Abstract
Genetic alterations in the carboxypeptidase A1 gene (CPA1) are associated with early onset chronic pancreatitis (CP). Besides CPA1, there are two other human pancreatic carboxypeptidases (CPA2 and CPB1). Here we examined whether CPA2 and CPB1 alterations are associated with CP in Japan and Germany. All exons and flanking introns of CPA2 and CPB1 were sequenced in 477 Japanese patients with CP (234 alcoholic, 243 nonalcoholic) and in 497 German patients with nonalcoholic CP by targeted next-generation sequencing and/or Sanger sequencing. Secretion and enzymatic activity of CPA2 and CPB1 variants were determined after transfection into HEK 293T cells. We identified six nonsynonymous CPA2 variants (p.V67I, p.G166R, p.D168E, p.D173H, p.R237W, and p.G388S), eight nonsynonymous CPB1 alterations (p.S65G, p.N120S, p.D172E, p.R195H, p.D208N, p.F232L, p.A317V, and p.D364Y), and one splice-site variant (c.687+1G>T) in CPB1. Functional analysis revealed essentially complete loss of function in CPA2 variants p.R237W and p.G388S and CPB1 variants p.R110H and p.D364Y. None of the CPA2 or CPB1 variants, including those resulting in a marked loss of function, were overrepresented in patients with CP. In conclusion, CPA2 and CPB1 variants are not associated with CP.
Keywords: next-generation sequencing, loss-of-function variant, misfolding, secretion defect
chronic pancreatitis (CP) is a progressive inflammatory disease that eventually leads to exocrine and/or endocrine dysfunction (15, 21). Genetic predisposition is a common etiological factor in CP, and affected individuals often carry mutations in multiple susceptibility genes, such as protease serine 1, cationic trypsinogen (PRSS1), serine protease inhibitor Kazal type 1 (SPINK1), chymotrypsin C (CTRC), and cystic fibrosis transmembrane conductance regulator (CFTR) (6–8, 12, 13, 20, 21, 22). Mutations in these risk genes alter activation or degradation of trypsinogen and result in elevated intrapancreatic trypsin activity (2, 6–8, 12, 13, 16, 20, 21, 23). However, in our nationwide survey of hereditary pancreatitis in Japan (7), about 30% of the families had no mutations in these genes, indicating that other, yet unknown, susceptibility genes exist. In 2013, an international study showed that functionally impaired carboxypeptidase A1 gene (CPA1) variants are strongly associated with early onset CP worldwide (22). Functional analysis revealed that loss of CPA1 activity or protein has no effect on trypsinogen activation, indicating that loss-of-function CPA1 variants exert their effect via a mechanism unrelated to trypsin activity. Further studies suggested that mutation-induced misfolding of CPA1 with subsequent endoplasmic reticulum (ER) stress in pancreatic acinar cells might be a relevant pathogenic mechanism (22). The discovery that CPA1 mutations predispose to CP raised the question whether variants in the two other pancreatic carboxypeptidases, CPA2 and CPB1, also contribute to pancreatitis risk. Thus, we examined whether CPA2 and CPB1 variants are associated with CP.
METHODS
Nomenclature and reference sequences.
Nucleotide numbering reflects coding DNA numbering with c.1 corresponding to the first nucleotide of the translation initiation codon. Amino acid numbering starts with the initiator methionine of the primary translation product. The open-reading frame of human CPA2 contains two methionines at its NH2-terminus (Met-Ala-Met-Arg-). In the present study amino acid numbering of CPA2 starts with the first methionine according to convention. Note that previously we designated the second methionine as the NH2-terminal amino acid of human CPA2, because only this methionine is conserved in other mammalian CPA2 enzymes (17). CPA2 (MIM600688) maps to chromosome 7q32.2, spans ∼23 kb, and contains 11 exons. CPB1 (MIM114852) maps to chromosome 3q24, spans ∼69 kb, and contains 11 exons. Genbank reference sequences used were NM_001869.2 for CPA2 and NM_001871.2 for CPB1.
Subjects.
In the Japanese cohort, 234 unrelated patients with alcoholic CP (216 male, 18 female; median age: 57.0 yr; mean age ± SD: 54.8 ± 13.7 yr; age range: 21–87 yr) and 243 patients with nonalcoholic CP (208 idiopathic, 22 hereditary, and 13 familial; 121 male, 122 female; median age: 35.5 yr; mean age ± SD: 39.3 ± 22.9 yr; age range: 1–93 yr) were enrolled. All patients were Japanese living in Japan. Patients were referred to us for the treatment of CP and/or genetic testing. In the German cohort, 497 unrelated patients with nonalcoholic CP (270 female, 227 male; median age: 13 yr; mean age ± SD: 17.5 ± 14.2 yr; age range: 1–71 yr) and 410 controls for CPB1 (or 221 controls for CPA2) (209 female, 201 male; median age: 28 yr; mean age ± SD: 34.7 ± 13.1 yr; age range: 19–68 yr) were enrolled. All patients and controls were of German descent. Patients were referred to us for genetic testing from virtually all parts of Germany, because the German research group serves as a genetic reference laboratory for pancreatic diseases. Healthy, unrelated German medical students/staff and blood donors were recruited as controls for the genetic association studies. The diagnosis of CP was based on at least two separate episodes of abdominal pain and radiological findings of pancreatic calcifications by computed tomography, endoscopic ultrasonography, and/or morphological findings such as pancreatic ductal irregularities and dilatations revealed by endoscopic retrograde cholangiopancreatography or by magnetic resonance imaging (3). Hereditary pancreatitis was diagnosed when one first-degree relative or two or more second-degree relatives had recurrent acute pancreatitis or CP without any apparent predisposing factor (4). Patients with CP in whom the criteria for hereditary pancreatitis were not met but who had at least two affected family members were classified as having familial pancreatitis. Idiopathic CP was diagnosed in the absence of a positive family history or possible predisposing factors such as alcohol abuse, trauma, medication, and anatomic abnormalities. Patients who consumed alcohol over 80 g/day (for men) or 60 g/day (for women) for more than two years were classified as alcoholic CP. All individuals gave written informed consent. If patients were under the age of 18 yr, parents gave informed consent. This study was approved by the Ethics Committee of the Tohoku University School of Medicine (2013-1-498 and 2014-1-221) and of the Technische Universität München (2525/09).
Targeted NGS.
After written informed consent was obtained, genomic DNA was extracted from peripheral blood leukocytes. We used the online design tool SureDesign to generate a customized HaloPlex Target Enrichment System (Agilent Technologies, Santa Clara, CA) targeting the regions including CPA2 and CPB1 exons and 50 nucleotides of flanking introns. The HaloPlex Target Enrichment System relies on a tailored cocktail of restriction enzymes and customized probes to capture genomic regions of interest, which are subsequently amplified by PCR. Sequencing libraries were prepared according to the manufacturer's instructions. Briefly, genomic DNA was digested with restriction enzymes, followed by hybridization to the biotinylated HaloPlex probe library in the presence of the indexing primer cassette. Hybridization results in the circularization of genomic DNA fragments and incorporation of indexes and Illumina sequencing motifs. Hybridized probes were captured with streptavidin-coated magnetic beads. Subsequently, libraries were amplified by PCR to produce a sequencing-ready, target-enriched sample. All libraries of target-enriched DNA were analyzed on a 2200 TapeStation (Agilent Technologies) to verify successful enrichment. All samples were sequenced on the Illumina Miseq platform (Illumina Japan, Tokyo, Japan) with paired-end 151-base pair reads according to the manufacturer's instruction. The sequencing data covered 99.9% of the coding regions of the CPA2 gene by >20 reads with a mean read depth of 621 and a median depth of 551. Similarly, the sequencing data covered 96.4% of the coding regions of the CPB1 gene by >20 reads with a mean read depth of 532 and a median depth of 479. These results demonstrated a high-resolution capability of our targeted NGS system for the identification of variants.
Sanger sequencing.
All exons and adjacent intronic regions of CPA2 and CPB1 were amplified by PCR using the primer sets listed in Table 1. Cycle conditions were as follows: preheating at 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and then a final extension at 72°C for 5 min. PCR products were cleaned up using the illustra ExoProStar S (GE Healthcare Life Sciences, Little Chalfont, UK). The PCR products were sequenced using an ABI Prism BigDye Terminator Cycle Sequencing Kit, V.3.1 on ABI3730xl DNA Analyzer (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions.
Table 1.
Primers used for PCR amplification in direct sequencing
| Exon | Forward | Reverse |
|---|---|---|
| CPA2 | ||
| 1 | AGTTCTCAGAAGGCCAGCAG | TCCTTTTTGATCATCCACTCC |
| 2 | TGCAGAAAGCAACAGGAAAC | GCCCAATGTCCTGTAGTTGG |
| 3 | CTGTCCATTCCCCAGAACAG | CTGCTGGAAGGTCCTAGTGG |
| 4 | AAAGCTTGGTACTGGGGACC | GCAGGAGTAGGTGTGAAGCC |
| 5 | AAGGGAATGGGGAAAATCAC | GGACCGGTTAGGAGGAAAAG |
| 6 | AGGGTAGAAAGGAGGCAACC | AGCCCGTGTGACAAGAGAAG |
| 7 | GATCCCTGGTTCACATCTGG | CTTCTCTAACCCTCATTTCCG |
| 8 | GATCAGGCTGAATGACCCTC | TGTGGGAAAACCAAGATGTG |
| 9 | TGGTGACCTCCTCCCTACAG | ATGGATGTTTGTTGCTTGGC |
| 10 | GGCAACTGATCTCCCAAGAG | TTTTGGCTCAAACTCACACG |
| 11 | AGAGGTTCCATTGCGTGAAG | AAACTGGAGAAACGGTGCTG |
| CPB1 | ||
| 1, 2 | TTGCAATCAGGAGAAGAAAGC | TGTCGGAATCACAGTGCG |
| 3 | CTTGGTTCCAACCCTTGTTC | AAGGCATTGGTCTTGGTCTC |
| 4 | CTCTGGAGCAGCTGAAGGTC | AGGGCTGGATTCTCAGTGG |
| 5 | AGTTTGATAGCCGGGTTCG | AGACATGGCAAGGTCATGG |
| 6 | ATCATGGGATCCAGCTCTTG | TTTCCAAAATTGGGCACTTG |
| 7 | CACCTCAATAGGAAATGCTGC | TCGGCTCTGAAAGTGGACTAC |
| 8 | TGATGGCTACATCTACACCTGG | AATGGATACATGCAAAATGGC |
| 9 | TTCACTAGGTGATCCTTGCC | TTGTATTTCTTTTAGCCAACTCCAG |
| 10 | TGGGGAGGGTATTAGGGAAG | TGCACAGTTCTGAAGCCAAG |
| 11 | TTTTGCACAGTGAAACATTTG | CACGGCTTGCTTAGTCATTC |
CPA1, carboxypeptidase A1 gene; CPB1, carboxypeptidase B1 gene.
Plasmid construction and mutagenesis.
Expression plasmids carrying the coding DNA of human CPA2 and CPB1 genes in the pcDNA3.1(−) vector were described previously (17). For this study, we constructed a new CPB1 expression plasmid in which we restored the native 3′-untranslated region [181 nucleotides (nt) without the polyA tail] missing from the original design. This was achieved by cloning a custom-synthesized 439 nt DNA fragment (GenScript, Piscataway, NJ) into the existing CPB1 expression plasmid using BsgI and BamHI restriction sites. Mutations were generated by overlap extension PCR mutagenesis and cloned into the expression plasmids.
Cell culture and transfection.
HEK 293T cells (GenHunter, Nashville, TN) were cultured in six-well tissue culture plates at a density of 1.5 × 106 cells/well, in Dulbecco's modified Eagle medium (DMEM) (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum, 4 mM glutamine, and 1% penicillin/streptomycin at 37°C. Transfections were carried out using 4 μg plasmid DNA and 10 μl Lipofectamine 2000 (Life Technologies) in 2 ml DMEM. After overnight incubation, the transfection medium was removed, and cells were rinsed and covered with 1.5 ml OptiMEM reduced serum medium (Life Technologies). Conditioned medium was harvested for analysis 48 h later.
Measurement of procarboxypeptidase secretion.
Secreted protein levels of CPA2 and CPB1 precursors in the conditioned medium were determined by SDS-PAGE and densitometry. An aliquot (200 μl) of the medium was precipitated with 10% trichloroacetic acid (final concentration), and the precipitate was recovered by centrifugation, dissolved in 20 μl Laemmli sample buffer containing 100 mM dithiothreitol, and heat denatured at 95°C for 5 min. Samples were electrophoresed on 15% SDS-polyacrylamide mini gels, and proteins were stained with Coomassie Blue (Brilliant Blue R-250). Quantitation of bands was carried out with the GelDocXR+ gel documentation system and Image Lab 3.0 software (Bio-Rad, Hercules, CA).
Measurement of carboxypeptidase activity.
Enzymatic activity of CPA2 and CPB1 in the conditioned medium was determined using N-[4-methoxyphenylazoformyl]-l-phenylalanine and N-[4-methoxyphenylazoformyl]-l-arginine substrates, respectively, as described previously (9, 17). Before the activity measurements, CPA2 was activated with 100 nM human cationic trypsin and 50 nM human CTRC, and CPB1 was activated with 100 nM human cationic trypsin (final concentrations) for 1 h at 37°C. The 40-μl activation mixture contained 20 μl conditioned medium, 0.1 M Tris·HCl (pH 8.0), 1 mM CaCl2, and 0.05% Tween 20 (final concentrations). Carboxypeptidase activity was measured by adding 50 μl assay buffer [0.1 M Tris·HCl (pH 8.0), 1 mM CaCl2, 0.05% Tween 20] and 10 μl of 600 μM substrate to the activation mix (100 μl final volume, 60 μM final substrate concentration). The decrease in absorbance was followed at 350 nm for 2 min. Rates of substrate cleavage were calculated from fits to the initial linear portion of the curves and were expressed as percent of the wild-type rate.
Statistical analysis.
The variant frequencies in the Japanese population were obtained from the Human Genetic Variation Database (HGVD) (www.genome.med.kyoto-u.ac.jp/SnpDB). HGVD aims to provide a central resource to archive and display Japanese genetic variations and association between the variations and transcription levels of genes. The database currently contains genetic variations of 1,208 individuals determined by exome sequencing. The significance of the differences in variant frequencies between patients and controls was tested by two-tailed Fisher's exact test. A P value of <0.05 was considered significant. All statistical analyses were performed using the SPSS version 17.0 statistical analysis software (SPSS, Chicago, IL). For the functional assays, data are shown as means + SD.
RESULTS
CPA2 and CPB1 variants in Japanese patients with CP.
We first performed a comprehensive analysis of CPA2 and CPB1 in 237 Japanese patients (167 idiopathic, 46 alcoholic, 15 hereditary, and 9 familial CP) by targeted NGS. The coding regions and the adjacent noncoding regions of the CPA2 gene were captured, sequenced, and analyzed. All exons in which variants were identified were then sequenced using Sanger sequencing in an additional 240 patients (41 idiopathic, 188 alcoholic, 7 hereditary, and 4 familial CP). Taken together, in 477 Japanese patients (234 alcoholic and 243 nonalcoholic), we identified six CPA2 variants (three synonymous and three nonsynonymous; Table 2) and six CPB1 variants (two synonymous and three nonsynonymous variants, and a variant affecting the 5′-splice site in intron 7; Table 3). There were no significant differences in the frequency of any of the CPA2 and CPB1 variants between the 477 Japanese CP patients and the control group (HGVD) (Tables 2 and 3). The frequencies were not different even if the patients were stratified by etiologies (Tables 2 and 3). Of note, the frequency of the CPA2 p.R237W variant in the Japanese population was 23/673 (3.4%) according to the HGVD. This is higher than observed in European American (0.1%) or in African American (0.2%) populations, according to the NHLBI Exome Sequencing Project (http://evs.gs.washington.edu/EVS/).
Table 2.
CPA2 variants identified in Japanese patients with CP
|
P Value |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon | Nucleotide Change | Amino Acid Change | Genotype | All CP (%) | NACP (%) | ACP (%) | HGVD (%) | All CP | NACP | ACP |
| Nonsynonymous | ||||||||||
| 6 | c.504C>G | p.D168E | CG | 1/477 (0.2) | 1/243 (0.4) | 0/234 (0) | 1/373 (0.3) | 1.00 | 1.00 | 1.00 |
| 8 | c.709C>T | p.R237W | CT | 19/477 (4.0) | 9/243 (3.7) | 10/234 (4.3) | 23/673 (3.4) | 0.63 | 0.84 | 0.55 |
| 11 | c.1162G>A | p.G388S | GA | 1/477 (0.2) | 1/243 (0.4) | 0/234 (0) | 0/673 (0) | 0.41 | 0.27 | 1.00 |
| Synonymous | ||||||||||
| 7 | c.633T>C | p.D211= | TC | 159/477 (33.3) | 71/243 (29.2) | 88/234 (37.6) | 417/1208 (34.5) | 0.75 | 0.28 | 0.42 |
| CC | 291/477 (61.0) | 159/243 (65.4) | 132/234 (56.4) | 732/1208 (60.6) | ||||||
| 9 | c.828C>T | p.H276= | CT | 5/477 (1.0) | 2/243 (0.8) | 3/234 (1.3) | 17/747 (2.3) | 0.13 | 0.19 | 0.44 |
| 11 | c.1161C>T | p.Y387= | CT | 5/477 (1.0) | 1/243 (0.4) | 4/234 (1.7) | 19/1140 (1.7) | 0.50 | 0.23 | 1.00 |
CP, chronic pancreatitis; NACP, nonalcoholic CP; ACP, alcoholic CP; HGVD, Human Genetic Variation Database. P values were determined between HGVD and all CP, NACP, or ACP by the Fisher's exact test.
Table 3.
CPB1 variants identified in Japanese patients with CP
|
P Value |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon | Nucleotide Change | Amino Acid Change | Genotype | All CP (%) | NACP (%) | ACP (%) | HGVD (%) | All CP | NACP | ACP |
| Nonsynonymous | ||||||||||
| 7 | c.622G>A | p.D208N | GA | 155/477 (32.5) | 80/243 (32.9) | 75/234 (32.1) | 409/1191 (34.3) | 0.42 | 0.54 | 0.56 |
| AA | 27/477 (56.6) | 14/243 (5.8) | 13/234 (5.6) | 51/1191 (4.3) | ||||||
| 8 | c.694T>C | p.F232L | TC | 29/477 (6.1) | 13/243 (5.3) | 16/234 (6.8) | 45/1132 (4.0) | 0.18 | 0.39 | 0.15 |
| CC | 1/477 (0.2) | 1/243 (0.4) | 0/234 (0) | 3/1132 (0.3) | ||||||
| 11 | c.950C>T | p.A317V | CT | 1/477 (0.2) | 0/243 (0) | 1/234 (0.4) | 3/803 (0.4) | 1.00 | 1.00 | 1.00 |
| Splice site | ||||||||||
| Intron 7 | c.687 + 1G>T | GT | 1/477 (0.2) | 1/243 (0.4) | 0/234 (0) | 0/673 (0) | 0.41 | 0.27 | 1.00 | |
| Synonymous | ||||||||||
| 9 | c.828C>T | p.A276= | CT | 2/477 (0.4) | 1/243 (0.4) | 1/234 (0.4) | 1/429 (0.2) | 1.00 | 1.00 | 1.00 |
| 11 | c.1182C>T | p.I394= | CT | 1/477 (0.2) | 1/243 (0.4) | 0/234 (0) | 2/429 (0.5) | 0.61 | 1.00 | 0.54 |
P values were determined between HGVD and all CP, NACP, or ACP by the Fisher's exact test.
CP is a complex multigenetic disease, and affected individuals often carry mutations in several susceptibility genes (7, 12). We investigated all 12 Japanese subjects with nonalcoholic CP carrying rare nonsynonymous or splice-site variants in CPA2 or CPB1 for mutations in PRSS1, SPINK1, CTRC, CFTR, and CPA1. One patient was homozygous for SPINK1 p.N34S, and another patient was homozygous for SPINK1 c.194+2T>C, but none carried a pathogenic genetic alteration in the other genes investigated.
CPA2 and CPB1 variants in German CP patients.
Geographical and ethnic differences exist in the genetics of CP (7, 12). Therefore, we examined German CP patients for CPA2 and CPB1 variants by direct sequencing of all exons and adjacent intronic regions. In 497 individuals with nonalcoholic CP we identified 4 nonsynonymous and 6 synonymous CPA2 variants (Table 4). The CPA2 p.R237W variant was found also in the Japanese subjects. In CPB1, we identified 11 nonsynonymous and one synonymous variant (Table 5). Five nonsynonymous variants were identified only in controls. We observed no significant differences in the frequency of any of these CPA2 and CPB1 variants between patients and controls. The frequency of the CPA2 p.R237W variant was 1/497 (0.2%) in patients and 0/221 (0%) in controls, suggesting that this variant, in contrast to Japan, is rare in Germany.
Table 4.
CPA2 variants identified in German patients with CP
| Exon | Nucleotide Change | Amino Acid Change | Genotype | Nonalcoholic CP (%) | Controls (%) | P Value |
|---|---|---|---|---|---|---|
| Nonsynonymous | ||||||
| 3 | c.199G>A | p.V67I | GA | 1/497 (0.2) | 0/221 (0) | 1.00 |
| 6 | c.496G>A | p.G166R | GA | 1/497 (0.2) | 0/221 (0) | 1.00 |
| 6 | c.523G>C | p.D175H | GC | 1/497 (0.2) | 0/221 (0) | 1.00 |
| 8 | c.709C>T | p.R237W | CT | 1/497 (0.2) | 0/221 (0) | 1.00 |
| Synonymous | ||||||
| 3 | c.198C>T | p.H66= | CT | 1/497 (0.2) | 0/221 (0) | 1.00 |
| 6 | c.507G>A | p.K169= | GA | 1/497 (0.2) | 0/221 (0) | 1.00 |
| 7 | c.633C>T | p.D211= | CT | 234/497 (47.1) | 117/221 (52.9) | 0.17 |
| p.D211= | TT | 100/497 (20.1) | 34/221 (15.4) | |||
| 9 | c.828C>T | p.H276= | CT | 5/497 (1.0) | 1/221 (0.5) | 0.67 |
| 9 | c.858G>A | p.V286= | GA | 2/497 (0.4) | 1/221 (0.5) | 1.00 |
| 9 | c.939G>A | p.M313= | GA | 1/497 (0.2) | 0/221 (0) | 1.00 |
Table 5.
CPB1 variants identified in German patients with CP
| Exon | Nucleotide Change | Amino Acid Change | Genotype | Nonalcoholic CP (%) | Controls (%) | P Value |
|---|---|---|---|---|---|---|
| Nonsynonymous | ||||||
| 3 | c.193A>G | p.S65G | AG | 6/497 (1.2) | 4/410 (1.0) | 1.00 |
| 4 | c.329G>A | p.R110H | GA | 0/497 (0) | 1/410 (0.2) | 0.45 |
| 4 | c.359A>G | p.N120S | AG | 1/497 (0.2) | 1/410 (0.2) | 1.00 |
| 5 | c.457G>A | p.A153T | GA | 0/497 (0) | 1/410 (0.2) | 0.45 |
| 6 | c.516C>A | p.D172E | CA | 12/497 (2.4) | 4/410 (1.0) | 0.13 |
| 7 | c.584G>A | p.R195H | GA | 12/497 (2.4) | 11/410 (2.7) | 0.83 |
| 7 | c.622G>A | p.D208N | GA | 160/497 (32.2) | 138/410 (33.7) | 0.82 |
| AA | 24/497 (4.8) | 17/410 (4.2) | ||||
| 8 | c.622G>A | p.F232L | GA | 0/497 (0) | 1/410 (0.2) | 0.45 |
| 9 | c.927G>T | p.M309I | GT | 0/497 (0) | 1/410 (0.2) | 0.45 |
| 11 | c.1090G>T | p.D364Y | GT | 1/497 (0.2) | 1/410 (0.2) | 1.00 |
| 11 | c.1207C>G | p.L403V | CG | 0/497 (0) | 1/410 (0.2) | 0.45 |
| Synonymous | ||||||
| 3 | c.192C>T | p.H64= | CT | 2/497 (0.4) | 2/410 (0.5) | 1.00 |
We investigated all German patients with nonalcoholic CP for mutations in PRSS1, SPINK1, CTRC, CFTR, and CPA1. The subject carrying the CPA2 p.G166R was heterozygous for the PRSS1 p.R122H, and the individual carrying the CPA2 p.R237W was heterozygous for the SPINK1 p.N34S. Four subjects carrying the CPB1 p.S65G had pathogenic alterations in the PRSS1 or SPINK1 genes: PRSS1 p.R122H (one heterozygote), SPINK1 p.N34S (one homozygote and one heterozygote), and SPINK1 c.194+2T>C (one heterozygote). The subject carrying the CPB1 p.D172E was heterozygous for the PRSS1 p.R122H, and another subject carrying this variant was heterozygous for the CFTR F508del. Three subjects carrying the CPB1 p.R195H were heterozygous for the CFTR F508del, and two subjects were heterozygous for the SPINK1 p.N34S. One subject carrying this variant was heterozygous for the SPINK1 p.V60YfsX35.
Functional analysis.
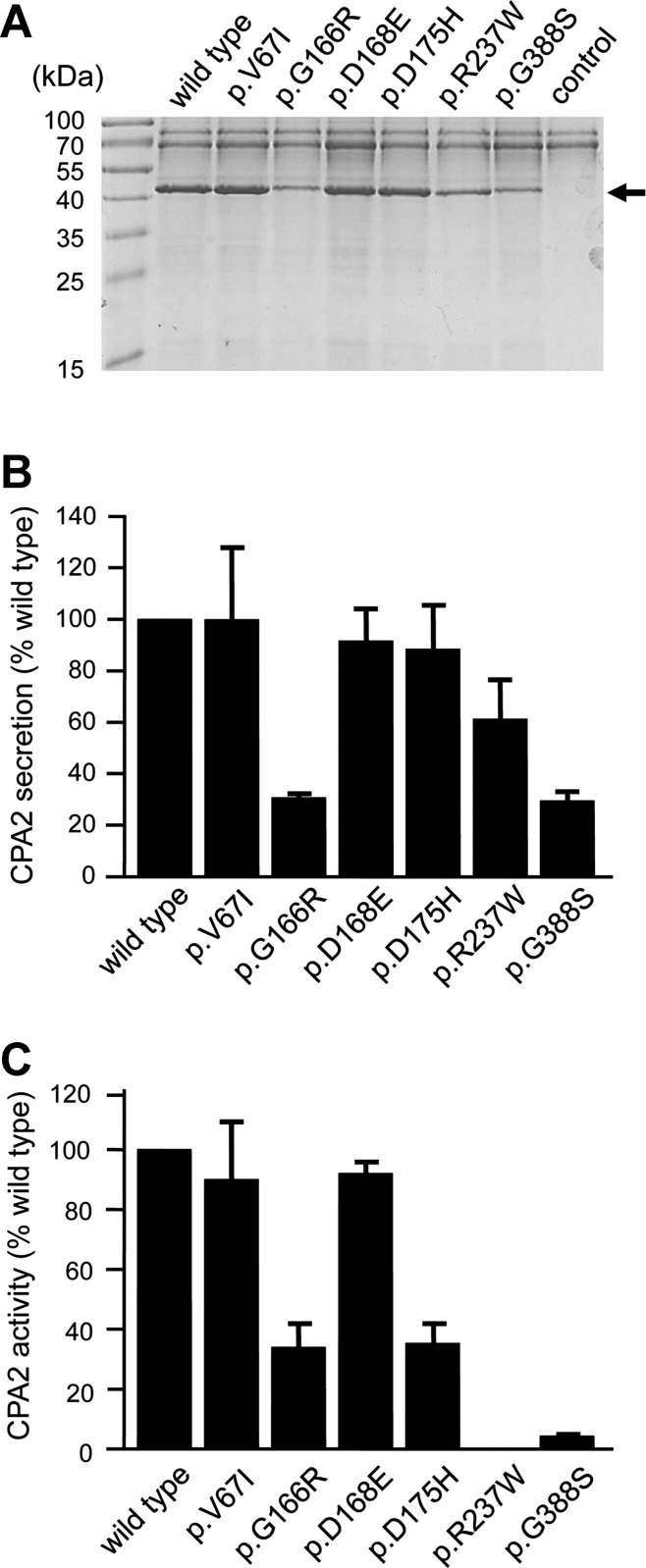
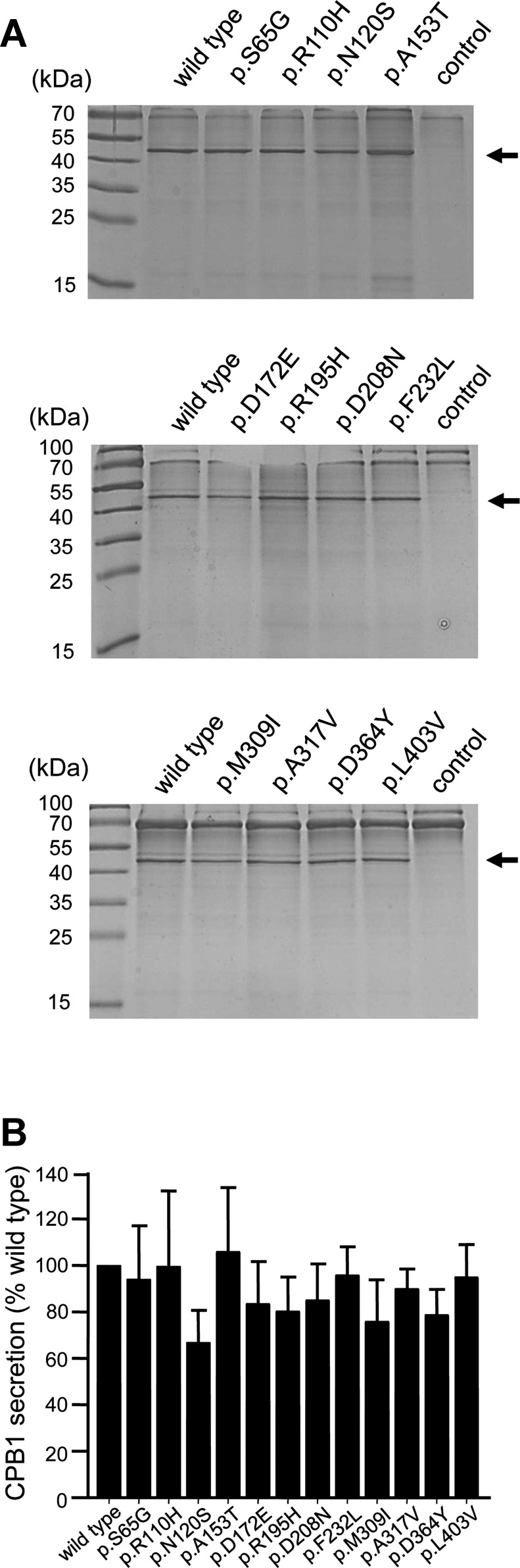
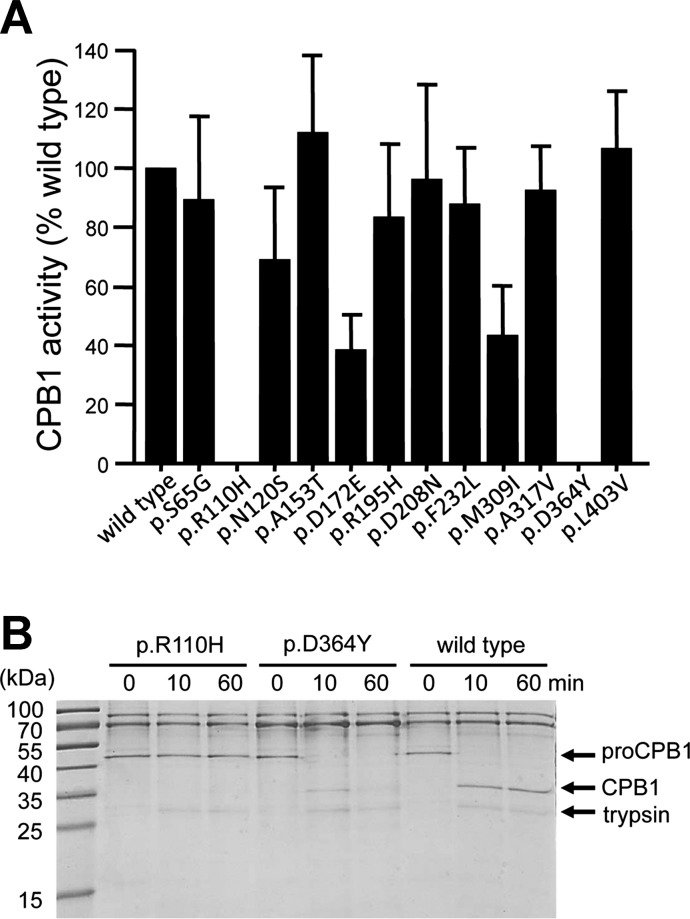
To clarify the functional consequences of the nonsynonymous CPA2 and CPB1 variants, we transfected HEK 293T cells and measured proenzyme secretion and carboxypeptidase activity in the conditioned medium. We found that secretion of CPA2 variants p.G166R and p.G388S was reduced to ∼30% of the wild type, whereas variant p.R237W was secreted to 60% of wild-type levels (Fig. 1, A and B). The lower carboxypeptidase activity (∼34% of wild type) of variant p.G166R in the conditioned medium was consistent with its reduced secretion levels; however, the diminished activity (5% of wild type) of variant p.G388S could not be explained by reduced secretion alone and pointed to a catalytic defect (Fig. 1C). As expected, variant p.R237W, which affects a catalytic residue in CPA2 (10, 11), was completely inactive (Fig. 1C). A similar mutation, p.R237H, was also identified in CPA1 previously in two control subjects (22). Secretion of CPB1 was not significantly affected by any of the variants studied (Fig. 2). Despite normal secretion, no CPB1 activity was measurable in the conditioned medium of variants p.R110H and p.D364Y (Fig. 3A). Variant p.R110H was resistant to activation by trypsin due to the destruction of the activating cleavage site, whereas variant p.D364Y was proteolytically degraded by trypsin (Fig. 3B). Finally, relative to their normal secretion levels, variants p.D172E and p.M309I exhibited lower activity (∼40% of wild type), indicating partial catalytic defects (Fig. 3A).
Fig. 1.
Secretion and activity of carboxypeptidase A2 (CPA2) variants. HEK 293T cells were transfected with the indicated constructs, and conditioned medium was collected 48 h after transfection. A and B: levels of secreted CPA2 were determined by SDS-PAGE and Coomassie Blue staining as described in Methods. A: a representative gel is shown. The molecular mass markers were PageRuler Prestained Protein Ladder (PI-26616; Thermo Scientific). The arrow indicates bands for CPA2. B: densitometric quantitation of secreted CPA2 levels. Average values from 3 transfections with SD are indicated, expressed as percent of wild-type levels. C: activity of the CPA2 variants. CPA2 in the conditioned medium was activated with trypsin and chymotrypsin C, and carboxypeptidase activity was determined as described in Methods. Activity (average values ± SD, n = 3) was expressed as percent of wild type.
Fig. 2.
Secretion of the nonsynonymous CPB1 variants. HEK 293T cells were transfected with the indicated constructs, and conditioned medium was collected 48 h after transfection. Levels of secreted CPB1 were determined by SDS-PAGE and Coomassie Blue staining as described in Methods. A: representative gels are shown. The molecular mass markers were PageRuler Prestained Protein Ladder (PI-26616; Thermo Scientific). The arrows indicate bands for CPB1. B: densitometric quantitation of secreted CPB1 levels. Average values from 4 transfections with SD are shown, expressed as percent of wild-type levels.
Fig. 3.
Activity and proteolytic stability of the CPB1 variants. A: activity of the CPB1 variants. CPB1 in the conditioned medium was activated with trypsin, and carboxypeptidase activity was measured as described in Methods. Activity was expressed as percent of the wild-type value. Averages from 4 transfections with SD are shown. B: proteolytic stability of CPB1 variants p.R110H and p.D364Y. Conditioned medium (110 μl/lane) was supplemented with 0.1 M Tris·HCl (pH 8.0) and 1 mM CaCl2 and treated with 50 nM human cationic trypsin for the indicated times (final concentrations). The reactions were stopped by precipitation with 10% trichloroacetic acid, and samples were analyzed by SDS-PAGE and Coomassie Blue staining. A representative gel from 2 experiments is shown.
Distribution of loss-of-function CPA2 and CPB1 variants between patients and controls.
Association of CPA1 variants with CP became only evident when variants with a functional defect were considered (22). In contrast to CPA1 variants, however, the loss-of-function CPA2 and CPB1 variants, individually or combined, were not enriched in CP patients relative to controls to a statistically significant extent (Tables 2–5).
DISCUSSION
Pancreatic carboxypeptidases are metalloproteases that hydrolyze COOH-terminal peptide bonds in dietary proteins and peptides (1, 19). Three different isoforms are secreted by the human pancreas. CPA1 and CPA2 act on COOH-terminal aromatic and aliphatic amino acid residues exposed by the action of chymotrypsins and elastases, whereas the CPB1 hydrolyzes COOH-terminal lysine and arginine residues generated by tryptic cleavages. Carboxypeptidases are secreted by the pancreas as inactive proenzymes containing a 94- to 96-amino acid-long propeptide. Proteolytic activation in the duodenum takes place by the sequential action of trypsin and chymotrypsin C (17).
Previously we found that CPA1 variants are strongly associated with early onset CP (22). In this study, we performed comprehensive mutational analysis of the two other human carboxypeptidases, CPA2 and CPB1, in CP patients and healthy controls. Because genetic risk factors for pancreatitis show geographical and ethnic heterogeneity (7, 12), we studied two independent populations. We identified several nonsynonymous variants in CPA2 and CPB1 and a splice site variant in CPB1. However, these variants were not overrepresented in patients with CP from Japan and Germany, suggesting that genetic variants in these carboxypeptidases do not alter disease risk. This stands in contrast to CPA1, in which functionally impaired variants are associated with CP (22).
Association of CPA1 variants with CP was revealed only after functional analysis of all variants, which demonstrated strong enrichment of variants with a functional defect in CP patients. Here we employed a similar approach and subjected all identified CPA2 and CPB1 variants to functional evaluation. We found that some variants indeed exhibited various functional defects, which included reduced secretion (CPA2 p.G166R and p.G388S), complete or partial loss of catalytic activity (CPA2 p.R237W, p.D157H, and p.G388S; CPB1 p.D172E and p.M309I), resistance to trypsin-mediated activation (CPB1 p.R110H), and degradation by trypsin (CPB1 p.D364Y). However, in contrast to CPA1 variants, these variants were found also in control subjects and were not overrepresented in patients. The relatively high frequency of the CPA2 p.R237W variant in Japan (∼4%) and the equal distribution between patients and controls offers particularly strong evidence that loss of CPA2 activity is immaterial to pancreatitis. Because all identified variants were heterozygous, loss of carboxypeptidase activity is unlikely to compromise normal food digestion either. In summary, a reduction in CPA2 or CPB1 activity seems to be unimportant for the pathogenesis of CP.
The majority of pathogenic CPA1 variants exhibit a secretion defect caused by mutation-induced misfolding and intracellular retention and degradation. With the use of the p.N256K CPA1 mutant as a test case, this misfolding phenotype was also shown to cause ER stress in acinar cells (22). A robust ER stress response can activate cell death pathways, which could potentially explain the increased risk to pancreatitis. This mechanism has been also suggested for certain misfolding variants of PRSS1 and CTRC (2, 5, 18). In this study, secretion of two private CPA2 variants found in CP patients (p.G166R and p.G388S) was reduced to ∼30% of the wild type, suggesting some degree of misfolding, although our experiments did not rule out other possibilities such as decreased mRNA expression or inefficient protein translation. The German patient carrying the CPA2 p.G166R variant was also heterozygous for PRSS1 p.R122H, which explains the development of CP and suggests that the CPA2 variant was a coincidental finding. After trypsinogens, CPA1 is the second major component of pancreatic juice, contributing about 17% of the total protein, as judged by radioactive amino acid incorporation and two-dimensional gel electrophoresis (14). In contrast, the amount of CPA2 is circa 8% while the two chromatographic forms of CPB1 constitute ∼7% of juice protein (14). Thus, compared with CPA1, there are less CPA2 and CPB1 secreted, which would render misfolding effects weaker. Nonetheless, we cannot rule out the possibility that rare CPA2 variants p.G166R and p.G388S act as weak risk factors for CP with minimal, if any, clinical relevance.
In summary, we demonstrated no association of CP with either CPA2 or CPB1 variants. Our findings highlight the important and specific role of CPA1 variants in the pathogenesis of CP.
GRANTS
This work was supported in part by the Pancreas Research Foundation of Japan (to E. Nakano), the HIROMI Medical Research Foundation (to A. Masamune), the Mother and Child Health Foundation (to A. Masamune), the Smoking Research Foundation (to A. Masamune), the Ministry of Health, Labor, Welfare of Japan (Principal investigators: Y Matsubara and Yoshifumi Takeyama), the Deutsche Forschungsgemeinschaft (Wi 2036/2–3) (to H. Witt), and National Institute of Diabetes and Digestive and Kidney Diseases Grants R01-DK-058088, R01-DK-082412, and R01-DK-095753 (to M. Sahin-Tóth).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: E.N., A.G., A.M., T.N., S.H., K.K., Y.K., Y.A., K.E., M.L., M.B., and D.A.G. performed experiments; E.N., A.G., A.M., T.N., Y.A., Y.M., K.E., M.L., M.B., D.A.G., M.S.-T., and H.W. analyzed data; E.N., A.G., A.M., K.E., M.L., M.B., D.A.G., M.S.-T., and H.W. interpreted results of experiments; E.N., A.G., A.M., T.N., S.H., K.K., Y.K., Y.A., Y.M., K.E., M.L., M.B., D.A.G., T.S., M.S.-T., and H.W. approved final version of manuscript; A.M., T.S., M.S.-T., and H.W. conception and design of research; A.M., M.S.-T., and H.W. prepared figures; A.M., M.S.-T., and H.W. drafted manuscript; A.M., M.S.-T., and H.W. edited and revised manuscript.
ACKNOWLEDGMENTS
We are grateful to Yoko Tateda for technical assistance and to Melinda Bence and Richárd Szmola for performing preliminary functional analysis of CPA2 variants.
REFERENCES
- 1.Avilés FX, Vendrell J, Guasch A, Coll M, Huber R. Advances in metallo-procarboxypeptidases. Emerging details on the inhibition mechanism and on the activation process. Eur J Biochem 211: 381–389, 1993. [DOI] [PubMed] [Google Scholar]
- 2.Beer S, Zhou J, Szabó A, Keiles S, Chandak GR, Witt H, Sahin-Tóth M. Comprehensive functional analysis of chymotrypsin C (CTRC) variants reveals distinct loss-of-function mechanisms associated with pancreatitis risk. Gut 62: 1616–1624, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etemad B, Whitcomb DC. Chronic pancreatitis: diagnosis, classification, and new genetic developments. Gastroenterology 120: 682–707, 2001. [DOI] [PubMed] [Google Scholar]
- 4.Howes N, Lerch MM, Greenhalf W, Stocken DD, Ellis I, Simon P, Truninger K, Ammann R, Cavallini G, Charnley RM, Uomo G, Delhaye M, Spicak J, Drumm B, Jansen J, Mountford R, Whitcomb DC, Neoptolemos JP. European Registry of Hereditary Pancreatitis, and Pancreatic Cancer (EUROPAC). Clinical and genetic characteristics of hereditary pancreatitis in Europe. Clin Gastroenterol Hepatol 2: 252–261, 2004. [DOI] [PubMed] [Google Scholar]
- 5.Kereszturi E, Szmola R, Kukor Z, Simon P, Weiss FU, Lerch MM, Sahin-Tóth M. Hereditary pancreatitis caused by mutation-induced misfolding of human cationic trypsinogen: a novel disease mechanism. Hum Mutat 30: 575–582, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kume K, Masamune A, Kikuta K, Shimosegawa T. [−215G>A; IVS3+2T>C] mutation in the SPINK1 gene causes exon 3 skipping and loss of the trypsin binding site. Gut 55: 1214, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masamune A. Genetics of pancreatitis: the 2014 update. Tohoku J Exp Med 232: 69–77, 2014. [DOI] [PubMed] [Google Scholar]
- 8.Masamune A, Nakano E, Kume K, Kakuta Y, Ariga H, Shimosegawa T. Identification of novel missense CTRC variants in Japanese patients with chronic pancreatitis. Gut 62: 653–654, 2013. [DOI] [PubMed] [Google Scholar]
- 9.Mock WL, Liu Y, Stanford DJ. Arazoformyl peptide surrogates as spectrophotometric kinetic assay substrates for carboxypeptidase A. Anal Biochem 239: 218–222, 1996. [DOI] [PubMed] [Google Scholar]
- 10.Phillips MA, Fletterick R, Rutter WJ. Arginine 127 stabilizes the transition state in carboxypeptidase. J Biol Chem 265: 20692–20698, 1990. [PubMed] [Google Scholar]
- 11.Phillips MA, Hedstrom L, Rutter WJ. Guanidine derivatives restore activity to carboxypeptidase lacking arginine-127. Protein Sci 1: 517–521, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosendahl J, Landt O, Bernadova J, Kovacs P, Teich N, Bödeker H, Keim V, Ruffert C, Mössner J, Kage A, Stumvoll M, Groneberg D, Krüger R, Luck W, Treiber M, Becker M, Witt H. CFTR, SPINK1, CTRC and PRSS1 variants in chronic pancreatitis: is the role of mutated CFTR overestimated? Gut 62: 582–592, 2013. [DOI] [PubMed] [Google Scholar]
- 13.Rosendahl J, Witt H, Szmola R, Bhatia E, Ozsvári B, Landt O, Schulz HU, Gress TM, Pfützer R, Löhr M, Kovacs P, Blüher M, Stumvoll M, Choudhuri G, Hegyi P, te Morsche RH, Drenth JP, Truninger K, Macek M Jr, Puhl G, Witt U, Schmidt H, Büning C, Ockenga J, Kage A, Groneberg DA, Nickel R, Berg T, Wiedenmann B, Bödeker H, Keim V, Mössner J, Teich N, Sahin-Tóth M. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat Genet 40: 78–82, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheele G, Bartelt D, Bieger W. Characterization of human exocrine pancreatic proteins by two-dimensional isoelectric focusing/sodium dodecyl sulfate gel electrophoresis. Gastroenterology 80: 461–473, 1981. [PubMed] [Google Scholar]
- 15.Steer ML, Waxman I, Freedman S. Chronic pancreatitis. N Engl J Med 332: 1482–1490, 1995. [DOI] [PubMed] [Google Scholar]
- 16.Szabó A, Sahin-Tóth M. Increased activation of hereditary pancreatitis-associated human cationic trypsinogen mutants in presence of chymotrypsin C. J Biol Chem 287: 20701–20710, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szmola R, Bence M, Carpentieri A, Szabó A, Costello CE, Samuelson J, Sahin-Tóth M. Chymotrypsin C is a co-activator of human pancreatic procarboxypeptidases A1 and A2. J Biol Chem 286: 1819–1827, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szmola R, Sahin-Tóth M. Pancreatitis-associated chymotrypsinogen C (CTRC) mutant elicits endoplasmic reticulum stress in pancreatic acinar cells. Gut 59: 365–372, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vendrell J, Querol E, Avilés FX. Metallocarboxypeptidases and their protein inhibitors. Structure, function and biomedical properties. Biochim Biophys Acta 1477: 284–298, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, Martin SP, Gates LK Jr, Amann ST, Toskes PP, Liddle R, McGrath K, Uomo G, Post JC, Ehrlich GD. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet 14: 141–145, 1996. [DOI] [PubMed] [Google Scholar]
- 21.Witt H, Apte MV, Keim V, Wilson JS. Chronic pancreatitis: challenges and advances in pathogenesis, genetics, diagnosis, and therapy. Gastroenterology 132: 1557–1573, 2007. [DOI] [PubMed] [Google Scholar]
- 22.Witt H, Beer S, Rosendahl J, Chen JM, Chandak GR, Masamune A, Bence M, Szmola R, Oracz G, Macek M Jr, Bhatia E, Steigenberger S, Lasher D, Bühler F, Delaporte C, Tebbing J, Ludwig M, Pilsak C, Saum K, Bugert P, Masson E, Paliwal S, Bhaskar S, Sobczynska-Tomaszewska A, Bak D, Balascak I, Choudhuri G, Nageshwar Reddy D, Rao GV, Thomas V, Kume K, Nakano E, Kakuta Y, Shimosegawa T, Durko L, Szabó A, Schnúr A, Hegyi P, Rakonczay Z Jr, Pfützer R, Schneider A, Groneberg DA, Braun M, Schmidt H, Witt U, Friess H, Algül H, Landt O, Schuelke M, Krüger R, Wiedenmann B, Schmidt F, Zimmer KP, Kovacs P, Stumvoll M, Blüher M, Müller T, Janecke A, Teich N, Grützmann R, Schulz HU, Mössner J, Keim V, Löhr M, Férec C, Sahin-Tóth M. Variants in CPA1 are strongly associated with early onset chronic pancreatitis. Nat Genet 45: 1216–1220, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witt H, Luck W, Hennies HC, Classen M, Kage A, Lass U, Landt O, Becker M. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet 25: 213–216, 2000. [DOI] [PubMed] [Google Scholar]