Abstract
Phytohormones are involved in diverse aspects of plant life including the regulation of plant growth, development and reproduction, as well as governing biotic and abiotic stress responses. We have generated a comprehensive transcriptional reference map of the early potato responses to exogenous application of the defence hormones abscisic acid, brassinolides (applied as epibrassinolide), ethylene (applied as the ethylene precursor aminocyclopropanecarboxylic acid), salicylic acid and jasmonic acid (applied as methyl jasmonate). Of the 39000 predicted genes on the microarray, a total of 2677 and 2473 genes were significantly differentially expressed at 1 h and 6 h after hormone treatment, respectively. Specific marker genes newly identified for the early hormone responses in potato include: a homeodomain 20 transcription factor (DMG400000248) for abscisic acid; a SAUR gene (DMG400016561) induced in epibrassinolide treated plants; an osmotin gene (DMG400003057) specifically enhanced by aminocyclopropanecarboxylic acid; a gene weakly similar to AtWRKY40 (DMG402007388) that was induced by salicylic acid; and a jasmonate ZIM-domain protein 1 (DMG400002930) which was specifically activated by methyl jasmonate. An online database has been set up to query the expression patterns of potato genes represented on the microarray that can also incorporate future microarray or RNAseq-based expression studies.
Potato is the third most important food crop in the world and sustainable production is currently dependent on the application of chemicals or the development of new cultivars to achieve tolerance or resistance to biotic and abiotic stresses1. The major pathogens of potato include the oomycete Phytophthora infestans, cause of late blight, Potato Cyst Nematodes (PCN), viruses such as potato virus Y (PVY) and bacteria including Ralstonia solanacearum (1 and references within). Abiotic pressures that impact on yield stability comprise, amongst others, heat, cold and water stresses2,3,4.
Phytohormones regulate diverse aspects of plant life5,6. For example, the hormones brassinosteroids (BR) including the most bioactive form brassinolide (BL), and abscisic acid (ABA) are predominantly involved with plant growth and development but can, as shown for BL, also feature in defence responses7,8,9,10. Indeed, phytohormones play important roles in regulation of stress and defence responses8,11. Salicylic acid (SA), ethylene (ET), jasmonic acid (JA), and its derivatives (called jasmonates), are typically associated with the regulation of biotic stress responses12,13. The SA pathway is associated with responses to biotrophic pathogens, whereas JA and ET, which are generally thought to act synergistically, are linked to defences against necrotrophic pathogens and herbivorous insects12,14.
Phytohormone signalling is conveyed through a complex network with considerable cross-talk to facilitate the required fine-tuning and modulation of responses to environmental cues5. The majority of hormone studies have been conducted in the model plant Arabidopsis thaliana and are based on mutants that are impaired in the biosynthesis of hormones or downstream signalling cascades. Recent studies on crops like rice suggest that responses can vary between species and unique complexities in defence regulatory networks have been revealed13,15,16,17. Moreover, compared to A. thaliana, both rice and potato have, for example, elevated basal levels of SA18. Although the impact of the different basal hormone levels remains elusive, it highlights the need for species-specific hormone studies.
The sequencing of the potato genome19 has enabled the design of a gene expression microarray that encompasses a representative probe set for transcripts of the predicted gene models20. The aim of this study was to generate a transcriptional reference map of the early potato responses to exogenous application of five major plant hormones ABA, BL (applied as epibrassinolide [Epi]), ET (applied as aminocyclopropanecarboxylic acid [ACC], a precursor in ET biosynthesis), JA (applied as methyl jasmonate [Me-JA]) and SA, and to identify potential marker genes for the individual hormone pathways in potato. An online database to query the expression patterns of potato genes represented on the microarray has been setup at http://ics.hutton.ac.uk/solarray.
Results
Overview of genes affected by the different plant hormones
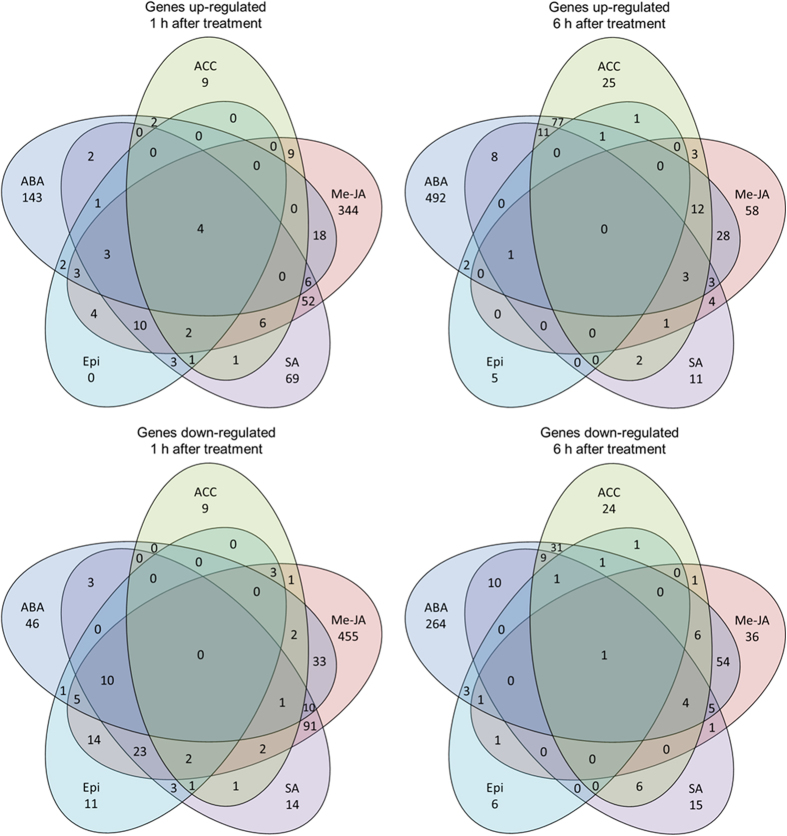
All five plant hormones had considerable effects on plant gene expression both at 1 h and 6 h after application. A total of 2677 genes showed significantly differential transcript abundance at 1 h after any of the hormone treatments and a total of 2473 genes at 6 h (p-value ≤ 0.05) (Table S1). At both time points, overlap in gene expression was observed, but large numbers of genes were uniquely up- or down-regulated by only one of the hormones (Fig. 1). Genes that were, dependent on the hormone treatment, induced or suppressed in their transcript abundance (referred to as affected in opposite direction) are highlighted in Table S1. The greatest influence on gene expression at 1 h after treatment was the application of Me-JA. In total, 344 genes were uniquely induced by Me-JA and 455 genes were uniquely down-regulated. Genes encoding members of the cytochrome P450 family were the highest (up to 25-fold) uniquely up-regulated group in Me-JA treated plants at 1 h after treatment (DMG400001098, DMG400001942, DMG400001944, DMG400015183, DMG400015184, DMG400015185, DMG400015242, DMG400016776, DMG401001941, DMG401010026, DMG402001903, DMG402001941). Genes involved in the JA biosynthesis and signalling pathways were also up-regulated 1 h after Me-JA application. These included a gene for lipoxygenase (DMG400022894) that was 2-fold induced compared to the water control and a JA-induced WRKY transcription factor (DMG400019824) that was 4-fold up-regulated. Genes for heat-shock proteins were the most (up to 13-fold) uniquely down-regulated group of transcripts in Me-JA treated plants at 1 h post treatment (DMG400000444, DMG400000398, DMG400005573, DMG400006271, DMG400009509, DMG400030405). Other genes uniquely down-regulated by Me-JA application encode EIX receptors (DMG400022978, DMG400022988, DMG400022990; 2-6-fold down-regulated) and ATP binding proteins (DMG400012203, DMG400015170, DMG400019514, DMG400030572, DMG400031081, DMG400044751, DMG401019239, DMG402003983, DMG402008483; 2-5-fold down-regulated).
Figure 1. Numbers of genes significantly affected in their expression in hormone treated vs. control plants (p-value ≤ 0.05).
Numbers of genes uniquely affected by one of the hormones and numbers of genes affected by more than one hormone are shown. Only genes that were consistently up- or down-regulated across the treatments were included in this figure. Genes that were effected in opposite direction (e.g. up-regulated by ABA but down-regulated by ACC) are not included. The figure displays genes up-regulated 1 h after treatment, genes down-regulated 1 h after treatment, genes up-regulated 6 h after treatment and genes down-regulated 6 h after treatment. ABA (abscisic acid); ACC (aminocyclopropanecarboxylic acid); Epi (epibrassinolide); Me-JA (methyl jasmonate); SA (salicylic acid).
At 1 hour post treatment, ABA, ACC and SA uniquely induced 143, 9 and 69 genes and specifically suppressed 46, 9 and 14 genes, respectively. Conversely, Epi did not up-regulate any genes uniquely but specifically down-regulated 11 genes. Me-JA and SA showed the greatest overlap at 1 h post treatment with 52 genes induced and 91 down-regulated by both hormone treatments. Examples include a transcript for a multi-drug resistance protein (DMG400024532) that was 5-fold up-regulated by Me-JA treatment and 8-fold induced by SA treatment, and an ABC transporter family protein for which the gene (DMG400009095) was induced 5-fold and 12-fold, respectively. Genes involved in heat stress were strongly down-regulated by both Me-JA and SA applications and include, for example, chloroplast small heat shock protein genes (DMG400011628, DMG400011630, DMG400038479) that were 5-20-fold down-regulated by both treatments. At 1 h after treatment, four genes were significantly up-regulated by all five hormone treatments (p-value ≤ 0.05). One of the genes encoded a beta-galactosidase (DMG400030954) which was induced 6-fold by ABA, 4-fold by ACC, 8-fold upon treatment with Epi, 17-fold by Me-JA and 9-fold by SA.
The greatest influence on transcript abundance at 6 h after treatment was the application of ABA. In total, 492 genes were uniquely induced and 264 genes were specifically down-regulated by ABA. Genes involved in the ABA pathway in plants were strongly up-regulated and include, for example, the ABA and environmental stress-inducible protein TAS14 (DMG400003530) that was up-regulated 1720-fold. An MLO1 gene (DMG400020605) involved in stress response was 113-fold up-regulated by ABA. Furthermore, non-specific lipid transfer protein genes (DMG400011951, DMG400011952, DMG400011953, DMG400011954, DMG400012837, DMG400012838, DMG400012839, DMG400016102, DMG400020131, DMG401031237) were 5-165-fold up-regulated and protein phosphatase genes (DMG400002573, DMG400009112, DMG400016742, DMG400019604, DMG400020517, DMG400027196, DMG400030332, DMG400030410) were 5-40-fold up-regulated.
Whilst fewer genes were affected by Me-JA and SA applications at 6 h after treatment if compared to 1 h, ACC had a greater effect on gene expression at 6 h than at 1 h (Fig. 1; Table S1). At this time point, ACC, Epi, Me-JA and SA uniquely up-regulated 25, 5, 58 and 11 genes and down-regulated 24, 6, 36 and 15 genes, respectively. ABA and ACC showed the greatest overlap in gene induction at 6 h after treatment and affected 77 genes jointly. Among the proteins encoded by these genes were, for example, a WRKY-type DNA binding protein (DMG400021895) that was 17-fold up-regulated by ABA and 11-fold induced by ACC as well as a tryptophan decarboxylase (DMG400018358) that was 33-fold up-regulated by both ABA and ACC.
Gene ontology classification of transcripts altered by plant hormones
A GO classification was performed to identify biological processes that are influenced by treatments with the plant hormones (Fig. 2). Generally, genes involved in the GO term annotations ‘cellular processes’, ‘single-organism processes’ and ‘metabolic processes’ were altered the most by hormone treatments.
Figure 2. Gene ontology classification of biological processes affected by any of the five hormone treatments at 1 h and 6 h are shown.
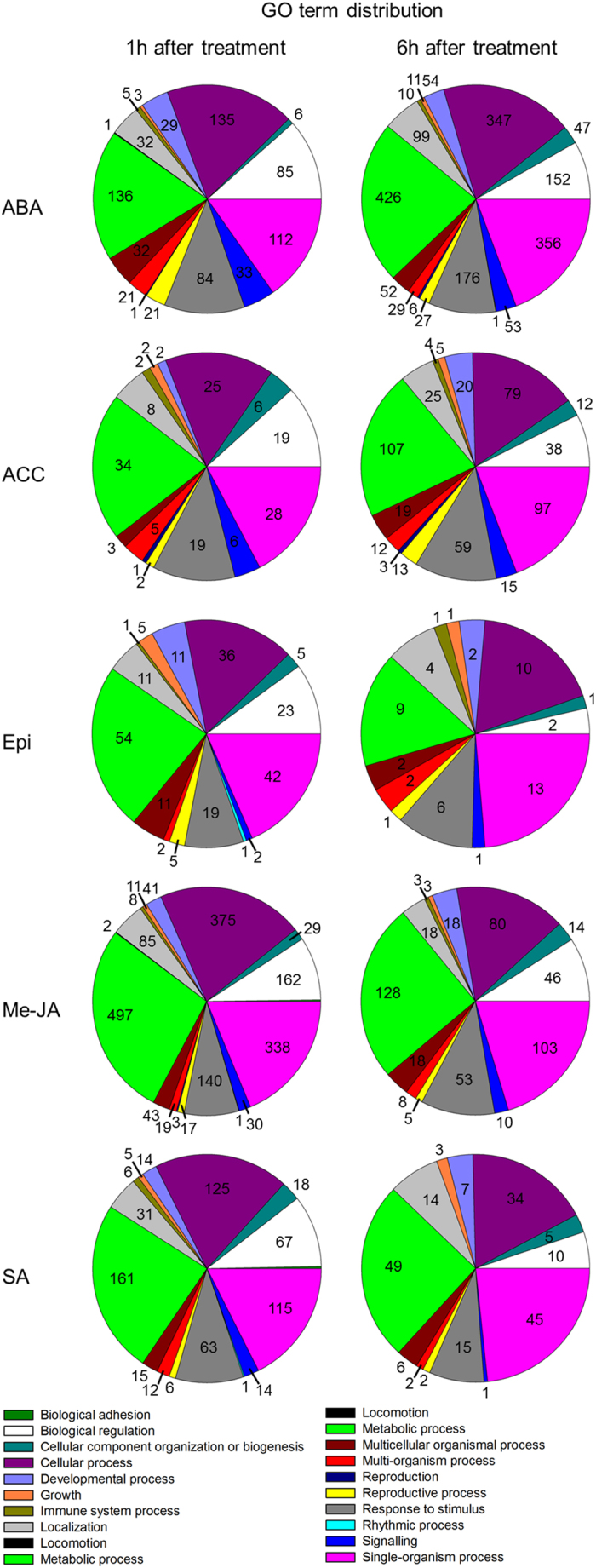
ABA (abscisic acid); ACC (aminocyclopropanecarboxylic acid); Epi (epibrassinolide); Me-JA (methyl jasmonate); SA (salicylic acid).
A GO term classification that was strongly influenced by all hormone treatments was ‘response to stimulus’. At 1 h after treatment, genes encoding for extracellular ligand-gated ion channels (DMG400013264, DMG400022304) were 3-fold up-regulated in ABA treated plants whereas SAUR family proteins (DMG400001614, DMG400001615, DMG400001667, DMG400001668) were 2-3-fold down-regulated by ABA and Me-JA. ACC treatment after 1 h led to 2-fold suppression of a multicystatin gene (DMG400005950). A peroxidase transcript (DMG401025083) was induced 5-fold by ACC, 2-fold by Epi, 9-fold by Me-JA and 3-fold upon SA treatment. Expression of oxidoreductase genes (DMG400011098, DMG400011929, DMG400019119) was 4-fold suppressed by Me-JA. At 1 h after treatment, SA strongly up-regulated UDP-glucose/glucosyltransferase genes (DMG400015579; 155-fold up-regulated and DMG400015601; 100-fold up-regulated).
Different gene families implicated in the GO term annotation ‘signalling’ were altered in transcript abundance by the hormone treatments at 1 h and 6 h. After 1 h, CBL-interacting protein kinase transcripts (DMG400028479, DMG400028491) were 3-fold up-regulated by ABA and ACC treatments, and 3-fold down-regulated by Me-JA. Me-JA application after 1 h also affected MAP kinase genes (DMG400007058, DMG400003528), which were 2-4-fold up-regulated. At 1 h and 6 h post-treatment, a total of 74 and 42 differentially transcribed transcription factor genes embedded in GO terms such as ‘signalling’, ’response to stimulus’, and ‘response to hormone’, were altered in their transcription, respectively (Table 1a,b). Seven of these genes were affected in opposite direction by the different hormone treatments at 1 h (Table 1a). AP2 domain-containing transcription factor 9 transcript (DMG400012154) was up-regulated by ABA and down-regulated by Me-JA whereas an AP2/ERF domain-containing transcription factor gene (DMG400008734) was up-regulated by SA and down-regulated by ABA. A gene encoding a COL domain class transcription factor (DMG400003711) was induced by Me-JA and supressed by ABA treatment. Transcripts for a MYB transcription factor (DMG400011250) and a MYB domain class transcription factor (DMG400019535) were strongly up-regulated in ABA treated leaves and down-regulated by Me-JA. Two WRKY transcription factor genes (DMG400001434, DMG400029207) were up-regulated by SA treatment and down-regulated in Me-JA treated potato leaves. At 6 h after treatment, only one gene was induced or suppressed by the different hormone treatments (Table 1b). This gene, encoding a COL domain class transcription factor (DMG400003711), was induced by Me-JA but repressed by ABA and these effects had also been seen at 1 h after treatment.
Table 1. Transcription factors (UniRef annotation) that are differentially expressed at 1 h (Table 1a) and 6 h (Table 1b) after hormone treatments are shown.

Values represent fold changes in gene expression in hormone treated plants in comparison to control plants. Up-regulated genes are highlighted in orange, down-regulated genes are highlighted in green. ABA (abscisic acid); ACC (aminocyclopropanecarboxylic acid); Epi (epibrassinolide); Me-JA (methyl jasmonate); SA (salicylic acid).
Database to query the expression patterns of potato genes represented on the microarray
The microarray signals (both raw and normalised) are stored in a database alongside information concerning the corresponding probes and the PGSC genes (DMGs) and transcripts (DMTs). The database is searchable through the associated website http://ics.hutton.ac.uk/solarray. The website allows users to access the information by searching for sequence IDs (PGSC transcript/gene, or probe name), by conducting a BLAST search of a nucleotide or protein sequence against the PGSC transcripts to identify homologous sequences, or via annotation-based keyword search (Figure S1). The microarray signals are displayed in both graphical and tabular format for each probe and significant fold changes (p-value ≤ 0.05) are highlighted (Figure S1). The database is linked to the Spud DB Genome Browser (http://potato.plantbiology.msu.edu/cgi-bin/gbrowse/potato/) to facilitate additional analysis.
Marker gene analysis
The closest homologs in potato of some previously described A. thaliana genes involved in hormone signalling or described as phytohormone markers were identified following a search for the reciprocal best hits or, if not discernible, the top potato protein blast hits and assessed for their gene expression profiles in potato (Table S2). The ET marker genes ETHYLENE INSENSITIVE 3 (AtEIN3) and ETHYLENE RESPONSE FACTOR 1 (AtER1) show closest similarities in potato to genes annotated as EIL1 (DMG400029908) and EFR10 (DMG400013402), respectively. The BR marker genes DWARF 4 (AtDWF4), CONSTITUTIVE PHOTOMORPHOGENIC DWARF (AtCPD) and EXPANSIN 8 (AtEXP8) are homologues in potato to cytochrome P450 genes (DMG400014902 and DMG400023819) and an expansin (DMG400016650), respectively. Genes involved in BR signalling BRASSINOSTEROID INSENSITIVE 1 (AtBRI1) and BRI1-ASSOCIATED KINASE 1 (AtBAK1) are homologous in potato to a BRI1 protein (DMG400019698) and the somatic embryogenesis receptor kinase 3B (DMG400012594), respectively. The JA marker genes encoding VEGETATIVE STORAGE PROTEIN 2 (AtVSP2) and PLANT DEFENSIN 1.2 (AtPDF1.2) show closest similarities to a glycoprotein (DMG400016494) and a defensin (DMG400008516), respectively. The SA marker genes PHENYLALANIN AMMONIA-LYASE (AtPAL1), PATHOGENESIS RELATED PROTEIN 1 (AtPR1) and NON-EXPRESSOR OF PR GENE 1 (AtNPR1) are homologous to genes annotated as a phenylalanine ammonia-lyase (DMG400031457), the basic PR-1 protein (DMG400005112) and PR1/NIM1 like defence protein (DMG401000923) in potato, respectively. All these genes were expressed in our experiments (Table S2), albeit the raw values of the homologs to ERF1, EXP8, PDF1.2, PR1 and NPR1 were below 10 and thus below the confidence threshold for detection. These genes were not differentially affected by any of the hormone treatments compared to the control (Table S2). Thus, none of these homologs of hormone-responsive marker genes in A. thaliana showed similar behaviour in potato. We therefore used the data presented here to develop marker genes for early, hormone-specific responses in potato.
Validation of expression patterns using quantitative RT-PCR
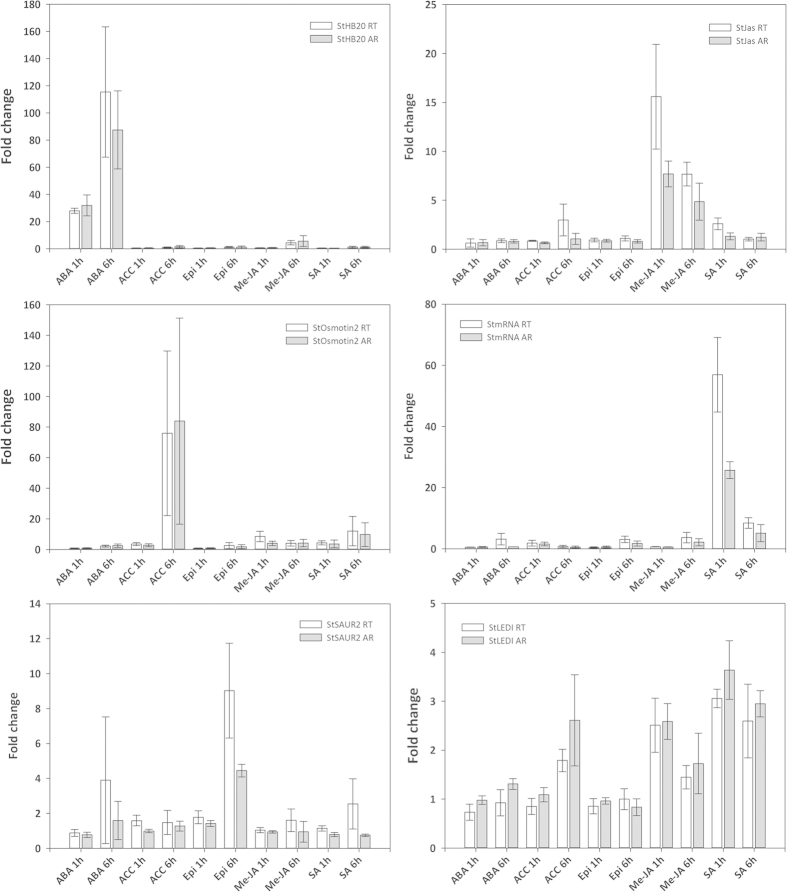
To identify potato phytohormone markers suitable for the early time points used in this study, genes that displayed hormone-specific expression profiles were selected for further analysis (Fig. 1). Genes that were specifically induced by only one hormone, either at both 1 h and 6 h post treatment or at just one of the time points, were sought. Genes that belong to large families of closely related sequences were avoided and priority was given to genes that displayed large expression changes upon hormone treatment. Quantitative RT-PCR (qRT-PCR) was used to validate the microarray-based expression patterns of these selected genes (Fig. 3). One gene, encoding a LEDI-5c protein (DMG402018777) and which was influenced in its expression by Me-JA at 1 h and SA at 1 and 6 h, was included to verify more complex expression patterns as seen by microarray analysis (Fig. 3). In all cases, qRT-PCR corroborated the expression patterns observed in the microarray analyses. The ABA specific gene Homeodomain 20 transcription factor (DMG400000248; previously annotated as HB1 [Table S1]) displayed approximately 30-fold induction at 1 h after ABA treatment in both the microarray and qRT-PCR study and about 80-fold and 110-fold induction at 6 h in the microarray and the qRT-PCR analysis, respectively. An osmotin gene (DMG400003057), specifically induced by ACC, was around 80-fold up-regulated at 6 h post treatment on both platforms. An auxin-induced SAUR gene (DMG400016561) was significantly higher expressed in Epi treated plants at 6 h than in any other hormone treated plant (p-value ≤ 0.05). The jasmonate ZIM-domain protein 1 (DMG400002930) displayed specific transcript induction by Me-JA and was around 7-fold and 15-fold induced in microarray and qRT-PCR-based analysis at 1 h and 4-fold and 7-fold up-regulated at 6 h, respectively. As measured by microarray and qRT-PCR, a gene annotated to be weakly similar to AtWRKY40 (mRNA, DMG402007388) was specifically induced by SA and displayed approximately 25-fold and 55-fold induction at 1 h after treatment with SA only.
Figure 3. Fold changes in gene expression for six genes (DMG400000248 (StHB20), DMG400003057 (StOsmotin2), DMG400016561 (StSAUR2), DMG400002930 (StJas), DMG402007388 (StmRNA), DMG402018777 (StLEDI)) in response to treatment with hormones as measured by qRT-PCR (white) and microarray (grey) technologies are shown.
Displayed are mean values (N = 3) and standard errors. ABA (abscisic acid); ACC (aminocyclopropanecarboxylic acid); Epi (epibrassinolide); Me-JA (methyl jasmonate); SA (salicylic acid).
Discussion
Potato yields are affected by a variety of abiotic and biotic stresses and current research is aimed at finding new approaches to alleviate the impact of these threats1. The importance of integrated disease management to reduce biotic stresses on crop plants is rising and the application of elicitor compounds that enhance plant immunity is one strategy to eventually reduce the need for chemical pesticides21. The molecular mode-of-action of many elicitor compounds is, however, not fully understood but it has been shown that plant hormone pathways are often stimulated20,22,23,24,25. Since elicitors impact on gene expression mostly in the first few hours after application22, we have generated a reference map for the early changes of hormone-dependent gene regulation in potato following exogenous applications of abscisic acid (ABA), aminocyclopropanecarboxylic acid (ACC) a precursor in ethylene synthesis, epibrassinolide (Epi), methyl jasmonate (Me-JA) and salicylic acid (SA). A. thaliana, the most studied model plant, is of limited use for hormone studies in unrelated species since it has been shown, for example in potato and rice, that marked differences can exist in pathways and responses to hormones in major crop plants16,17,20.
Generally, more genes were influenced in their expression at 1 h than at 6 h after treatment, showing that in potato, responses to external applications of plant hormones are very rapid and therefore in agreement with previous studies15,26. However, each plant hormone differed in their temporal influences. The strongest response in potato at 1 h after treatment was alteration by Me-JA. SA also differentially regulated more genes at 1 h than at 6 h, but fewer genes were altered than by Me-JA. In contrast, ABA and ACC impacted most on gene expression at 6 h after hormone application. Most genes were uniquely affected by a single phytohormone and relatively few were affected by more than one treatment. This is in agreement with a previous study in A. thaliana that focused on the effects of hormones involved in growth regulation and concluded that hormones, if applied individually and in isolation, mostly affect gene expression independently26.
However, upon perception of environmental cues, phytohormone signalling pathways are strongly interconnected to allow plants to quickly and efficiently respond to biotic and abiotic stresses5. The crosstalk between different plant hormones has been well documented in A. thaliana5,14,27. The best-known examples are reported crosstalk between the defence hormones JA and SA. For example, in A. thaliana, the JA-responsive genes PDF1.2 and VSP2 are very sensitive to suppression by SA and the transcription factors WRKY50 and WRKY51 play roles in SA-induced suppression of the JA pathway5,14. However, parallel effects of hormone treatments have also been reported5,12,27 and have also been observed in this study. For example, 52 genes were up-regulated by both Me-JA and SA at 1 h after treatment and 91 were down-regulated. The transcript abundance of only 9 genes was affected in opposite direction. This equates to 94% of the genes being similarly affected and 6% in opposite directions. Similar results have been observed in rice, where 88% of genes were similarly affected by JA and SA15. In addition, in our potato study, 77 genes were up-regulated by both ABA and ACC at 6 h after treatment and 31 were down-regulated by both hormones at this time point. Only 7 genes were affected in the opposite direction. Therefore, 94% of the genes were similarly affected and 6% in opposite direction. This contrasts to a similar study in rice, where 50% of the genes were affected in opposite direction by ABA and ACC15.
The second objective of this study was to identify marker genes for the early influences of ABA, ACC, Epi, Me-JA and SA hormone treatments in potato. Most of the marker genes commonly used for hormone synthesis and signalling pathways are derived from A. thaliana. Marker genes for ABA response include ABI genes in A. thaliana and genes containing the ABA-responsive elements ABRE9,10,28. In this study in potato, an ABRE binding factor (DMG400008011) was uniquely 30-fold up-regulated by ABA at both 1 h and 6 h after treatment (Table S1). In addition to the ABRE binding factor, a transcript encoding Homeodomain 20 transcription factor (DMG400000248) was specifically induced by ABA and displayed approximately 30-fold induction at 1 h after treatment and 80-fold induction at 6 h in the microarray (Fig. 3).
Marker genes for ET signalling in A. thaliana encode ETHYLENE INSENSITIVE 3 (EIN3) and EIN3-like transcription factors (EIL) that further regulate other marker genes like ETHYLENE RESPONSE FACTOR 1 (ERF1) and ETHYLENE RESPONSE DNA BINDING FACTORs 1 to 4 (EDF1 – EDF4)14,29,30,31. In this study, the potato homolog of EIN3 (DMG400029908), whilst detectable, was not differentially expressed. The expression of the ERF1 homolog in potato (DMG400013402) remained below the limit of detection (Table S2). However, a suitable alternative marker gene, osmotin OSML15 (DMG400003057), was identified that displayed strong (up to 80-fold) and specific induction at 6 h (Fig. 3).
The three most commonly used marker genes for BR in A. thaliana are DWARF4 (DWF4) and CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD), both of which are down-regulated by application of BR32, and EXPANSIN8 (EXP8) which is induced by BR treatment33. The closest potato homologs to DWF4 and CPD are annotated as cytochrome P450 genes (DMG400014902 and DMG400023819, respectively). Both genes were expressed although expression of the DWF4 homolog was close to the detection limit, but neither of the genes was differentially affected by Epi treatment. An expansin (DMG400016650) is the potato homolog to AtEXP8 but the raw expression values were below the confidence threshold for detection (Table S2). Genes involved in BR signalling are the leucine-rich repeat receptor-like kinase BRASSINOSTEROID-INSENSITIVE 1 (BRI1) and BRI1-ASSOCIATED KINASE 1 (BAK1)13,34,35,36. The potato BRI1 gene (DMG400019698) was detectable, although not differentially expressed following Epi application at 1 h after treatment, but not expressed at 6 h (Table S2). Similarly, expression of the BAK1 potato homolog (DMG400012594) was detectable but no differential regulation was apparent (Table S2). Nevertheless, a gene annotated as auxin-induced SAUR gene (DMG400016561) was significantly more highly expressed in Epi treated plants at 6 h than in any other hormone treated plants (p-value ≤ 0.05; Fig. 3). Up-regulation of auxin-induced SAUR genes by treatment with brassinolides had been demonstrated previously37.
Typical JA marker genes encode JASMONATE ZIM domain (JAZ) proteins that, in the absence of jasmonate, repress transcription of jasmonate-inducible genes by binding to their promoters14,38. The JA signalling pathway has two major branches, the MYC and the ERF branch and JA-responsive marker genes for these branches encode VEGETATIVE STORAGE PROTEIN 2 (VSP2) and PLANT DEFENSIN 1.2 (PDF1.2), respectively5,39,40. Application of JA also leads to activation of APETALA 2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factors, which regulate the transcription of JA-responsive defence genes41,42. In this study, we also identified a jasmonate ZIM-domain protein 1 (DMG400002930) that was uniquely 7-fold up-regulated by Me-JA at 1 h after treatment and uniquely 4-fold up-regulated 6 h after treatment. The JA-specific expression was confirmed by qRT-PCR (Fig. 3). The potato homolog of the VSP2 gene (DMG400016494) was expressed but did not reveal differential expression at 1 h or 6 h post treatments in this experiment, whereas the raw expression values of the potato homolog to PDF1.2 (DMG400008516) were below the confidence threshold for detection (Table S2).
Typical A. thaliana SA marker genes encode PHENYLALANINE AMMONIA LYASE (PAL) involved in SA synthesis and PATHOGENESIS-RELATED (PR) genes involved in SA defence signalling5,14,43,44. The regulatory protein NONEXPRESSOR OF PR GENES1 (NPR1) plays a role in both JA and SA signalling because it is activated by SA and supresses JA responses5,14,45. In this study, the potato PAL1 homolog (DMG400031457) was expressed but was not significantly affected by any treatment (Table S2). The expression levels of the potato gene homolog of PR1 (DMG400005112) and of NPR1 (DMG401000923) were below the confidence threshold of detection (Table S2). A gene similar to AtWRKY40 (DMG402007388), however, responded specifically to SA treatment (Fig. 3). The involvement of WRKY transcription factors in SA signalling in A. thaliana had been shown before5,42, but involvement of WRKYs is more complex in defence responses in rice17.
This study has provided novel insight into hormone signalling pathways in potato and represents a state-of-the-art reference database for the transcriptional regulation of genes by the major defence hormones. The hormones in our study did not specifically influence all the traditionally used hormone marker genes from A. thaliana. We therefore used the data presented in this study to develop new marker genes for the different plant hormones in potato. These newly identified marker genes will, in the future, be useful to ascertain the mode-of-action of novel elicitor compounds or to elucidate functions of pathogen effectors. In addition, the data generated in this study have been used to generate a new potato gene expression database which is also linked to the Spud DB Genome Browser to enable more comprehensive analysis. The potato gene expression database will accommodate future gene expression studies and will allow users to quickly determine expression changes for current potato gene models.
Material and Methods
Plant material, growth conditions and experimental setup
Single stem potato plants (Solanum tuberosum cultivar Desiree) were grown from tubers in individual pots in a glasshouse set at 18 °C for 16 h day length supplemented with artificial lighting and 14 °C for 8 h in the dark. Plants were grown for five weeks prior to treatments to obtain leaves from comparable developmental stages and maturity46. Plants were treated with 1 mM abscisic acid (ABA), 1 mM aminocyclopropanecarboxylic acid (ACC), 0.05 mM epibrassinolide (Epi), 1 mM methyl jasmonate (Me-JA) or 1 mM salicylic acid (SA). A water control was included. All solutions contained 10 μl DMSO ml-1 and all chemicals were obtained from Sigma Aldrich. For each treatment, six plants were sprayed with a total of 100 ml per hormone solution until runoff. After 1 h and 6 h post treatments, six compound leaves from the middle part of three plants per time point each were harvested and snap frozen in liquid nitrogen. Three independent biological replicates were conducted.
RNA extraction
Total RNA was extracted from 100 mg of frozen ground leaf material using 0.5 ml of TRIzol Reagent and 0.2 ml of chloroform according to the manufacturer’s protocol. RNA was further purified using a phenol-chloroform step at pH 4.3 to eliminate DNA carryover. RNA concentration and quality (260/280 nm > 2.0) were measured using a Nanodrop 2000 UV-Vis spectrophotometer (Thermo Scientific). The integrity of the samples was checked using a Bioanalyzer (Agilent Technologies).
Microarray analysis
High-throughput gene expression analysis of three independent biological replicates was performed using custom Agilent microarrays designed to represent predicted transcripts of assembly version 3.4 of the potato DM genome19: A-MEXP-2272 was designed to multiple transcript isoforms (53, 000 in total). Microarray experimental design and datasets are available in ArrayExpress (ebi.ac.uk/arrayexpress/; accession E-MTAB-3542). All microarrays were processed essentially as described by Hancock et al.47, with the following exception: a single-channel design was used throughout, with all RNA samples labelled individually with Cy3 as recommended (One-Color Microarray-Based Gene Expression Analysis protocol v.6.5; Agilent). All other hybridisation, washing and scanning procedures, along with feature extraction (FE) and microarray data normalisation in GeneSpring (v. 7.3; Agilent) were as described. Unreliable data flagged as absent in all replicate samples by the FE software were discarded. Statistical filtering of data was performed for each time point separately (1 h, 6 h) using analysis of variance (ANOVA; p-value ≤ 0.05) with Benjamini & Hochberg multiple testing correction. Volcano plots on the ANOVA-filtered gene-lists were used to identify significantly changing probes between control and each hormone treatment (≥2x fold-change; p-value ≤ 0.05). In some cases, different probes on the array are representative of the same gene. For consistency, gene names (PGSC0003DMG numbers abbreviated to DMG4XXXXXXXX) are given throughout the entire manuscript. For the identification of housekeeping genes that are suitable for downstream qRT-PCR validation, only genes that were consistently detected in all samples and displayed a normalised expression ratio between 0.8 and 1.2 were considered.
Gene ontology analysis
For gene ontology classification, Blast2GO (version 2.8)48 was used to annotate the gene transcripts. For each treatment all overrepresented gene ontology (GO) terms (biological process, level 2) were analysed.
Marker gene analysis
Previously described A. thaliana genes involved in hormone signalling or described as phytohormone markers were assessed for their gene expression profiles and specificity in potato (Table S2). Protein sequences from the A. thaliana marker genes were blasted against potato proteins using the Spud DB website (http://potato.plantbiology.msu.edu). The DMG numbers corresponding to the potato proteins (DMPs) were used to identify the corresponding genes on the microarray49. Where readily identifiable, the reciprocal best hits (ERF1, DWF4, CPD, EXP8, BRI1, BAK1, PAL1) were selected or, if not discernible, the top potato protein blast hits (EIN3, VSP2, PDF1.2, PR1, NPR1) were utilised.
Expression validation via qRT-PCR
For gene expression validation, cDNA was synthesised from 2 μg of total RNA from hormone treated leaf tissue using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare Life Sciences) in conjunction with oligodT and random hexamer primers mixed in a 1:1 ratio (Thermo Fisher Scientific). Amplification of six target and two housekeeping genes (StNuclear [DMG400023981 a small nuclear ribonucleoprotein G] and St40S [DMG400020803]) was assessed (Table 2). All reactions were setup in a total volume of 25 μl using 2x FastStart Universal Probe Master (Rox) (Roche Diagnostics Corporation) and Universal ProbeLibrary probes (Roche Diagnostics Corporation) as specified by the manufacturer. Primer and probe combinations are listed in Table 2. A StepOnePlus™ Real-Time PCR System (Life Technologies) was used with the following conditions for all genes: initial denaturation for 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Relative expression of the target genes was calculated using a modified Pfaffl method50 with the geometric mean of the two housekeeping genes as the reference.
Table 2. Primers and Universal Probe Library identities used for the amplification of two housekeeping genes (StNuclear and St40S) and six target genes (StHB20, StOsmotin2, StSAUR2, StJas, StmRNA, StLEDI).
| Name | Accession | UPL probe | Left primer | Right primer |
|---|---|---|---|---|
| StNuclear | DMG400023981 | #114 | tggtgacaattgaagcgttg | tggggcataaacaaagatcc |
| St40S | DMG400020803 | #69 | gccactggtggcaagaag | ctggcggccaagttcata |
| StHB20 | DMG400000248 | #143 | gctgcttcagcagtctgtca | acctcctcgcgttgttattg |
| StOsmotin2 | DMG400003057 | #10 | ctgcccctacaccgtttg | caccaactctgacctctctcg |
| StSAUR2 | DMG400016561 | #150 | cacaaagcattgctcctatcaa | tgttggtttccactttcttgg |
| StJas | DMG400002930 | #145 | ctcaacaaacagctaccaccac | cgatgaatcacttgatttctcaat |
| StmRNA | DMG402007388 | #133 | aaaatatggtcaaaaagtgacaagag | catgttggtgcaaatgaacac |
| StLEDI | DMG402018777 | #122 | ggatgaaaacagttggggtaaa | ccttcctcatgggtacaagg |
StNuclear is annotated in potato as a small nuclear ribonucleoprotein G and the closest homolog in A. thaliana is AT2G23930.1 with the same annotation. ST40S, annotated as a 40 S ribosomal protein S8 in potato, has similarity to A. thaliana AT5G59240.1, also a ribosomal protein S8e family protein. The closes A. thaliana homolog to potato StHB20, a homedomain 20 transcription factor, is AT3G61890.1 encoding for homeobox 12. StOsmotin2 has been annotated as Osmotin OSML15 in potato and the highest sequence identity in A. thaliana is to AT4G11650.1, annotated as osmotin 34. StSAUR2, an auxin-induced SAUR, shows homology to A. thaliana gene AT1G29510.1, a SAUR-like auxin-responsive protein. StJas, annotated as jasmonate ZIM-domain protein 1 in potato, displays homology to AT1G19180.1 with the same annotation. StmRNA is a 1346 bp sequence with no further annotation in potato with the highest sequence similarity to A. thaliana AT1G80840.1, a WRKY DNA-binding protein 40. StLEDI is annotated as LEDI-5c with the highest sequence similarity to AT1G76690.1, encoding for 2-oxophytodienoate reductase 2.
Additional Information
How to cite this article: Wiesel, L. et al. A transcriptional reference map of defence hormone responses in potato. Sci. Rep. 5, 15229; doi: 10.1038/srep15229 (2015).
Supplementary Material
Acknowledgments
This work was funded by the Rural & Environment Science & Analytical Services Division of the Scottish Government and the BBSRC through the joint projects CRF/2009/SCRI/SOP 0929, BB/L008025/1, BB/K018299/1 (IH, BH) and BB/H018697/1 (PRJB). Additional funding was obtained through the James Hutton Institute SEEDCORN initiative (LW, JLD, LM, VRF, MBH, JMW, JM, PEH, ACN, EMG).
Footnotes
The authors declare no competing financial interests.
Author Contributions L.W., P.E.H., A.C.N., P.R.J.B., E.M.G. and I.H. designed the experimental approach. L.W., J.L.D., V.R.F., J.M.W., J.M. and B.H. conducted the practical work. L.W., J.L.D., M.B.H., P.E.H. and I.H. analysed the data and produced the figures. L.M. designed the potato database. L.W. and I.H. wrote the manuscript. All authors reviewed and approved the manuscript.
References
- Birch P. R. J. et al. Crops that feed the world 8: Potato: are the trends of increased global production sustainable? Food Sec. 4, 477–508 (2012). [Google Scholar]
- Battisti D. S. & Naylor R. L. Historical warnings of future food insecurity with unprecedented seasonal heat. Science 323, 240–244 (2009). [DOI] [PubMed] [Google Scholar]
- Jefferies R. A. Responses of potato genotypes to drought. 1. Expansion of individual leaves and osmotic adjustment. Ann. Appl. Biol. 122, 93–104 (1993). [Google Scholar]
- Porter G. A., Bradbury W. B., Sisson J. A., Opena G. B. & McBurnie J. C. Soil management and supplemental irrigation effects on potato: I. Soil properties, tuber yield, and quality. Agron. J. 91, 416–425 (1999). [Google Scholar]
- Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A. & Van Wees S. C. M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521 (2012). [DOI] [PubMed] [Google Scholar]
- Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 40, 219–229 (2015). [DOI] [PubMed] [Google Scholar]
- Bishop G. J. & Koncz C. Brassinosteroids and plant steroid hormone signaling. Plant Cell Online 14, S97–S110 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashita H. et al. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898 (2003). [DOI] [PubMed] [Google Scholar]
- Finkelstein R. R., Gampala S. S. L. & Rock C. D. Abscisic acid signaling in seeds and seedlings. Plant Cell Online 14, S15–S45 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S. et al. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115, 4891–4900 (2002). [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R. & Abrams S. R. Abscisic acid: emergence of a core signaling network. Ann. Rev. Plant Biol. 61, 651–679 (2010). [DOI] [PubMed] [Google Scholar]
- Bari R. & Jones J. D. G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488 (2009). [DOI] [PubMed] [Google Scholar]
- De Bruyne L., Höfte M. & De Vleesschauwer D. Connecting growth and defense: The emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant 7, 943–959 (2014). [DOI] [PubMed] [Google Scholar]
- Derksen H., Rampitsch C. & Daayf F. Signaling cross-talk in plant disease resistance. Plant Sci. 207, 79–87 (2013). [DOI] [PubMed] [Google Scholar]
- Garg R., Tyagi A. K. & Jain M. Microarray analysis reveals overlapping and specific transcriptional responses to different plant hormones in rice. Plant Signal Behav. 7, 951–956 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D., Gheysen G. & Höfte M. Hormone defense networking in rice: tales from a different world. Trends Plant Sci. 18, 555–565 (2013). [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Xu J. & Höfte M. Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front. Plant Sci. 5, 611 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Liu Y., Fan B., Klessig D. F. & Chen Z. Is the high basal level of salicylic acid important for disease resistance in potato? Plant Physiol. 115, 343–349 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Potato Genome Consortium. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 (2011). [DOI] [PubMed] [Google Scholar]
- Bengtsson T. et al. Proteomics and transcriptomics of the BABA-induced resistance response in potato using a novel functional annotation approach. BMC Genomics 15, 315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel L. et al. Molecular effects of resistance elicitors from biological origin and their potential for crop protection. Front. Plant Sci. 5, 655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burra D. D. et al. Phosphite-induced changes of the transcriptome and secretome in Solanum tuberosum leading to resistance against Phytophthora infestans. BMC Plant Biol. 14, 254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros F. C. L., Resende M. L. V., Medeiros F. H. V., Zhang H. M. & Paré P. W. Defense gene expression induced by a coffee-leaf extract formulation in tomato. Physiol. Mol. Plant Pathol. 74, 175–183 (2009). [Google Scholar]
- Yin H., Li S., Zhao X., Du Y. & Ma X. cDNA microarray analysis of gene expression in Brassica napus treated with oligochitosan elicitor. Plant Physiol. Biochem. 44, 910–916 (2006). [DOI] [PubMed] [Google Scholar]
- Zuluaga A. P., Vega-Arreguín J. C. & Fry W. E. Transcriptome profile of acibenzolar-S-methyl-induced genes in tomato suggests a complex polygenic effect on resistance to Phytophthora infestans. Physiol. Mol. Plant Pathol. 81, 97–106 (2013). [Google Scholar]
- Nemhauser J. L., Hong F. & Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475 (2006). [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M. & Jones J. D. G. Hormone crosstalk in plant disease and defense: More than just JASMONATE-SALICYLATE antagonism. Ann. Rev. Phytopathol. 49, 317–343 (2011). [DOI] [PubMed] [Google Scholar]
- Liu Y., Ji X., Zheng L., Nie X. & Wang Y. Microarray analysis of transcriptional responses to abscisic acid and salt stress in Arabidopsis thaliana. Int. J. Mol. Sci. 14, 9979–9998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q. et al. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144 (1997). [DOI] [PubMed] [Google Scholar]
- Muday G. K., Rahman A. & Binder B. M. Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17, 181–195 (2012). [DOI] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q. & Ecker J. R. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. et al. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138, 1117–1125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gro Malinovsky F. et al. Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH INCREASED LEAF INCLINATION1 BINDING bHLH1. Plant Physiol. 164, 1443–1455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D. M., Joazeiro C. A. P., Li J. M., Hunter T. & Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1255 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 (2002). [DOI] [PubMed] [Google Scholar]
- She J. et al. Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., Shimada Y., Asami T., Fujioka S. & Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130, 1319–1334 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007). [DOI] [PubMed] [Google Scholar]
- Dombrecht B. et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A. & Pieterse C. M. J. Cross talk in defense signaling. Plant Physiol. 146, 839–844 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert W. F., Delauré S. L., De Bolle M. F. C. & Cammue B. P. A. The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416 (2006). [DOI] [PubMed] [Google Scholar]
- Lyons R., Manners J. M. & Kazan K. Jasmonate biosynthesis and signaling in monocots: a comparative overview. Plant Cell Rep. 32, 815–827 (2013). [DOI] [PubMed] [Google Scholar]
- Antoniw J. F., Ritter C. E., Pierpoint W. S. & Van Loon L. C. Comparison of three pathogenesis-related proteins from plants of two cultivars of tobacco infected with TMV. J. Gen. Virol. 47, 79–87 (1980). [Google Scholar]
- Mauch-Mani B. & Slusarenko A. J. Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8, 203–212 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S. A., Gordon A. S. & Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired-resistance. Plant Cell 6, 1583–1592 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomiets M. V., Chen H., Gladon R. J., Braun E. J. & Hannapel D. J. A leaf lipoxygenase of potato induced specifically by pathogen infection. Plant Physiology 124, 1121–1130 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. D. et al. Physiological, biochemical and molecular responses of the potato (Solanum tuberosum L.) plant to moderately elevated temperature. Plan, Cell Environ. 37, 439–450 (2014). [DOI] [PubMed] [Google Scholar]
- Conesa A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- Hirsch C. D. et al. Spud DB: A resource for mining sequences, genotypes, and phenotypes to accelerate potato breeding. Plant Genome. doi: 10.3835/plantgenome2013.12.0042 (2014). [DOI] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.