canonical ATM-regulated SQ motif (33). To investigate the functional relevance of this posttranslational modification and the role of ATM in the PPM1G-7SK RNA interaction, we further evaluated the mechanisms. First, to test whether PPM1G is phosphorylated at S183 (pS183) in response to etoposide, we transfected HeLa cells with Strep-tagged WT PPM1G or the S183A mutant and examined their expression and relative site-specific phosphorylation by Western blotting. Interestingly, we found that etoposide triggers rapid (30 min) phosphorylation of PPM1G at residue S183 using a phospho-specific antibody (pS183), but pretreatment with an ATMi completely eliminated this event (Fig. 8E). As a control, we transfected a point mutant (S183A) and observed no phosphorylation, thus revealing the specificity of the antibody used. To further evaluate the role of ATM in this pathway, we performed an in vitro kinase assay where the Strep-tagged WT PPM1G or S183A mutant protein was incubated with ATM and monitored PPM1G phosphorylation by Western blotting with the phospho-specific PPM1G antibody (pS183). In agreement with the data from the pharmacological inhibition experiment, ATM indeed mediated PPM1G phosphorylation at S183, and mutation of this residue (S183A) abrogated detection with the phospho-specific antibody (Fig. 8F).
Our above-described findings suggested that ATM regulates the PPM1G-7SK RNA interaction in response to DNA damage (Fig. 8D). To evaluate the functional role of ATM-mediated site-specific PPM1G phosphorylation, we transfected HeLa cells with the Strep-tagged WT PPM1G or S183A mutant protein, treated the cells with etoposide or DMSO, and performed X-RIP assays to monitor levels of PPM1G-7SK RNA interactions. We found that PPM1G-7SK RNA levels increase ∼8-fold upon short-term etoposide treatment and that the S183A mutation decreased the protein-RNA interaction to ∼2.5-fold, demonstrating that ATM-mediated S183 phosphorylation on PPM1G is an important regulatory step in the mechanism of interaction between PPM1G and 7SK RNA in cells (Fig. 8G). To further examine the role of this mutation in the activation of NF-κB transcription, we performed an RNAi rescue assay in which the siRNA-resistant WT PPM1G and S183A mutant proteins were transfected after knockdown of endogenous PPM1G. While the ectopic expression of WT PPM1G rescued IL-8 induction, the S183A mutant played a dominant negative role in blocking the activation of this NF-κB target gene (Fig. 8H). Together, we provide compelling evidence that ATM kinase promotes the PPM1G-7SK snRNP interaction through site-specific PPM1G phosphorylation to facilitate NF-κB gene activation in response to DNA damage.
To further investigate the requirement for the S183 residue in the context of P-TEFb dephosphorylation/release from the snRNP complex and 7SK RNA binding, we affinity purified the WT and the S183A mutant (Fig. 8I). Using an in vitro binding assay and gel shift analysis, we observed that both the WT and S183A proteins bind 7SK RNA with nearly identical affinities (Kdapp values of 0.49 and 0.48 μM, respectively) (Fig. 8J and K), similar to the catalytically dead D496A mutant (see Fig. S2 at http://www.utsouthwestern.edu/labs/dorso/research/lab-projects.html). Taken together, while the S183 residue does not play any direct role in PPM1G's ability to bind 7SK RNA in vitro, it serves as a site of ATM phosphorylation to regulate 7SK snRNP binding in vivo in response to DNA damage.
Since PPM1G catalytic activity is required for 7SK RNA binding post-P-TEFb release from the snRNP complex (Fig. 4D), we next further examined whether the reduced binding of the S183A mutant to 7SK RNA in response to etoposide is due to its compromised catalytic activity (Fig. 8G). To test this, we performed kinetic experiments using para-nitrophenyl phosphate (pNPP), a widely used phosphatase substrate, to determine whether the S183 mutation has altered kinetic parameters (Km and Vmax). We observed that both the WT and S183A proteins have very similar, if not identical, Km and Vmax values (Fig. 8L), in contrast to the D496A mutant, demonstrating that S183 does not contribute to the catalytic activity of the phosphatase on this substrate.
Because pNPP is an artificial substrate, we examined the efficiency of both the WT and the S183A mutant on a natural substrate (Cdk9) as part of the P-TEFb complex and found that both proteins can dephosphorylate the T-loop (pT186) of Cdk9 after 30 min of incubation with the substrate (Fig. 8M).
Given that Cdk9 T-loop dephosphorylation is required for P-TEFb's release from the 7SK snRNP, we examined whether the S183A mutant can couple the dephosphorylation of Cdk9's T-loop with its release from the inhibitory snRNP. To test this possibility, we performed an in vitro dephosphorylation-release assay as previously described (26). Purified snRNP was incubated with the WT or the S183A mutant under dephosphorylation conditions, and P-TEFb was then purified by using FLAG beads to separate bound and unbound Cdk9 (pellet and supernatant, respectively). Using this assay, we found that mutation of the S183 residue in PPM1G does not alter its ability to dephosphorylate/release the P-TEFb kinase from the snRNP complex (Fig. 8N) or its 7SK RNA-binding potential (Fig. 8J and K).
Collectively, the above-described data support a model in which ATM-mediated phosphorylation of PPM1G at S183 is required for the recruitment of PPM1G to the 7SK snRNP to regulate NF-κB-mediated Pol II transcription elongation in response to DNA damage.
Functional interplay between PPM1G, the 7SK snRNP, and the ATM kinase during activation of the NF-κB transcriptional program in response to DNA damage.
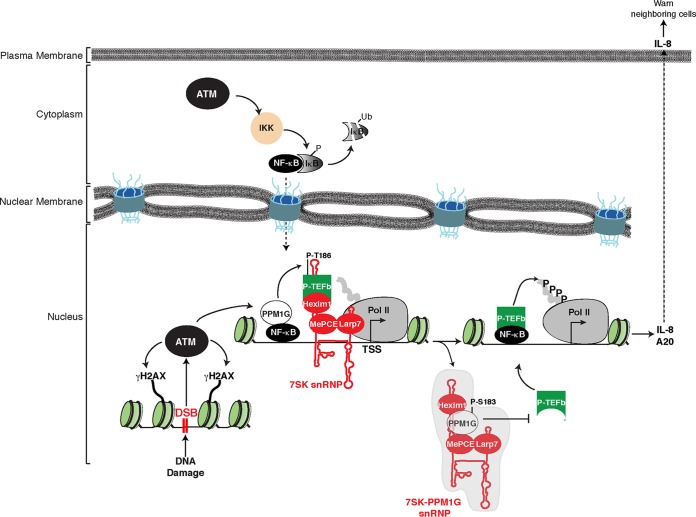
Our findings are consistent with a model in which an initial DNA damage insult (DSB) is recognized and the ATM kinase is activated, which in turn phosphorylates a number of substrates, including the histone variant H2AX (γH2AX), at the site of DNA damage (Fig. 9). Concurrently, ATM phosphorylates and activates IKK in the cytoplasm, which in turn phosphorylates the NF-κB inhibitor (IκB), triggering its ubiquitination (Ub) and proteasomal degradation and the translocation of NF-κB to the nucleus (56). In this subcellular compartment, NF-κB recruits its transcriptional coactivator PPM1G to target gene promoters (such as the IL-8 and A20 promoters) to dephosphorylate the 7SK snRNP-bound Cdk9 (P-T186) (26), thus releasing P-TEFb to induce phosphorylation of the Pol II CTD, which is paused in the vicinity of the transcription start site (TSS). Simultaneously, once P-TEFb is released from the 7SK snRNP complex and the inhibitory subunits are ejected from the promoter, PPM1G undergoes site-specific phosphorylation (P-S183), which promotes the assembly of the 7SK-PPM1G snRNP. In this protein-RNA complex, PPM1G interacts with Hexim1 (at its P-TEFb-binding domain), thereby preventing the kinase-inhibitory function of Hexim1. These interactions directly antagonize the P-TEFb reassociation with 7SK RNA and snRNP complex formation, thereby sustaining the transcriptional program (multiple rounds of elongation) until the insult is resolved. Importantly, a combination of the these molecular events leads to transcriptional activation and maintenance of several NF-κB proinflammatory genes, resulting in the secretion of proinflammatory cytokines, such as IL-8, which alert neighboring cells of the damage, and the expression of factors such as A20/TNFAIP3, which participate in a negative-feedback loop, leading to an attenuation of the NF-κB signaling/transcription pathway (59).
FIG 9.
Model depicting the functional interplay between PPM1G, the 7SK snRNP, and ATM kinase during the activation of the NF-κB transcriptional program in response to DNA damage. In response to double-strand breaks (DSB), ATM activates the NF-κB signaling pathway through phosphorylation of the NEMO subunit of the IKK complex, which in turn phosphorylates the NF-κB inhibitor (IκB), leading to its ubiquitination (Ub) and proteasomal degradation. NF-κB translocates to the nucleus, where it recruits ATM-phosphorylated PPM1G to target genes such as the IL-8 gene, dephosphorylating and releasing the P-TEFb kinase from the promoter-bound 7SK snRNP complex. Upon its release, P-TEFb phosphorylates (P) paused Pol II in proximity to the transcription start site (TSS) to promote transcriptional pause release. After releasing P-TEFb, the inhibitory snRNP subunits are evicted from chromatin, and phosphorylated PPM1G binds 7SK RNA along with Hexim1 to prevent the reassociation of P-TEFb back into the snRNP to sustain transcription elongation. Once the damage is resolved, this regulatory circuitry subsides, PPM1G is dislodged from the snRNP, and P-TEFb is recycled back to promote the formation of the inhibitory 7SK snRNP at the promoter, thereby blocking Pol II pause release.
Collectively, we have identified a transcriptional mechanism involving a master regulator (the ATM kinase), a substrate of ATM (the PPM1G phosphatase), and a noncoding RNA-protein complex (the 7SK snRNP), which participate in a novel transcriptional regulatory network activated in response to DNA damage.
DISCUSSION
The relevance of P-TEFb in the activation of transcription elongation programs is undisputed, yet the mechanisms through which it maintains the active transcriptional state are poorly understood. In particular, it is unclear how P-TEFb is kept in the kinase-active, 7SK-unbound state, once it becomes released from the inhibitory snRNP complex during transcription activation. In this article, we provide genetic and biochemical evidence of a molecular mechanism in which the PPM1G phosphatase (a novel transcriptional coactivator [26]) regulates P-TEFb availability for transcription elongation after mediating the disassembly of the inhibitory 7SK snRNP complex. While other factors functioning to promote transcription elongation through the 7SK snRNP were recently identified, such as the JMJD6 demethylase, the bromodomain-containing protein BRD4, SR splicing factors, and the RNA helicase DDX21 (27–29), our findings provide the first evidence that a cellular enzyme functions to both activate and maintain transcription elongation programs. In addition to the previously reported roles for PPM1G in histone exchange, splicing, and DNA damage repair (32, 40, 54), our findings indicate that PPM1G also plays a critical role in the transcriptional regulation of NF-κB-mediated Pol II transcription.
We demonstrated that in response to DNA damage, PPM1G is rapidly recruited to NF-κB target gene promoters with kinetics similar to those of the transcription factor NF-κB and Pol II. Upon its recruitment, PPM1G dephosphorylates residue T186 in the activating T-loop of Cdk9 (26), thereby releasing P-TEFb from its endogenous inhibitors (Hexim1 and 7SK RNA) and allowing Pol II phosphorylation and rapid activation of the transcription elongation program. Interestingly, we found that once P-TEFb is released from the 7SK snRNP, PPM1G remains bound with Hexim1 on 7SK RNA (7SK-PPM1G snRNP complex) to prevent the reassociation of P-TEFb on 7SK RNA, thereby blocking the formation of the inhibitory snRNP complex. We propose that this step maintains high levels of active, free P-TEFb to sustain transcription elongation. Remarkably, supporting this model, the PPM1G-7SK RNA interaction is direct, kinetically follows the recruitment of PPM1G to target gene promoters, and temporally precedes transcription activation. It is noteworthy that this protein-RNA interaction is reversible, since PPM1G dissociates from the RNA upon resolution of the transcriptional program. At this point, the equilibrium shifts back to P-TEFb binding to Hexim1-7SK RNA and reassembly of the inhibitory 7SK snRNP complex, resetting the transcriptional circuitry to the off state. Although the RIP assay measures bulk levels of PPM1G-7SK RNA, and not gene-specific interactions, using a temporal analysis of PPM1G recruitment to gene promoters, in combination with RIP and gene expression data, we provide compelling evidence for the proposed model.
Despite our previous report that PPM1G binds 7SK RNA (26), the molecular and structural determinants involved in this interaction remained unknown. In this study, we showed that PPM1G binds to a distal portion in 7SK RNA stem I, a critical element through which Hexim1 interacts with P-TEFb to assemble the inhibitory 7SK snRNP complex (42, 45, 46, 60, 61). Strikingly, PPM1G bears a Lys-rich region that resembles the RNA-binding motif of Hexim1 to recognize 7SK RNA (45). While Hexim1 binds 7SK RNA as a homodimer to sequester the P-TEFb kinase (21), Hexim1 forms a heterodimer with PPM1G to block Hexim1-mediated P-TEFb recruitment to 7SK RNA and thus interfere with the assembly of the inhibitory snRNP complex. Consistent with this model, we showed that PPM1G interacts with coiled-coil region 1 (CR1) on the Hexim1 C-terminal domain, which is responsible for P-TEFb binding, 7SK snRNP assembly, and inhibition of transcription elongation (36, 37).
Since PPM1G by itself appears to bind the full-length RNA with much higher affinity than that for stem I alone, it is possible that several PPM1G monomers contact multiple high-affinity (stem I) and low-affinity (stem III) binding sites and oligomerize on the RNA. In addition, given that PPM1G also binds other 7SK snRNP components (Larp7 and MePCE) (26), in vitro reconstitution experiments with this ribonucleoprotein complex are required to precisely define the stoichiometry of all subunits as well as the assembly mechanism.
If the in vitro PPM1G-7SK interactions were relevant, we would expect the assembly mechanism to be regulated in vivo during the activation of inducible transcription elongation programs. Strikingly, we found that in response to DNA damage, PPM1G is phosphorylated by the ATM kinase at S183, which is part of a canonical SQ motif. Importantly, we showed that this posttranslational modification is required for the recruitment of PPM1G to the 7SK snRNP complex to facilitate NF-κB-mediated Pol II transcription in response to DNA damage. Since inhibition of ATM blocks the PPM1G-7SK RNA interaction as well as the activation of NF-κB transcription in response to DNA damage (48, 57, 62), it appears that both molecular events are intimately coupled and that the role of ATM in promoting the PPM1G-7SK RNA interaction relies on NF-κB signaling. However, further investigation is needed to define whether ATM-mediated PPM1G phosphorylation requires NF-κB translocation to the nucleus and whether S183 phosphorylation is needed for PPM1G recruitment to the 7SK snRNP assembled at gene promoters. Notwithstanding, we provide evidence for a previously unprecedented role for the ATM-PPM1G-7SK snRNP network in NF-κB-mediated Pol II transcription elongation during DNA damage signaling.
Previous studies have implicated several coactivators in the regulation of transcription elongation (1, 63). However, mechanisms controlling both the activation and maintenance of transcription elongation programs were largely unexplored. We have described the molecular basis by which the transcriptional coactivator PPM1G couples both steps of the transcriptional process. Our results thus provide, to the best of our knowledge, the first example of the establishment of a full cycle of transcriptional activation and maintenance through the formation of a protein-RNA complex with a regulatory enzyme.
ACKNOWLEDGMENTS
We thank Nozomi Kamimatuzi and Sandeep Burma for sharing reagents and protocols for ATM kinase purification and kinase assays. We also thank Steve Jackson (University of Cambridge) for personal communications about the PPM1G pS183 antibody and Matjaz Barboric (University of Helsinki) for the Hexim1 domain constructs and personal communications.
Research reported in this publication was supported by an intramural grant from the American Cancer Society (ACS-IRG-02-196); the National Cancer Institute of the NIH under award number 5P30CA142543; the National Institute of Allergy and Infectious Diseases of the NIH under award numbers R00AI083087, R56AI106514, and R01AI114362; and Welch Foundation grant I-1782 to Iván D'Orso. Ryan P. McNamara was supported by NIH training grant T32 2T32AI007520-16.
REFERENCES
- 1.Smale ST. 2010. Selective transcription in response to an inflammatory stimulus. Cell 140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith E, Shilatifard A. 2013. Transcriptional elongation checkpoint control in development and disease. Genes Dev 27:1079–1088. doi: 10.1101/gad.215137.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonkers I, Lis JT. 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuda NJ, Ardehali MB, Lis JT. 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J, Price DH. 2013. RNA polymerase II transcription elongation control. Chem Rev 113:8583–8603. doi: 10.1021/cr400105n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng J, Zhu Y, Milton JT, Price DH. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev 12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu TJ, Peng J, Lee G, Price DH, Flores O. 1999. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem 274:34527–34530. doi: 10.1074/jbc.274.49.34527. [DOI] [PubMed] [Google Scholar]
- 11.Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Marshall NF, Peng J, Xie Z, Price DH. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 13.Bres V, Yoh SM, Jones KA. 2008. The multi-tasking P-TEFb complex. Curr Opin Cell Biol 20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 8:327–337. doi: 10.1016/S1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 16.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 18.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462. doi: 10.1016/S0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 19.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byers SA, Price JP, Cooper JJ, Li Q, Price DH. 2005. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem 280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. 2005. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem 280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 22.Ott M, Geyer M, Zhou Q. 2011. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe 10:426–435. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R, Yang Z, Zhou Q. 2004. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem 279:4153–4160. [DOI] [PubMed] [Google Scholar]
- 24.Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, Zhou Q. 2008. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev 22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Orso I, Frankel AD. 2010. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol 17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNamara RP, McCann JL, Gudipaty SA, D'Orso I. 2013. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep 5:1256–1268. doi: 10.1016/j.celrep.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. 2013. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. 2013. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. 2015. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518:249–253. doi: 10.1038/nature13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol Cell Biol 20:6958–6969. doi: 10.1128/MCB.20.18.6958-6969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. 2012. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 19:1108–1115. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura H, Takizawa N, Allemand E, Hori T, Iborra FJ, Nozaki N, Muraki M, Hagiwara M, Krainer AR, Fukagawa T, Okawa K. 2006. A novel histone exchange factor, protein phosphatase 2Cgamma, mediates the exchange and dephosphorylation of H2A-H2B. J Cell Biol 175:389–400. doi: 10.1083/jcb.200608001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. 2012. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holowachuk EW, Ruhoff MS. 1995. Efficient gene synthesis by Klenow assembly/extension-Pfu polymerase amplification (KAPPA) of overlapping oligonucleotides. PCR Methods Appl 4:299–302. doi: 10.1101/gr.4.5.299. [DOI] [PubMed] [Google Scholar]
- 35.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell 12:971–982. doi: 10.1016/S1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 36.Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. 2005. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res 33:7000–7010. doi: 10.1093/nar/gki997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. 2005. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J 24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Conrad NK. 2008. Chapter 15. Co-immunoprecipitation techniques for assessing RNA-protein interactions in vivo. Methods Enzymol 449:317–342. doi: 10.1016/S0076-6879(08)02415-4. [DOI] [PubMed] [Google Scholar]
- 40.Allemand E, Hastings ML, Murray MV, Myers MP, Krainer AR. 2007. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat Struct Mol Biol 14:630–638. doi: 10.1038/nsmb1257. [DOI] [PubMed] [Google Scholar]
- 41.Wassarman DA, Steitz JA. 1991. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol 11:3432–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egloff S, Van Herreweghe E, Kiss T. 2006. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol 26:630–642. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, Coulombe B, Price DH. 2008. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res 36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muniz L, Egloff S, Kiss T. 2013. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res 41:4686–4698. doi: 10.1093/nar/gkt159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. 2010. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog 6:e1001152. doi: 10.1371/journal.ppat.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujinaga K, Luo Z, Peterlin BM. 2014. Genetic analysis of the structure and function of 7SK small nuclear ribonucleoprotein (snRNP) in cells. J Biol Chem 289:21181–21190. doi: 10.1074/jbc.M114.557751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biton S, Ashkenazi A. 2011. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-alpha feedforward signaling. Cell 145:92–103. doi: 10.1016/j.cell.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 48.Li N, Banin S, Ouyang H, Li GC, Courtois G, Shiloh Y, Karin M, Rotman G. 2001. ATM is required for IkappaB kinase (IKKk) activation in response to DNA double strand breaks. J Biol Chem 276:8898–8903. doi: 10.1074/jbc.M009809200. [DOI] [PubMed] [Google Scholar]
- 49.Hande KR. 1998. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur J Cancer 34:1514–1521. doi: 10.1016/S0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 50.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 51.Cheung-Ong K, Giaever G, Nislow C. 2013. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem Biol 20:648–659. doi: 10.1016/j.chembiol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Hande KR, Wedlund PJ, Noone RM, Wilkinson GR, Greco FA, Wolff SN. 1984. Pharmacokinetics of high-dose etoposide (VP-16-213) administered to cancer patients. Cancer Res 44:379–382. [PubMed] [Google Scholar]
- 53.Mili S, Steitz JA. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Parsons JL, Dianov GL. 2012. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell 45:801–813. doi: 10.1016/j.molcel.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 56.Shiloh Y, Ziv Y. 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14:197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 57.Piret B, Schoonbroodt S, Piette J. 1999. The ATM protein is required for sustained activation of NF-kappaB following DNA damage. Oncogene 18:2261–2271. doi: 10.1038/sj.onc.1202541. [DOI] [PubMed] [Google Scholar]
- 58.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 59.Ruland J. 2011. Return to homeostasis: downregulation of NF-kappaB responses. Nat Immunol 12:709–714. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 60.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol 23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. 2004. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J 23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang L, Choudhary S, Zhao Y, Edeh CB, Yang C, Boldogh I, Brasier AR. 2014. ATM regulates NF-kappaB-dependent immediate-early genes via RelA Ser 276 phosphorylation coupled to CDK9 promoter recruitment. Nucleic Acids Res 42:8416–8432. doi: 10.1093/nar/gku529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diamant G, Dikstein R. 2013. Transcriptional control by NF-kappaB: elongation in focus. Biochim Biophys Acta 1829:937–945. doi: 10.1016/j.bbagrm.2013.04.007. [DOI] [PubMed] [Google Scholar]