Abstract
Tristetraprolin (TTP) regulates the expression of AU-rich element-containing mRNAs through promoting the degradation and repressing the translation of target mRNA. While the mechanism for promoting target mRNA degradation has been extensively studied, the mechanism underlying translational repression is not well established. Here, we show that TTP recruits eukaryotic initiation factor 4E2 (eIF4E2) to repress target mRNA translation. TTP interacted with eIF4E2 but not with eIF4E. Overexpression of eIF4E2 enhanced TTP-mediated translational repression, and downregulation of endogenous eIF4E2 or overexpression of a truncation mutant of eIF4E2 impaired TTP-mediated translational repression. Overexpression of an eIF4E2 mutant that lost the cap-binding activity also impaired TTP's activity, suggesting that the cap-binding activity of eIF4E2 is important in TTP-mediated translational repression. We further show that TTP promoted eIF4E2 binding to target mRNA. These results imply that TTP recruits eIF4E2 to compete with eIF4E to repress the translation of target mRNA. This notion is supported by the finding that downregulation of endogenous eIF4E2 increased the production of tumor necrosis factor alpha (TNF-α) protein without affecting the mRNA levels in THP-1 cells. Collectively, these results uncover a novel mechanism by which TTP represses target mRNA translation.
INTRODUCTION
Tristetraprolin (TTP) plays important roles in immunity, development, and tumorigenesis by posttranscriptionally regulating the expression of a variety of genes (1). For example, TTP regulates the expression of tumor necrosis factor alpha (TNF-α) (2). Knockout of TTP in mice results in TNF-α excess and causes severe immune disorders (2, 3).
TTP specifically recognizes AU-rich elements (AREs), which are cis elements with tandem AUUUA-like motifs in a context rich in A and/or U, mainly located in the 3′ untranslated region (3′ UTR) of the mRNAs of stringently regulated genes, such as cytokine and growth factor genes and proto-oncogenes (4, 5). TTP directly binds to ARE motifs and recruits cellular mRNA decay enzymes, including the deadenylase complex CCR4-NOT, decapping complex DCP1a/DCP2, 3′-5′ exoribonuclease complex exosome, and 5′-3′ exoribonuclease Xrn1, to degrade target mRNAs (6–14). This activity of TTP is regulated by phosphorylation modification (6–14). There is increasing evidence suggesting that TTP also regulates the translation of target mRNAs. It has been reported elsewhere that TTP associates with polysomes (15, 16) and that a TTP-interacting protein, cullin 4B, promotes the loading of TTP-associated TNF-α mRNA complex onto polysomes (17). Recently, Qi et al. reported that TTP inhibits the translation of TNF-α in an ARE-dependent manner and the RNA helicase RCK is involved in this process (18). In addition, Tiedje et al. reported that TTP represses the translation of TNF-α through competing with HuR to bind to ARE (19). Nonetheless, detailed mechanisms underlying TTP-mediated translational repression are not well established.
Translation initiation is the main target of translational repression, although other mechanisms also exist (20). Most eukaryotic translation initiation is cap dependent. The cap-binding factor eukaryotic initiation factor 4E (eIF4E) binds to the cap structure of an mRNA and recruits eIF4G, a scaffold protein which further recruits eIF4A, eIF4B, and the 43S preinitiation complex for the assembly of a translation initiation complex (20). This process can be disturbed by a variety of means (20, 21). For example, the zinc finger antiviral protein (ZAP) represses the translation of target mRNA by interfering with the interaction between eIF4A and eIF4G (22).
eIF4E2, a homologue of eIF4E, can bind to the cap structure of an mRNA but does not interact with eIF4G (23, 24) and thus can act as a translational repressor by competing with eIF4E. eIF4E2-knockout mice died within a few hours after birth, with an increase in general translation in brain tissues (25). eIF4E2 knockdown in HeLa cells also increased general protein synthesis (25). In addition to regulating general translation, eIF4E2 has been reported to repress the translation of specific mRNAs. Since the binding affinity of eIF4E2 for the cap structure is estimated to be much lower than that of eIF4E (26), eIF4E2 needs to be recruited by a specific RNA binding protein to target mRNA to compete with eIF4E for cap binding. In Drosophila melanogaster, the eIF4E2 homologue, d4EHP, is recruited by Bicoid to repress the translation of caudal mRNA to drive embryo development (27). In oocytes developing in mice, eIF4E2 interacts with Prep1 to repress the translation of Hoxb4 mRNA (28). eIF4E2 has also been reported to be able to initiate translation under some circumstances. A recent study showed that in hypoxia eIF4E2 is recruited by the oxygen-regulated protein hypoxia-inducible factor alpha and the RNA binding protein RBM4 to initiate the translation of hypoxia response genes (29). This activity of eIF4E2 is exploited by cancer cells to produce hypoxia response proteins to drive tumor progression (30). The underlying molecular mechanism is not clear yet.
Research on TTP-mediated translational repression and mRNA decay is hampered by the low efficiency of each process. The total inhibition is often no more than 5-fold (18). As a result, it is difficult to quantitatively evaluate the magnitudes of mRNA decay and translational repression separately. Multiplication of ARE motifs has been shown to amplify ARE-mediated inhibition of reporter expression (31). In the present study, using a reporter that contains eight ARE motifs, we investigated the mechanism underlying TTP-mediated translational repression. We provide evidence showing that TTP recruits eIF4E2 to repress target mRNA translation.
MATERIALS AND METHODS
DNA constructs and siRNAs.
Fragments of AREm, ARE, AREm*2, and ARE*2 were generated by annealing paired oligonucleotides. The sequences of the oligonucleotides are as follows: AREm oligonucleotides, 5′-CTAGAATCGATTATGTATTATGTATGTATTATGTATGTATTTGGC-3′ and 5′-GGCCGCCAAATACATACATAATACATACATAATACATAATCGATT-3′; ARE oligonucleotides, 5′-CTAGAATCGATTATTTATTATTTATTTATTATTTATTTATTTAGC-3′ and 5′-GGCCGCTAAATAAATAAATAATAAATAAATAATAAATAATCGATT-3′; AREm*2 oligonucleotides, 5′-CTAGTATGATGTATCATGTATCTATGATCTATGTACTTGTCTAGAATCGGCGGCCGCATGATGTATCATGTATCTATGATCTATGTACTTGCG-3′ and 5′-GGCCCGCAAGTACATAGATCATAGATACATGATACATCATGCGGCCGCCGATTCTAGACAAGTACATAGATCATAGATACATGATACATCATA-3′; ARE*2 oligonucleotides, 5′-CTAGTATTATTTATTATTTATTTATTATTTATTTATTTATCTAGAATCGGCGGCCGCATTATTTATTATTTATTTATTATTTATTTATTTACG-3′ and 5′-GGCCCGTAAATAAATAAATAATAAATAAATAATAAATAATGCGGCCGCCGATTCTAGATAAATAAATAAATAATAAATAAATAATAAATAATA-3′. pFL-CMV was generated through replacing the renilla luciferase (RL) coding sequence in phRL-CMV (Promega) with the firefly luciferase (FL) coding sequence from pGL3-Luc-linker (32). pFL-CMV-AREm, pFL-CMV-AREm*2, pFL-CMV-ARE, and pFL-CMV-ARE*2 were generated through inserting the fragments of AREm, ARE, AREm*2, and ARE*2 into pFL-CMV using restriction sites XbaI and NotI. pFL-CMV-AREm*4 and pFL-CMV-ARE*4 were generated by inserting fragments of AREm*2 and ARE*2 into pFL-CMV-AREm*2 and pFL-CMV-ARE*2 using restriction sites XbaI and NotI. pFL-CMV-AREm*6 and pFL-CMV-ARE*6 were generated by inserting fragments of AREm*2 and ARE*2 into pFL-CMV-AREm*4 and pFL-CMV-ARE*4 using restriction sites XbaI and NotI. pFL-CMV-AREm*8 and pFL-CMV-ARE*8 were generated by inserting fragments of AREm*2 and ARE*2 into pFL-CMV-AREm*6 and pFL-CMV-ARE*6 using restriction sites XbaI and NotI. pcDNA4-TTP-myc has been described previously (32). To generate pCMV-HF-eIF4E and pCMV-HF-eIF4E2a/b/c/d/e, the coding sequences of eIF4E and eIF4E2a/b/c/d/e were each PCR amplified from cDNAs from HeLa cells using specific primers and cloned into pCMV-HF (33) using restriction sites EcoRI and XhoI. Sequences of the primers are as follows: eIF4E primers, 5′-GCACGAATTCATGGCGACTGTCGAAC-3′ and 5′-GCACCTCGAGTTAAACAACAAACCT-3′; eIF4E2a/b/c/d/e primers, 5′-GCACGAATTCATGAACAACAAGTTCGAC-3′ (forward primer for all the isoforms), 5′-GCACCTCGAGTCATGGCACATTCAAC-3′ (reverse primer for isoforms a and e), 5′-GCACCTCGAGTCAGCGGCCGCTGCTGTTC-3′ (reverse primer for isoform b), and 5′-GCACCTCGAGCTTGTGTACTCTCACAATGTG-3′ (reverse primer for isoforms c and d). The coding sequence of the N-terminal fragment of eIF4E2 (eIF4E2N) was generated by PCR from pCMV-HF-eIF4E2 using primers 5′-GCACGAATTCATGAACAACAAGTTCGAC-3′ and 5′-GCACCTCGAGTCATTCTTTGAAGAGATGGAAG-3′. The coding sequence of the C-terminal fragment of eIF4E2 (eIF4E2C) was generated by PCR from pCMV-HF-eIF4E2 using primers 5′-GCACGAATTCGGAATTAAACCCATGTG-3′ and 5′-GCACCTCGAGTCATGGCACATTCAAC-3′. The Y78A mutation was generated by overlapping PCR from pCMV-HF-eIF4E2 using primers 5′-CAGAGCGCTGAACAGAATATCAAAC-3′ and 5′-CTGTTCAGCGCTCTGTGAGCTCGTG-3′. The short hairpin RNA (shRNA)-expressing vectors, pSR-Ctrli, pSR-TTPi, pSR-eIF4E2i-302, and pSR-eIF4E2i-977, were generated by annealing paired oligonucleotides and subsequently cloning them into pSR vector (OligoEngine) using restriction sites BglII and HindIII. Sequences of the oligonucleotides are as follows: Ctrli oligonucleotides, 5′-GATCCCCGCGCGCTTTGTAGGATTCGTTCAAGAGACGAATCCTACAAAGCGCGCTTTTTA-3′ and 5′-AGCTTAAAAAGCGCGCTTTGTAGGATTCGTCTCTTGAACGAATCCTACAAAGCGCGCGGG-3′; TTPi oligonucleotides, 5′-GATCCCCCGCTGCCACTTCATCCACATTCAAGAGATGTGGATGAAGTGGCAGCGTTTTTA-3′ and 5′-AGCTTAAAAACGCTGCCACTTCATCCACATTCAAGAGATGTGGATGAAGTGGCAGCGGGG-3′; eIF4E2i-302 oligonucleotides, 5′-GATCCCCCACAGAGCTATGAACAGAATATTCAAGAGATATTCTGTTCATAGCTCTGTGTTTTTA-3′ and 5′-AGCTTAAAAACACAGAGCTATGAACAGAATATCTCTTGAATATTCTGTTCATAGCTCTGTGGGG-3′; eIF4E2i-977 oligonucleotides, 5′-GATCCCCAGCTGAGATCACTTAATAATTCAAGAGATTATTAAGTGATCTCAGCTTTTTTA-3′ and 5′-AGCTTAAAAAAGCTGAGATCACTTAATAATCTCTTGAATTATTAAGTGATCTCAGCTGGG-3′. Small interfering RNAs (siRNAs) were purchased from GenePharma. The sense sequences are as follows: Ctrl, 5′-UUCUCCGAACGUGUCACGUTT-3′; eIF4Ei, 5′-GGAUGGUAUUGAGCCUAUGTT-3′; eIF4E2i, 5′-CGAGACAAGAAUCAGAGCAGUTT-3′.
Cell culture and transfection.
HeLa, HEK293, and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Gibco). THP-1 cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (Gibco). THP-1 cells stably expressing a control shRNA or the shRNAs directed against TTP or eIF4E2 were established through infecting THP-1 cells with vesicular stomatitis virus G protein (VSV-G)-pseudotyped shRNA-expressing retroviral vectors and subsequent puromycin selection. To induce TNF-α expression in THP-1 cells, cells were treated with phorbol myristate acetate (PMA; Sigma) at a final concentration of 200 nM for 2 days, followed by resting in fresh medium for 1 day and then lipopolysaccharide (LPS) (Sigma; final concentration of 100 ng/ml) treatment for 4 h. To knock down eIF4E or eIF4E2 in HeLa cells, siRNA was transfected into the cells using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, the cells were transfected with the reporters and a TTP-expressing plasmid using the Neofectin DNA transfection reagent (SciLight). Another 24 h later, cells were harvested.
Luciferase assay and RNA extract.
Luciferase activities were measured with the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions, except that the cells were lysed with RNase-free RLN lysis buffer (50 mM Tris-Cl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol [DTT], 1,000 U/ml RNase inhibitor), which keeps the nuclei intact. Cytoplasmic RNA was extracted from clarified cell lysate using TRIzol (Invitrogen) according to the manufacturer's instructions.
Polysome profiling analysis.
The polysome profiling analysis procedure was similar to that described previously (22). Briefly, cells were treated with fresh medium containing 100 μg/ml cycloheximide to fix the polysomes for 30 min. The cells were lysed with the lysis buffer (10 mM HEPES, pH 8.0, 300 mM KCl, 5 mM MgCl2, 0.5% NP-40, 100 μg/ml cycloheximide), 1 ml for each 10-cm dish. The clarified cell lysate was separated through ultracentrifugation in a 10% to 50% sucrose gradient. RNA concentrations of the samples were measured with a continuous 254-nm absorbance detector to indicate the positions of ribosome subunits and polysomes. Twelve fractions, 1 ml each, were collected for each sample. The RNA in each fraction was isolated by two rounds of extraction with phenol-chloroform-isopentanol (25:24:1) and one round of phenol removal with chloroform-isopentanol (49:1), followed by alcohol precipitation. The FL-ARE and renilla luciferase control reporter mRNA levels were measured by quantitative reverse transcription PCR (RT-qPCR).
Protein-protein coimmunoprecipitation assay.
Cells from a 35-mm dish were lysed in 300 μl lysis buffer (30 mM HEPES [pH 7.5], 100 mM NaCl, 0.5% NP-40, 1 mM NaF, 1 mM NaVO3, and a protease inhibitor cocktail) for 30 min on a roller at 4°C. The lysate was clarified by centrifugation at 12,000 rpm for 10 min at 4°C. The clarified cell lysate was incubated with antibodies and protein G-Sepharose (Amersham Pharmacia) for 2 h on a roller at 4°C. For RNase A treatment, RNase A was added to a final concentration of 100 μg/ml just before incubation. Immunoprecipitates were washed three times with phosphate-buffered saline (PBS), resuspended in SDS loading buffer, boiled for 5 min, resolved by SDS-PAGE, and detected by Western blotting.
Cap-binding assay.
The cap-binding assay procedure was the same as the protein-protein coimmunoprecipitation assay except that the antibodies and protein G-Sepharose were replaced with M7-GTP–Sepharose (GE Healthcare Biosciences).
RNA-protein coimmunoprecipitation assay.
Cells were lysed with optimized RLN lysis buffer (50 mM Tris-Cl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM DTT, 1,000 U/ml RNase inhibitor) for 30 min on a roller at 4°C. The lysate was clarified by centrifugation at 2,000 rpm for 5 m at 4°C. The clarified lysate was incubated with antibodies against eIF4E (A-10, sc-271480; Santa Cruz) or eIF4E2 (YB-18, sc-100731; Santa Cruz) and protein G-Sepharose (Amersham Pharmacia) for 2 h on a roller at 4°C. Immunoprecipitates were washed three times with PBS. Half of the immunoprecipitate was resuspended in TRIzol (Invitrogen) for RNA extraction. The remaining immunoprecipitate was resuspended in SDS loading buffer for protein detection by Western blotting.
Quantitative RT-PCR.
RNA was reverse transcribed in a 20-μl reaction mixture [1 to 5 μg RNA, 1 μg oligo(dT)18, 10 mM deoxynucleoside triphosphates (dNTPs), Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM DTT, 1 μl RT]. The cDNA levels were measured by SYBR green real-time PCR in the Rotor-Gene 6000 system (Corbett Life Science). Sequences of the primers are as follows: qLuc-FP, 5′-TGAGGCACTGGGCAGGTGTC-3′ (walking over the intron sequence, for both firefly luciferase and renilla luciferase reporter); qFL-RP, 5′-ATGCAGTTGCTCTCCAGCGG-3′; qRL-RP, 5′-ATGAAGGAGTCCAGCACGTTC-3′; qGAPDH-FP, 5′-TCGGAGTCAACGGATTTG-3′; qGAPDH-RP, 5′-GCATCGCCCCACTTGATT-3′; qTNF-FP, 5′-CCTCTCTCTAATCAGCCCTCTG-3′; qTNF-RP, 5′-GAGGACCTGGGAGTAGATGAG-3′; qTTP-FP, 5′-GACTGAGCTATGTCGGACCTT-3′; qTTP-RP, 5′-GAGTTCCGTCTTGTATTTGGGG-3′.
Statistics.
The mean values ± standard deviations (SD) were calculated from three independent experiments or measurements. P values were calculated using the Student t test.
RESULTS
TTP represses the translation of ARE reporters.
In an attempt to increase the responsiveness of ARE reporters to TTP, reporters containing different numbers of a typical TNF-α ARE motif in the 3′ UTR of firefly luciferase were constructed (Fig. 1A). To avoid plasmid DNA contamination of the RNA samples during quantitative RT-PCR (RT-qPCR) measurement of the reporter mRNA levels, an intron was inserted into the 5′ UTR (Fig. 1A). PCR using a forward primer walking over the intron sequence (Fig. 1A) amplifies the cDNA reverse transcribed from the spliced mRNAs but not the fragment in the plasmids. A no-RT control was used in the RT-qPCR assay to monitor plasmid contamination, and no plasmid contamination was observed.
FIG 1.
TTP represses the translation of ARE reporters. (A) Schematic representation of ARE-containing firefly luciferase (FL) reporters and a control renilla luciferase (RL) reporter. In the firefly reporter, each black box in the 3′ UTR represents a wild-type or mutated ARE motif. The indicated primers were used for quantitative PCR analyses. (B) An empty vector or a plasmid expressing myc-tagged TTP was cotransfected into HeLa cells with the FL reporters containing the indicated number of ARE motifs (ARE*n) or mutated ARE motifs (AREm*n). The plasmid expressing control reporter RL was included to serve as a control for transfection efficiency and sample handling. At 48 h posttransfection, cells were harvested and lysed. One-tenth of the cell lysate was used to measure luciferase activities, and the rest was used to extract cytoplasmic RNA. Reporter mRNA levels were measured by RT-qPCR. Translational efficiency was calculated as luciferase activity divided by mRNA level. FL activity, mRNA level, and translational efficiency were normalized by RL activity, mRNA level, and translational efficiency, respectively. Fold inhibition was calculated as the normalized FL activity, mRNA level, or translational efficiency in the absence of TTP divided by that in the presence of TTP. (C and D) A plasmid expressing an shRNA directed against TTP or a control shRNA was transfected into HEK293 cells with the FL-ARE*8 and RL reporters. At 48 h posttransfection, cells were harvested and lysed. One-tenth of the cell lysate was used to detect protein expression by Western blotting, 1/10 of the cell lysate was used to measure luciferase activities, and the rest was used to extract cytoplasmic RNA. The mRNA levels were measured by RT-qPCR. (C) The mRNA level of TTP was normalized with that of glyceraldehyde-3-phosphate dehydrogenase. The relative mRNA level in the control cells was set as 1. (D) FL activity, mRNA level, and translational efficiency were normalized by RL activity, mRNA level, and translational efficiency, respectively. (E) Indicated amounts of a plasmid expressing TTP-myc were transfected into HeLa cells with the FL-ARE*8 and RL reporters. A plasmid expressing green fluorescent protein (GFP)-myc was included to serve as a control for transfection efficiency and sample handling. At 48 h posttransfection, cells were lysed, luciferase activities were measured, and protein expression levels were detected by Western blotting. Fold inhibition of reporter expression was calculated as described for panel B. Data presented are means ± SD from three independent experiments. *, P < 0.05; **, P < 0.01.
The reporters and a plasmid expressing TTP were transfected into HeLa cells. The sensitivity of a reporter to TTP was indicated by fold inhibition, calculated as the luciferase activity expressed from the reporter in the absence of TTP divided by the luciferase activity in the presence of TTP. Increased numbers of ARE motifs significantly increased the sensitivity of the reporter to TTP (Fig. 1B). Increased sensitivity to TTP was not likely caused by the increased length of the 3′ UTR since the reporter containing eight mutated ARE motifs (AREm*8) was not sensitive to TTP (Fig. 1B).
TTP inhibited the expression of the FL-ARE*8 reporter by about 16-fold on the protein level, as indicated by the luciferase activity (Fig. 1B). However, the mRNA level of the reporter was reduced by only about 3-fold, as determined by RT-qPCR (Fig. 1B), implying that translational repression exists in TTP inhibition of the reporter expression, which is consistent with the results reported previously by Qi et al. (18). Relative translational efficiency was calculated as the luciferase activity divided by the relative mRNA level to evaluate the inhibitory effect of TTP on reporter mRNA translation. Data showed that TTP inhibited the translational efficiency of the reporter by about 5-fold (Fig. 1B). The reporter containing eight ARE motifs was used in the following studies and named FL-ARE.
We next examined whether the endogenous TTP inhibits the expression of the FL-ARE reporter. TTP was downregulated in HEK293 cells with an shRNA, and the reporter was expressed in these cells. Data showed that moderate downregulation of TTP (Fig. 1C) increased the protein expression level, the mRNA level, and the translational efficiency of the FL-ARE reporter (Fig. 1D). These results indicate that the endogenous TTP regulates the mRNA abundance and translation efficiency of the reporter, which is consistent with the results reported previously by Qi et al., wherein a single ARE motif-containing reporter was used (18).
To determine whether TTP functions in a dose-dependent manner, the FL-ARE reporter was transfected into cells together with increasing amounts of the TTP-expressing plasmid. Increased expression levels of TTP displayed increased fold inhibition on the ARE reporter (Fig. 1E). Notably, when 5 ng of or more of the TTP-expressing plasmid per well of the 12-well plate was used to transfect cells, cytotoxicity was significant, as indicated by the decreased expression of the control renilla luciferase reporter (Fig. 1E). To avoid the cytotoxicity, we used 2 ng of the TTP-expressing plasmid per well in the following studies.
TTP interacts with eIF4E2 but not eIF4E.
A previous study suggested that ARE-mediated translational repression may be cap dependent (34). We speculated that TTP might repress cap-dependent translation initiation. To test this hypothesis, we analyzed the interactions of TTP with cap-binding proteins eIF4E and eIF4E2 by coimmunoprecipitation assays. To prevent nonspecific RNA tethering, the cell lysate was treated with RNase A before immunoprecipitation. Data showed that TTP did not interact with eIF4E but interacted with eIF4E2 (Fig. 2A). To validate the efficiency and the specificity of the coimmunoprecipitation assay, the interaction of endogenous eIF4G with eIF4E or eIF4E2 was detected. Consistent with the previously reported results (24), eIF4G interacted with eIF4E but not with eIF4E2 (Fig. 2A).
FIG 2.
TTP interacts with eIF4E2 but not with eIF4E. (A) TTP-myc was transiently expressed in HEK293 cells together with FLAG-tagged eIF4E or eIF4E2a. At 48 h posttransfection, cells were harvested and lysed. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody in the presence of RNase A. The precipitates were resolved by SDS-PAGE followed by Western blotting. (B) Schematic representation of eIF4E2 isoforms. (C) TTP-myc was transiently expressed in HEK293 cells together with each FLAG-tagged eIF4E2 isoform. At 48 h posttransfection, cells were harvested and lysed. Cell lysates were immunoprecipitated with anti-FLAG antibody in the presence of RNase A. The precipitates were resolved by SDS-PAGE followed by Western blotting.
Sequence analysis recently revealed that there are four additional eIF4E2 isoforms (NCBI; http://www.ncbi.nlm.nih.gov/gene/9470). Compared with prototype eIF4E2, isoform a, isoforms b, c, and d have different C termini, and isoforms c and d lack a fragment close to the N terminus (Fig. 2B). We explored whether these eIF4E2 isoforms interact with TTP. Coimmunoprecipitation assays revealed that all the eIF4E2 isoforms interacted with TTP (Fig. 2C).
Overexpression of eIF4E2 enhances TTP-mediated translational repression.
To probe whether eIF4E2 is involved in TTP-mediated translational repression, we analyzed the effect of overexpression of eIF4E2a on TTP inhibition of reporter expression. Overexpression of eIF4E was included as a control. While overexpression of eIF4E modestly reduced TTP-mediated translational repression, overexpression of eIF4E2a at a comparable expression level enhanced TTP-mediated translational repression (Fig. 3A and B). The fold inhibition on the mRNA levels was not significantly affected by overexpression of eIF4E or eIF4E2a (Fig. 3A). Notably, only the overexpression of eIF4E2a at a relatively high level enhanced TTP-mediated translational repression (Fig. 3C and D).
FIG 3.
Overexpression of eIF4E2 enhances TTP-mediated translational repression. (A and B) A plasmid expressing FLAG-tagged eIF4E or eIF4E2 or an empty vector (EV) was transfected into HeLa cells with the luciferase reporters, with or without a plasmid expressing TTP-myc. At 48 h posttransfection, cells were harvested and lysed. (A) Luciferase activities and mRNA levels of the reporters were measured, and fold inhibition was calculated as described in the legend to Fig. 1B. (B) One-tenth of the cell lysate was resolved by SDS-PAGE followed by Western blotting. (C and D) Indicated amounts of the plasmid expressing FLAG-tagged eIF4E2a or an empty vector were transfected into HeLa cells with the luciferase reporters, with or without a plasmid expressing TTP-myc. At 48 h posttransfection, cells were harvested and lysed. (C) Luciferase activities and mRNA levels of the reporters were measured, and fold inhibition was calculated as described in the legend to Fig. 1B. (D) One-tenth of the cell lysate was resolved by SDS-PAGE followed by Western blotting. (E and F) A plasmid expressing a FLAG-tagged eIF4E2 isoform as indicated or an empty vector was transfected into HeLa cells with the luciferase reporters, with or without a plasmid expressing TTP-myc. At 48 h posttransfection, cells were harvested and lysed. (E) Luciferase activities and mRNA levels of the reporters were measured, and fold inhibition was calculated as described in the legend to Fig. 1B. (F) One-tenth of the cell lysate was resolved by SDS-PAGE, followed by Western blotting. Data presented are means ± SD from three independent experiments. *, P < 0.05.
We next tested whether other eIF4E2 isoforms also function in TTP-mediated translational repression. Overexpression of any one of the isoforms had no significant effect on the fold inhibition on the mRNA levels (Fig. 3E). While overexpression of isoforms a, b, c, and d significantly enhanced TTP-mediated translational repression, overexpression of isoform e had little effect (Fig. 3E). Notably, the expression level of isoform e was considerably lower than those of the other isoforms (Fig. 3F), which may account for its lack of activity in this assay. In the following studies, we focused on isoform a (here referred to as “eIF4E2”).
Downregulation of eIF4E2 impairs TTP-mediated translational repression.
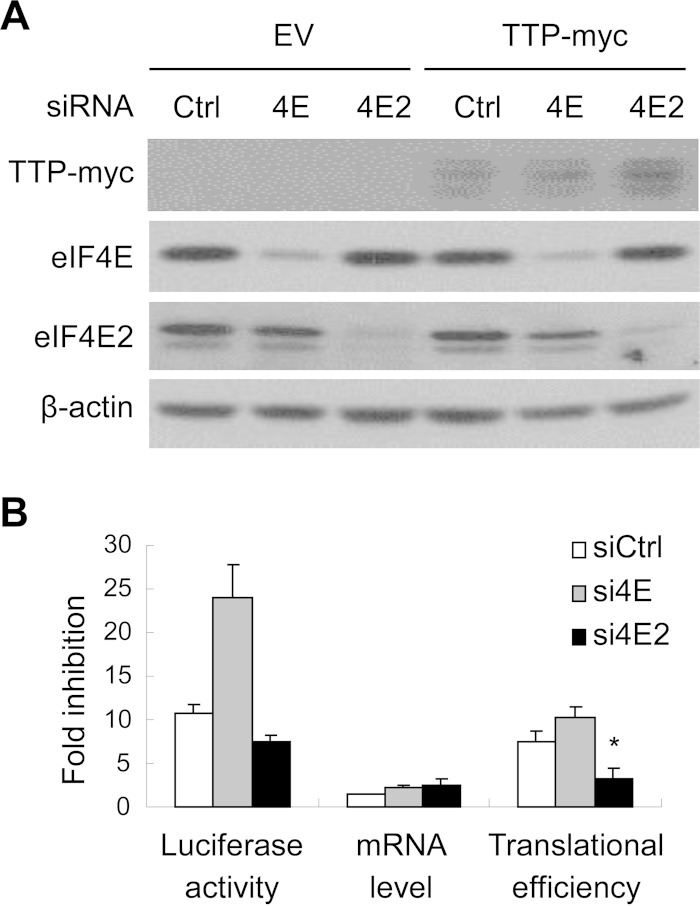
We next examined the effect of downregulating the endogenous eIF4E2 on TTP-mediated translational repression. An siRNA directed against the common sequence of all the isoforms of eIF4E2 was employed. An siRNA directed against eIF4E was used as a control. The siRNAs significantly reduced the expression levels of eIF4E and eIF4E2 (Fig. 4A). While downregulation of eIF4E modestly enhanced TTP-mediated translational repression, downregulation of eIF4E2 significantly reduced TTP-mediated translational repression (Fig. 4B).
FIG 4.
Downregulation of eIF4E2 impairs TTP-mediated translational repression. A control siRNA (Ctrl) or an siRNA targeting eIF4E or eIF4E2 was transfected into HeLa cells. At 24 h posttransfection, reporter plasmids were transfected into these cells with or without a plasmid expressing TTP-myc. At 24 h after the second round of transfection, cells were harvested and lysed. (A) One-tenth of the cell lysate was resolved by SDS-PAGE followed by Western blotting. (B) Luciferase activities and mRNA levels of the reporters were measured, and fold inhibition was calculated as described in the legend to Fig. 1B. Data presented are means ± SD from three independent experiments. *, P < 0.05. EV, empty vector.
Overexpression of an eIF4E2 truncation mutant impairs TTP-mediated translational repression.
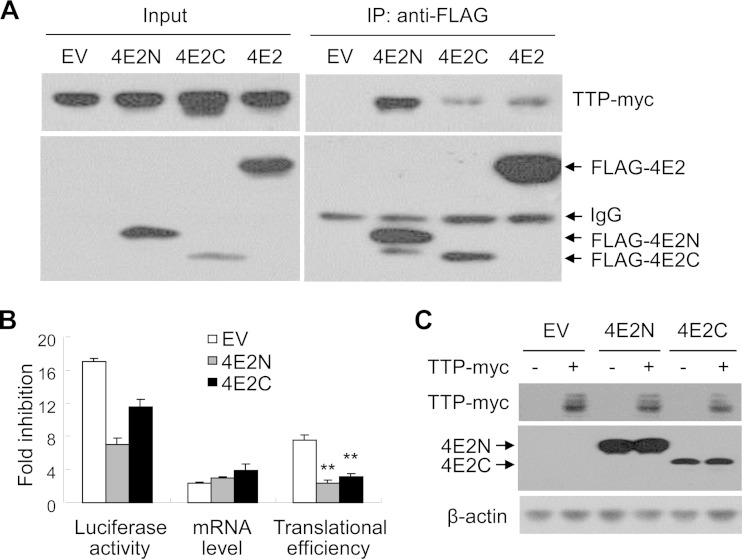
We next mapped the domains of eIF4E2 involved in its interaction with TTP. The full-length eIF4E2 was divided into two fragments, the N-terminal domain eIF4E2N and the C-terminal domain eIF4E2C. The interaction was analyzed by coimmunoprecipitation assays. To prevent nonspecific RNA tethering, the cell lysate was treated with RNase A before immunoprecipitation. Data showed that both eIF4E2N and eIF4E2C interacted with TTP (Fig. 5A). Interestingly, for unknown reasons, the interaction of TTP with eIF4E2N seemed to be stronger than that with the full-length eIF4E2 (Fig. 5A).
FIG 5.
Overexpression of eIF4E2 truncation mutants reduces TTP-mediated translational repression. (A) TTP-myc was transiently expressed in HEK293 cells together with FLAG-tagged eIF4E2N (4E2N), eIF4E2C (4E2C), or the full-length eIF4E2 (4E2). Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody in the presence of RNase A. The precipitates were resolved by SDS-PAGE followed by Western blotting. EV, empty vector. (B and C) An empty vector or a plasmid expressing FLAG-4E2N or -4E2C was transfected into HeLa cells with the reporters, with or without a plasmid expressing TTP-myc. At 48 h posttransfection, cells were harvested and lysed. (B) Luciferase activities and mRNA levels of the reporters were measured, and fold inhibition was calculated as described in the legend to Fig. 1B. Data presented are means ± SD from three independent experiments. **, P < 0.01. (C) The cell lysates were resolved by SDS-PAGE followed by Western blotting.
Based on the above results, we speculated that overexpression of the eIF4E2 truncation mutants might inhibit the activity of TTP in a dominant negative manner. Indeed, overexpression of both truncation mutants significantly impaired TTP-mediated translational repression without a significant effect on the fold inhibition of the mRNA levels (Fig. 5B) or on TTP expression (Fig. 5C). These results further suggest that eIF4E2 is involved in TTP-mediated translational repression.
Overexpression of eIF4E2N relieves TTP-mediated reduction of target mRNA association with polysomes.
To further validate the roles of TTP and eIF4E2 in translation repression of ARE-containing mRNAs, we analyzed the effect of overexpression of TTP and eIF4E2N on the association of target mRNA with polysomes by the polysome profiling assay. Overexpression of TTP or eIF4E2N did not significantly affect the pattern of ribosome distribution (Fig. 6A), suggesting that overexpression of TTP or eIF4E2N did not affect general translation under this condition. Overexpression of TTP significantly reduced the percentage of FL-ARE reporter mRNA in the polysome fractions, and overexpression of eIF4E2N partially relieved the reduction (Fig. 6B and C). These effects were specific since the distribution of the renilla luciferase control reporter mRNA was not significantly affected (Fig. 6B and C). These results further indicate that TTP represses the translation of ARE-containing mRNAs and that eIF4E2 is involved in this process.
FIG 6.
TTP-mediated reduction of target mRNA association with polysomes is relieved by overexpression of eIF4E2N. (A) An empty vector (EV) or a plasmid expressing FLAG-tagged eIF4E2N (4E2N) was transfected into HeLa cells with the reporters, with or without a plasmid expressing TTP-myc. At 48 h posttransfection, cells were treated with cycloheximide for 30 min and then lysed. The clarified cell lysates were fractionated through sucrose gradient centrifugation. Polysome profiles were analyzed and plotted. (B) The RNA in each fraction was isolated, and the FL-ARE and RL reporter mRNAs were measured by RT-qPCR. The percentage of the mRNA in each fraction was calculated as the mRNA level in each fraction divided by the total mRNA level in all the fractions. (C) Percentage of the mRNA in the polysome fractions was calculated as the mRNA level in fractions 1 to 6 divided by the total mRNA level in all the fractions. Data presented are means ± SD from three independent measurements, representative of two independent experiments. *, P < 0.05.
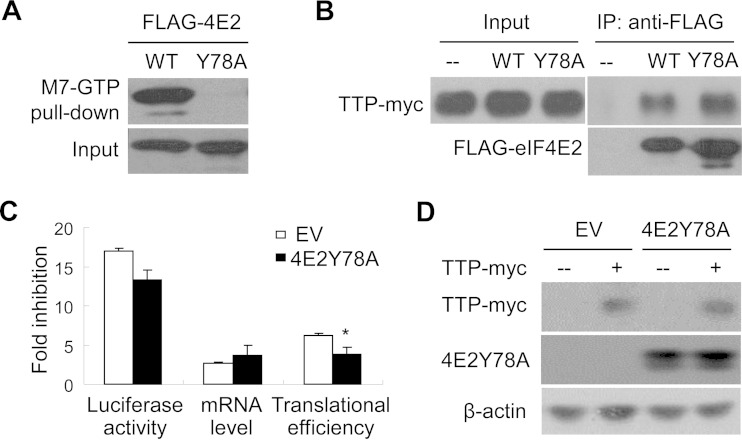
Overexpression of an eIF4E2 mutant that loses the cap-binding activity impairs TTP-mediated translational repression.
It is well established that eIF4E2 binds to the cap structure of an mRNA but does not interact with eIF4G (35). Hence, when eIF4E2 binds to the cap structure, it interferes with the assembly of the translation initiation complex. Residue Y78 is essential for cap binding of eIF4E2a (36). To investigate the role of the cap-binding activity of eIF4E2 in TTP-mediated repression, the FLAG-tagged eIF4E2Y78A mutant was constructed. The mutant was transiently expressed in HeLa cells and assayed for its ability to bind to the cap structure analog M7-GTP. Consistent with the previous results reported by others (36), the eIF4E2Y78A mutant failed to bind M7-GTP (Fig. 7A). However, the mutation did not affect the interaction of eIF4E2 with TTP (Fig. 7B). Overexpression of the eIF4E2Y78A mutant impaired TTP-mediated translational repression (Fig. 7C) without a significant effect on fold inhibition of the mRNA levels (Fig. 7C) or on TTP expression (Fig. 7D). These results suggest that the eIF4E2Y78A mutant inhibited TTP-mediated translational repression in a dominant negative manner and that the cap-binding activity of eIF4E2 is required for its function in TTP-mediated translational repression.
FIG 7.
Overexpression of an eIF4E2 mutant that loses the cap-binding activity impairs TTP-mediated translational repression. (A) FLAG-tagged eIF4E2 or its Y78A mutant was transiently expressed in HEK293 cells and pulled down with beads conjugated to cap structure analog M7-GTP. The precipitates (top) and cell lysates (bottom) were resolved by SDS-PAGE followed by Western blotting. WT, wild type. (B) TTP-myc was transiently expressed in HEK293 cells together with FLAG-tagged wild-type eIF4E2 or the Y78A mutant. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody in the presence of RNase A. The immunoprecipitates and total cell lysates were resolved by SDS-PAGE followed by Western blotting. (C) An empty vector (EV) or a plasmid expressing FLAG-tagged eIF4E2 Y78A mutant was transfected into HeLa cells with the reporters, with or without a plasmid expressing TTP-myc. At 48 h posttransfection, cells were harvested and lysed. Luciferase activities and mRNA levels of the reporters were measured, and fold inhibition was calculated as described in the legend to Fig. 1B. Data presented are means ± SD from three independent experiments. *, P < 0.05. (D) The cell lysates were resolved by SDS-PAGE followed by Western blotting.
TTP promotes eIF4E2 to bind to target mRNA.
Based on the above results, we hypothesized that TTP promotes eIF4E2 to bind to the cap structure of target mRNA and thus compete with eIF4E to inhibit the assembly of the translation initiation complex. To test the hypothesis, we measured the relative levels of the FL-ARE reporter mRNA associated with eIF4E2 or eIF4E2 in the presence or absence of TTP. The reporter and TTP were transiently expressed in HeLa cells. eIF4E or eIF4E2 was immunoprecipitated, and the associated reporter mRNA levels were measured. TTP expression decreased the association of the FL-ARE reporter mRNA with eIF4E (Fig. 8A) but increased its association with eIF4E2 (Fig. 8B). These effects were specific; the association of the renilla luciferase control reporter mRNA with eIF4E2 was minimal and little affected by TTP expression (Fig. 8A and B). Western blotting revealed that comparable amounts of eIF4E2 were immunoprecipitated (Fig. 8C). Collectively, these results imply that TTP specifically promotes eIF4E2 association with target mRNA.
FIG 8.
TTP promotes eIF4E2 binding to ARE-containing mRNAs. An empty vector (EV) or a plasmid expressing TTP-myc was transfected into HeLa cells with the FL-ARE and RL reporters. At 48 h posttransfection, one-fourth of the cell lysate was immunoprecipitated with anti-eIF4E or anti-eIF4E2 antibody. (A and B) RNA was extracted from one-eighth of the cell lysates and half of the precipitates. The reporter mRNA levels were measured by RT-qPCR. The percentage of input was calculated as the reporter mRNA level in the precipitates divided by the reporter mRNA level in the cell lysates. Data presented are means ± SD from three independent measurements, representative of two independent experiments. *, P < 0.05; **, P < 0.01. (C) The cell lysates and the remaining precipitates were resolved by SDS-PAGE followed by Western blotting. IP, immunoprecipitation.
Downregulation of eIF4E2 increases the production of TNF-α in THP-1 cells.
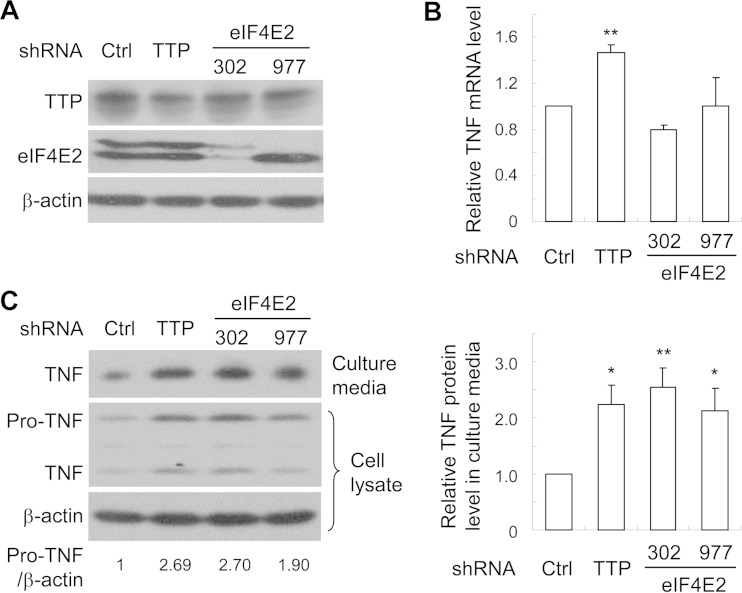
In the above-described experiments, we used an ARE reporter to explore the function of eIF4E2 in TTP-mediated translational repression. We next investigated whether eIF4E2 is involved in TTP-mediated translational repression under physiological conditions. Macrophages are the main source of TNF-α, and TTP regulates the production of TNF-α in these cells (3). Thus, we used macrophage-like PMA-treated THP-1 cells to analyze the roles of TTP and eIF4E2 in the regulation of TNF-α production. For this purpose, a THP-1 cell line was needed in which eIF4E2 is stably downregulated. Considering the fact that eIF4E2 knockout caused general translation dysregulation and perinatal lethality in mice (25), we reasoned that stably downregulating all the isoforms of eIF4E2 might be toxic to the cells. We thus chose to downregulate only a subset of the isoforms of eIF4E2. Two shRNAs, one (sh302) targeting isoforms a, b, and c and the other (sh977) targeting isoforms a and e, were stably expressed in THP-1 cells. An shRNA targeting TTP was stably expressed in THP-1 cells to serve as a positive control. Western analysis revealed that eIF4E2 was efficiently downregulated but TTP was only moderately downregulated (Fig. 9A). These cells were treated with PMA and then stimulated with LPS to induce TNF-α production. Downregulation of TTP modestly enhanced the mRNA level of TNF-α (Fig. 9B). In contrast, downregulation of eIF4E2 had little effect on the mRNA levels (Fig. 9B). Downregulation of TTP increased the production of TNF-α in the culture medium, as well as the pro-TNF and TNF-α in the cell lysate (Fig. 9C). In comparison, downregulation of eIF4E2 increased the production of TNF-α and the expression of pro-TNF as well as the downregulation of TTP (Fig. 9C). These results indicate that eIF4E2 is indeed involved in the expression of TNF-α in THP-1 cells.
FIG 9.
eIF4E2 is involved in the regulation of TNF-α protein synthesis in THP-1 cells. THP-1 cells stably expressing an shRNA indicated were treated with PMA for 2 days, followed by stimulation with LPS for 4 h. The culture media were collected, and cells were lysed. (A) Cell lysates were resolved by SDS-PAGE followed by Western blotting. (B) Cytoplasmic RNA was extracted from the cell lysates, and mRNA levels were measured by RT-qPCR. The mRNA level of TNF-α was normalized with that of glyceraldehyde-3-phosphate dehydrogenase. The relative TNF mRNA level in the control cells was set as 1. (C) The culture media and cell lysates were resolved by SDS-PAGE followed by Western blotting. The relative intensities of the bands of TNF-α in the culture media and pro-TNF and β-actin in the cell lysates were quantified using the ImageJ software. The relative intensity of the band of pro-TNF was normalized with that of β-actin (left panel, bottom). Data presented are representative of three independent experiments. The relative intensity of the band of TNF-α in the medium of the control cells was set as 1 (right panel). Data presented are means ± SD from three independent experiments. *, P < 0.05; **, P < 0.01.
DISCUSSION
TTP plays important roles in immunology, metabolism, and differentiation by regulating the expression of multiple target genes that contain type II AREs in the 3′ UTR (37). TTP regulates the expression of ARE-containing mRNAs by both promoting the degradation and repressing the translation of target mRNA (38). While the mechanisms by which TTP promotes the degradation of ARE-containing mRNAs have been extensively studied (39), the mechanism underlying TTP-mediated translational repression is not so well established. A difficulty in the studies on TTP repression of translation is that the magnitude of inhibition is relatively small. Reporters containing tandem multiple responsive elements have been widely used to increase reporter sensitivity in studies of microRNA-mediated gene silencing (40, 41). Tandem multiple AREs also exist in the 3′ UTR of many mRNAs, such as interleukin-6 (IL-6), IL-8, and IL-10 mRNA (http://rna.tbi.univie.ac.at/cgi-bin/AREsite.cgi). Here, we developed a reporter containing eight ARE motifs, which is much more sensitive to TTP than the reporter containing a single ARE motif (Fig. 1). Using this reporter, we probed the mechanism underlying TTP repression of translation and provide evidence showing that TTP recruits eIF4E2 to compete with eIF4E to block translational initiation.
TTP interacted with all the five isoforms of eIF4E2 but not with eIF4E in the coimmunoprecipitation assays (Fig. 2A and C). Sequence analyses revealed that isoforms b and c differ from isoform a at the C-terminal domain. Isoforms d and e lack an exon containing the residue Y78, which is essential for the cap-binding activity of isoform a (Fig. 2B and 7A). However, our preliminary results showed that isoforms d and e also bound the cap structure analog M7-GTP (X. Tao and G. Gao, unpublished data), implying that they bind the cap structure in a manner distinct from that of isoform a. This possibility is supported by the finding that eIF4E3 binds to the cap in a manner that does not involve the Y78 equivalent residue (42). Overexpression of eIF4E2a enhanced TTP-mediated translational repression in a dose-dependent manner (Fig. 3). Compared with the other isoforms, overexpression of isoform e appeared to fail to enhance TTP's activity (Fig. 3C). One plausible explanation is that the protein was expressed at a lower level than were the other isoforms (Fig. 3F), although the cells were transfected with a large amount of the expressing plasmid.
TTP is a phosphorylated protein with multiple phosphorylation status. It has been reported that phosphorylation modification inactivates TTP-mediated mRNA decay but does not affect TTP-mediated repression in another step(s), probably translation (43). eIF4E2 seemed to interact with multiple species of phosphorylated TTP (Fig. 2). It will be interesting to investigate whether phosphorylation modification of TTP affects its interaction with eIF4E2 and eIF4E2-mediated TTP repression of target mRNA translation.
Downregulation of the endogenous eIF4E2 compromised TTP inhibition of reporter expression (Fig. 4), indicating that eIF4E2 is involved in TTP-mediated translational repression at endogenous levels. We further showed that overexpression of truncation mutants of eIF4E2 significantly relieved TTP-mediated translational repression (Fig. 5). Overexpression of TTP reduced the association of target mRNA with the polysomes, and expression of the N-terminal domain of eIF4E2 to some extent restored target mRNA association with the polysomes (Fig. 6). In addition, overexpression of an eIF4E2 mutant that lost the cap-binding activity inhibited TTP-mediated translational repression (Fig. 7C). These results imply that TTP represses the translation of target mRNA by promoting eIF4E2 to compete with eIF4E to bind to the cap structure. This notion is supported by the observation that overexpression of TTP increased the association of target mRNA with eIF4E2 but decreased target mRNA association with eIF4E (Fig. 8). Based on these results, we propose that TTP inhibits the assembly of translation initiation complex on target mRNA through recruiting eIF4E2 to block the cap structure.
To validate the function of TTP and eIF4E2 in TNF-α translational regulation under physiological conditions, we measured the effect of downregulation of TTP or eIF4E2 on TNF-α production in THP-1 cells. While downregulation of TTP increased the production of TNF-α at both mRNA and protein levels, downregulation of eIF4E2 increased the production of TNF-α protein without affecting the mRNA levels (Fig. 9).
In addition to recruiting eIF4E2 to repress target mRNA translation, TTP may have other mechanisms to repress translation. It has been reported previously that TTP competes with HuR to bind to the ARE motif to inhibit translation (19). This mechanism seems to be distinct from the eIF4E2-mediated translational repression, although further investigation is needed to explore the relationship between these two processes. The RNA helicase RCK has been reported to interact with TTP and be required for TTP-mediated translational repression (18). Our preliminary results showed that RCK interacted with eIF4E2a (Tao and Gao, unpublished). It will be interesting to investigate whether RCK is involved in eIF4E2-mediated translational repression.
In summary, we developed a reporter with increased sensitivity to TTP-mediated translational repression. Using this system, we investigated the mechanism underlying TTP-mediated translational repression. We provide evidence supporting a model that TTP binds to target mRNA and recruits eIF4E2 to the cap structure of the target mRNA to block translational initiation. This mechanism is further supported by the observation that downregulation of endogenous eIF4E2 enhanced TNF-α production.
ACKNOWLEDGMENTS
This work was supported by grants to Guangxia Gao from the Ministry of Science and Technology of China (973 Program 2012CB910203) and the Chinese Academy of Sciences (KJZD-EWL10-01).
REFERENCES
- 1.Gruber AR, Fallmann J, Kratochvill F, Kovarik P, Hofacker IL. 2011. AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res 39:D66–D69. doi: 10.1093/nar/gkq990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. 1996. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445–454. doi: 10.1016/S1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 3.Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 4.Barreau C, Paillard L, Osborne HB. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khabar KS. 2010. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci 67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. 2004. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J Biol Chem 279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 7.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. 2011. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol 31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchese FP, Aubareda A, Tudor C, Saklatvala J, Clark AR, Dean JL. 2010. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J Biol Chem 285:27590–27600. doi: 10.1074/jbc.M110.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai WS, Kennington EA, Blackshear PJ. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol 23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lykke-Andersen J, Wagner E. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. 2007. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem 100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 12.Frasca D, Romero M, Landin AM, Diaz A, Riley RL, Blomberg BB. 2010. Protein phosphatase 2A (PP2A) is increased in old murine B cells and mediates p38 MAPK/tristetraprolin dephosphorylation and E47 mRNA instability. Mech Ageing Dev 131:306–314. doi: 10.1016/j.mad.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson BA, Stehn JR, Yaffe MB, Blackwell TK. 2002. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J Biol Chem 277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- 14.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks SA, Connolly JE, Diegel RJ, Fava RA, Rigby WF. 2002. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthritis Rheum 46:1362–1370. doi: 10.1002/art.10235. [DOI] [PubMed] [Google Scholar]
- 16.Rigby WF, Roy K, Collins J, Rigby S, Connolly JE, Bloch DB, Brooks SA. 2005. Structure/function analysis of tristetraprolin (TTP): p38 stress-activated protein kinase and lipopolysaccharide stimulation do not alter TTP function. J Immunol 174:7883–7893. doi: 10.4049/jimmunol.174.12.7883. [DOI] [PubMed] [Google Scholar]
- 17.Pfeiffer JR, Brooks SA. 2012. Cullin 4B is recruited to tristetraprolin-containing messenger ribonucleoproteins and regulates TNF-alpha mRNA polysome loading. J Immunol 188:1828–1839. doi: 10.4049/jimmunol.1102837. [DOI] [PubMed] [Google Scholar]
- 18.Qi MY, Wang ZZ, Zhang Z, Shao Q, Zeng A, Li XQ, Li WQ, Wang C, Tian FJ, Li Q, Zou J, Qin YW, Brewer G, Huang S, Jing Q. 2012. AU-rich-element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol Cell Biol 32:913–928. doi: 10.1128/MCB.05340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. 2012. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet 8:e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershey JW, Sonenberg N, Mathews MB. 2012. Principles of translational control: an overview. Cold Spring Harb Perspect Biol 4:a011528. doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Wang X, Goff SP, Gao G. 2012. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J 31:4236–4246. doi: 10.1038/emboj.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rom E, Kim HC, Gingras AC, Marcotrigiano J, Favre D, Olsen H, Burley SK, Sonenberg N. 1998. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem 273:13104–13109. doi: 10.1074/jbc.273.21.13104. [DOI] [PubMed] [Google Scholar]
- 24.Ptushkina M, Berthelot K, von der Haar T, Geffers L, Warwicker J, McCarthy JE. 2001. A second eIF4E protein in Schizosaccharomyces pombe has distinct eIF4G-binding properties. Nucleic Acids Res 29:4561–4569. doi: 10.1093/nar/29.22.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita M, Ler LW, Fabian MR, Siddiqui N, Mullin M, Henderson VC, Alain T, Fonseca BD, Karashchuk G, Bennett CF, Kabuta T, Higashi S, Larsson O, Topisirovic I, Smith RJ, Gingras AC, Sonenberg N. 2012. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol 32:3585–3593. doi: 10.1128/MCB.00455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuberek J, Kubacka D, Jablonowska A, Jemielity J, Stepinski J, Sonenberg N, Darzynkiewicz E. 2007. Weak binding affinity of human 4EHP for mRNA cap analogs. RNA 13:691–697. doi: 10.1261/rna.453107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. 2005. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Villaescusa JC, Buratti C, Penkov D, Mathiasen L, Planaguma J, Ferretti E, Blasi F. 2009. Cytoplasmic Prep1 interacts with 4EHP inhibiting Hoxb4 translation. PLoS One 4:e5213. doi: 10.1371/journal.pone.0005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S. 2012. An oxygen-regulated switch in the protein synthesis machinery. Nature 486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uniacke J, Perera JK, Lachance G, Francisco CB, Lee S. 2014. Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer Res 74:1379–1389. doi: 10.1158/0008-5472.CAN-13-2278. [DOI] [PubMed] [Google Scholar]
- 31.Wiklund L, Sokolowski M, Carlsson A, Rush M, Schwartz S. 2002. Inhibition of translation by UAUUUAU and UAUUUUUAU motifs of the AU-rich RNA instability element in the HPV-1 late 3′ untranslated region. J Biol Chem 277:40462–40471. doi: 10.1074/jbc.M205929200. [DOI] [PubMed] [Google Scholar]
- 32.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo X, Ma J, Sun J, Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci U S A 104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wax SD, Nakamura H, Anderson PJ. 2005. The tumor necrosis factor-alpha AU-rich element inhibits the stable association of the 40S ribosomal subunit with RNA transcripts. Biochem Biophys Res Commun 333:1100–1106. doi: 10.1016/j.bbrc.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 35.Rhoads RE. 2009. eIF4E: new family members, new binding partners, new roles. J Biol Chem 284:16711–16715. doi: 10.1074/jbc.R900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosettani P, Knapp S, Vismara MG, Rusconi L, Cameron AD. 2007. Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms. J Mol Biol 368:691–705. doi: 10.1016/j.jmb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 37.Ciais D, Cherradi N, Feige JJ. 2013. Multiple functions of tristetraprolin/TIS11 RNA-binding proteins in the regulation of mRNA biogenesis and degradation. Cell Mol Life Sci 70:2031–2044. doi: 10.1007/s00018-012-1150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks SA, Blackshear PJ. 2013. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta 1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanduja S, Blanco FF, Dixon DA. 2011. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA 2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. 2013. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 41.Fukaya T, Tomari Y. 2012. MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol Cell 48:825–836. doi: 10.1016/j.molcel.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 42.Osborne MJ, Volpon L, Kornblatt JA, Culjkovic-Kraljacic B, Baguet A, Borden KLB. 2013. eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proc Natl Acad Sci U S A 110:3877–3882. doi: 10.1073/pnas.1216862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kratochvill F, Gratz N, Qualls JE, Van De Velde LA, Chi H, Kovarik P, Murray PJ. 2015. Tristetraprolin limits inflammatory cytokine production in tumor-associated macrophages in an mRNA decay-independent manner. Cancer Res 75:3054–3064. doi: 10.1158/0008-5472.CAN-15-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]