Abstract
There are few better examples of the need for data sharing than in the rare disease community, where patients, physicians, and researchers must search for “the needle in a haystack” to uncover rare, novel causes of disease within the genome. Impeding the pace of discovery has been the existence of many small siloed datasets within individual research or clinical laboratory databases and/or disease-specific organizations, hoping for serendipitous occasions when two distant investigators happen to learn they have a rare phenotype in common and can “match” these cases to build evidence for causality. However, serendipity has never proven to be a reliable or scalable approach in science. As such, the Matchmaker Exchange (MME) was launched to provide a robust and systematic approach to rare disease gene discovery through the creation of a federated network connecting databases of genotypes and rare phenotypes using a common application programming interface (API). The core building blocks of the MME have been defined and assembled. Three MME services have now been connected through the API and are available for community use. Additional databases that support internal matching are anticipated to join the MME network as it continues to grow.
Keywords: Matchmaking, rare disease, genomic API, gene discovery, Matchmaker Exchange, GA4GH, IRDiRC
Introduction
The content of genetic tests has gradually expanded over the years, with major leaps happening recently with the introduction of exome and genome sequencing. Although the rate of solving monogenic ‘Mendelian’ disorders has increased with the ability to query all genes, a large fraction of patients still remain without a diagnosis. A portion of these unsolved cases harbor suspicious variants in candidate disease genes. For such cases, finding just a single additional unrelated case with a deleterious variant in the same gene and overlapping phenotype may provide sufficient evidence to causally implicate the gene, enabling a diagnosis for the patient. Methods for identifying these additional cases have evolved over time. From word of mouth between colleagues to sharing published case reports, laboratory diagnosticians and clinicians have worked to uncover connections between patients (Loucks et al, 2015, this issue). In a world of rapidly evolving information technologies, however, a more efficient solution is needed that can scale with the exploding growth in genomic sequencing.
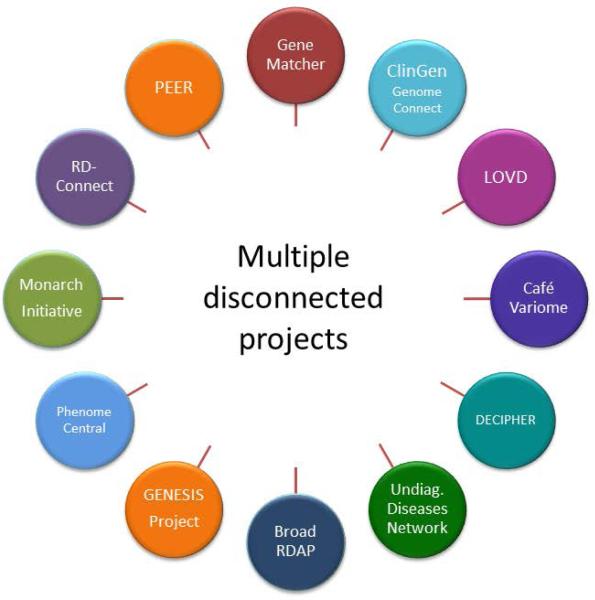
Multiple projects have addressed this need by developing platforms that use genotype and phenotype driven matching algorithms to identify cases with common phenotypes and disrupted genes (Washington et al., 2009; Gonzalez et al., 2012, Swaminathan et al., 2012, Gonzalez et al., 2013, Robinson et al., 2014; Zemojtel et al., 2014; Buske et al., 2015a (this issue), Lancaster et al., 2015 (this issue), Sobreira et al., 2015(this issue)). However, no organized system existed to facilitate the interaction between these multiple disconnected projects (Figure 1) before the Matchmaker Exchange (MME). To unify these efforts and harness the collective data across all of the databases, groups representing rare disease repositories held a meeting in October 2013 to launch an open collaboration later named the Matchmaker Exchange (http://www.matchmakerexchange.org). This collaborative effort has launched a federated platform (exchange) to facilitate the identification of cases with similar phenotypic and genotypic profiles (matchmaking) through a standardized application programming interface (API) and procedural conventions. The MME enables searches of multiple databases (matchmaker services) from another, connected matchmaker service, without having to separately query all services, or deposit data in each one. The queries are designed to allow a gene or genotype, combined with a condition or phenotypic features, to be sent as a query in order to get a returned response containing any similar or “matched” cases. Matching algorithms are defined by the matchmaker services and will evolve over time as described below.
Figure 1. Databases and programs that gathered to form the basis for the Matchmaker Exchange.
The MME includes representatives from the founding organizations and databases supporting or intending to support matchmaking services (Tables 1 and 2). Collaborative work has focused on both the technical aspects of data sharing, as well as policy considerations. This work has resulted in version 1.0 of a MME API (Buske et al., 2015b, this issue), a set of requirements for qualifying as a MME service, and a user agreement for querying the MME. The MME has been identified as a demonstration project for the Global Alliance for Genomics and Health (GA4GH) and the MME has been leveraging the expertise of the GA4GH working groups for guidance on pertinent aspects of the project.
Federated vs. Centralized Approaches to Data Sharing
Historically, most genetic and genomic data sharing has been accomplished through the aggregation of data in a single “centralized” site, such as the National Center for Biotechnology Information's (NCBI) Database of Genotypes and Phenotypes (dbGaP) (Tryka et al., 2013) or other large data centers such as those employed for the International Cancer Genome Consortium (ICGC) (Zhang et al., 2011) and the Cancer Genome Atlas (TCGA) (Weinstein et al., 2013). This approach allows for easy data analysis given that a data holder is in complete control of the entire dataset; however, a higher regulatory burden must be overcome to allow data to be shared with another entity, putting its security and privacy management entirely in the hands of the database owner. In addition, users may only wish to share certain datasets with others and only under certain circumstances which can be better controlled by the use of an API to enable data access. Finally, data annotations such as phenotype are dynamic within a patient, but static within a disconnected database, where they can be difficult to capture longitudinally. A federated system makes it easier to support longitudinal connections to patient phenotype and updated genomic interpretations.
An alternative approach is the use of a federated network in which multiple distributed databases are connected through APIs, whereby each database supports queries of other databases in the network. This allows each database to be autonomous with respect to its own data schema, maintain ongoing control of its own data, and continuously innovate at its own pace. In this model, no single database acts as the “central” database, nor does a single database take on the privacy and security requirements of the whole network.
It is this latter federated model that was chosen to support the MME, though some data contributors may prefer to deposit data into an existing matchmaker service for participation in the MME instead of setting up their own matchmaker. This initial approach allows each participating matchmaker service to maintain their autonomy and primary purpose, while contributing valuable data to the MME and the genomics community. Data contributors no longer need to deposit the same datasets into multiple databases in order to find matches, and they will have more options for databases in which to deposit data, including databases in their own jurisdiction if certain regulations prohibit data from leaving a region. Also, data contributors may decide to put some cases into one database and other cases into another database depending on the focus of each database. The decision of where to start may be based upon a variety of factors as described below, including the database's supported content and algorithms for matching. However, in the MME, data contributors are discouraged from depositing the same dataset into multiple databases in order to minimize data duplication.
Building Blocks to Support the Matchmaker Exchange
To promote responsible data sharing, the founding members of the MME have established a set of requirements for participating matchmaking services, a user agreement for those wishing to use the MME, and a steering committee (SC) to govern the program. The SC is composed of a representative from each approved MME service, as well as program organizers and representation from Global Alliance for Genomics and Health (GA4GH) and the International Rare Diseases Research Consortium (IRDiRC). The steering committee is charged with maintaining the service requirements, user agreement, and oversight of the API to ensure the MME meets the needs of the rare disease community and reflects consensus standards and best practices as set forth by the GA4GH and IRDiRC. The MME also supports a monthly conference call and periodic in-person meetings, most of which are open to the community to encourage active participation by all stakeholders.
Matchmaker Exchange Service Requirements
To become a MME service, each new site must achieve the following:
Require users to deposit case data to undertake a federated query across the MME service providers
Establish a minimum of two point-to-point API connections to other MME services
Contain content that is considered by the MME steering committee to be useful for matching, including the flagging of, or ability to prioritize, candidate genes
Successfully implement matching algorithms using test data
During user queries, enable dual notification of data requester (i.e. the querier) and prior data depositor (i.e. the queried) including sharing the identities and contact information for each
For each database to which a MME service is connected by an API, the connected database's disclaimers should be posted on the MME service's website and displayed with query results. Disclaimers can be found on GitHub (https://github.com/ga4gh/mme-apis)
Store queries sent and received between MME sites only for the purpose of auditing, defining query statistics, and following up queries to understand rates of validated gene discovery
Attest to database security requirements as defined by the GA4GH Security WG (forthcoming)
Advance the goals of the MME project through active participation in meetings and conference calls including defining a representative for the MME steering committee
Matchmaker Exchange End User Agreement
To use the MME, each data querier agrees to the following:
To make no attempt to identify individual patients in any MME database
To enable all cases submitted for querying to be stored in the query-initiating database for future matching
To obtain permission from the source of the matching data before publishing or presenting the results of queries
To acknowledge the MME, and the specific MME service that supported any discoveries in publications, as appropriate
Matchmaker Exchange API for Genotypes and Phenotypes
Application programming interfaces (APIs) define protocols for how components of computer systems communicate, and are a crucial part of the modern information technology landscape. In particular, web APIs have enabled the creation of our modern ecosystem of automatic communication between computer programs or services. APIs represent a defined protocol between technology services, such that a given input results in an expected output in a standardized format.
Participating matchmaker services are required to implement a standardized API, consistent with standards developed by the GA4GH Data Working Group, for exchanging genotypic and phenotypic information. The API supports queries, where a query is a patient record, and where the receiving system decides how best to process a specific query. Thus, the system does not support queries such as “Do you have any patients with a deleterious variant in CASQ2?” or “Do you have any patients with hypertelorism and arachnodactyly?,” but instead supports a query of “Do you have any patients similar to one who has hypertelorism and arachnodactyly with a deleterious variant in CASQ2”, where the definition of similarity is at the discretion of the receiving system. This API is described in greater detail in a companion article of this journal issue (Buske et al., 2015b). In brief, the core elements of each query that are transferred through the API include several mandatory elements: case ID, submitter information, and candidate gene(s) and/or phenotype terms. The API also accommodates additional fields to increase the specificity of queries including gender, age of onset, mode of inheritance, condition name (e.g. OMIM or Orphanet ID), chromosome, chromosome region, zygosity, and variant type (e.g. frameshift, missense, etc.).
Federated Authentication and Authorization
The MME recognizes the importance of authentication (validation of a user) and authorization (approval of a user to initiate a query) and has begun working closely with the GA4GH Security Working Group to define minimum standards to which each MME service must adhere in order to participate. Currently, these practices are defined by systems developed by the initial set of linked matchmaker services but is expected to develop more formally over time and in collaboration with the GA4GH Security WG.
Informed Consent Policy
The MME worked closely with the GA4GH's Regulatory and Ethics Working Group and Consent Task Team on developing a proposal for informed consent for data sharing in the context of genomic matchmaking within the MME. We have distinguished two levels of matchmaking and different consent requirements based on the data shared and the probability of re-identifying the patient:
Level 1
No additional consent required - This level of matchmaking involves a data requester querying on a broad phenotype description or disease name using standardized terms or codes (Human Phenotype Ontology (HPO), OMIM, Orphanet) and/or candidate gene names +/− variant type. This level of sharing is consistent with current clinical practice with low risk of possible re-identification and therefore specific patient consent for this activity is not required.
Level 2
Consent required - This level of matchmaking involves a data requester querying on a unique or sensitive phenotype description and/or sequence level and related information, such as defined variants and/or genomic datasets. This level of sharing requires consent from the patient. If the patient had previously consented to data being shared in an open or registered access database whose declared purpose involves data sharing for purposes consistent with those of this matchmaking, no additional consent is required.
The MME service in which data is deposited is responsible for ensuring patient data used in matchmaking is consented appropriately.
Matching Algorithms: Optimizing for Success
A key component of the success of the MME is implementing matching algorithms that balance sensitivity with specificity when executing matching algorithms. For example, if a case is annotated with a single candidate gene (Gene X) and a defined condition (Disease Y), a highly specific matching algorithm would require the gene and condition to be an exact match to return the result. However, matching algorithms could increase their sensitivity by allowing a case with any phenotype term that is a component of disease Y to also be returned. At the start of this program, when the number of MME services is few and the number of cases in each database is still limited, data contributors who are querying the MME may prefer matching algorithms that are less specific in hopes of having the highest sensitivity. However, as the MME scales and the number of cases deposited into each participating MME database grows, increased specificity and sophistication of matching algorithms will become critical.
It also is likely that data contributors will have different tolerances for being notified of matches on their data, with some only wishing to be notified of high-probability matches and others more tolerant of a range of results. To achieve this balance, some MME services have developed algorithms that have associated scores that can quantitate the specificity of a match. This allows contributors to specify their own threshold for notification of matches. It also allows the query results to be provided in a rank order.
It should be noted that the more detailed the query sent by the requester, the more information the recipient services will have at their disposal to sort cases in their database by relevance to the patient under query. With this additional detail, the query is more likely to result in successful and accurate matches, leading to a virtuous cycle that incentivizes data requestors to provide the greatest level of detail on their samples.
At the start of the program, MME services have defined their own algorithms for matching. This allows groups to constantly innovate on approaches to matching, yet MME services will be able to provide their algorithms on GitHub for other sites to adopt. In addition, allowing each site to control their own algorithms is necessary given the unique data schemas that support each MME database. For example, some MME databases have not yet implemented the flagging of candidate genes and instead simply store variant call format (vcf) files containing all variation on each case. In this scenario, most cases would result in a match with any executed query given the presence of variation in most genes in the genome. As such, matching algorithms can be further specified, for example, to require the optional field of variant type that would only return matches if a gene contains a predicted truncating or de novo variant.
Launching the Matchmaker Exchange
Defining the key approaches and requirements for supporting the initial intended purpose of the MME has been a critical step in launching this program. However, equally important is the execution of the project to launch a functionally connected federated network of matchmaker services that can demonstrate the identification and return of useful and successful matches in response to user-initiated queries. Such success enables the ongoing discovery of novel genetic causes of disease. Listed here, and detailed in the Supporting Information, are the steps that have been achieved in launching the MME: (1) goals of the MME defined, (2) MME API developed, (2) MME core policies developed, (3) MME website launched, (4) matching algorithm principles defined, (5) API test phase, (6) MME test dataset developed, and (7) user interfaces developed to support queries.
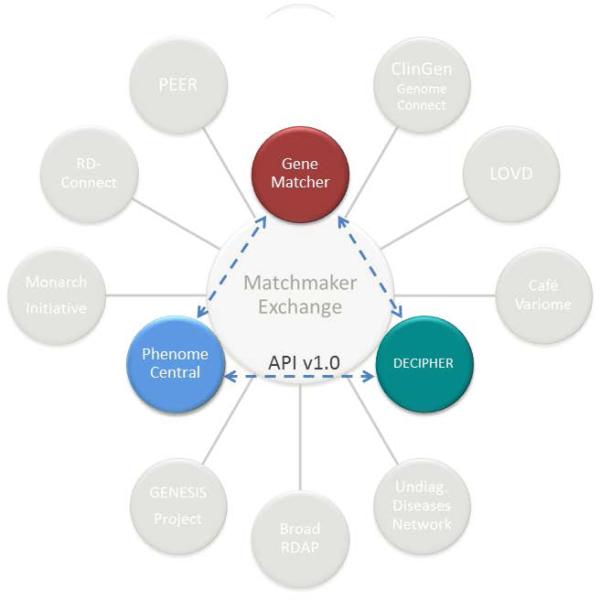
These steps have resulted in the current status of the MME in which three of the participating databases PhenomeCentral, GeneMatcher and DECIPHER are now capable of returning the results of queries from API-supported connections to other MME services (Table 1, Figure 2). The next areas of focus for the MME are to aid in bringing new MME services onto the network (Table 2) and promoting use of the MME by the broader community. In addition, MME services will continue refining the matching algorithms and integrate additional supporting evidence for why a candidate gene has been flagged in a given case.
Table 1.
Matchmaker Exchange Services
| DECIPHER | https://decipher.sanger.ac.uk/ |
| GeneMatcher | https://genematcher.org/ |
| PhenomeCentral | https://phenomecentral.org/ |
Figure 2. The current state of API-connected MME services.
The figure depicts those databases that have implemented the Matchmaker Exchange API and satisfied the MME service requirements as described above. Additional databases are in the process of implementing the API and other MME service requirements. Progress can be monitored via the MME website (http://www.matchmakerexchange.org).
Table 2.
Databases Intending to Launch the MME API
| Cafe Variome based networks | http://www.cafevariome.org/ |
| Broad Institute Rare Disease Analysis Portal | https://atgu.mgh.harvard.edu/xbrowse |
| ClinGen's GenomeConnect | http://genomeconnect.org/ |
| GENESIS (GEM.app) | https://genomics.med.miami.edu/ |
| Leiden Open Variation Database (LOVD) | http://www.lovd.nl/3.0 |
| Monarch Initiative | http://monarchinitiative.org/ |
| Platform for Engaging Everyone Responsibly (PEER) | http://www.geneticalliance.org/peer |
| RD-Connect | http://rd-connect.eu |
Guiding Community Use of the MME
The MME is a true federated system and as such, there is no single centralized entry point. Instead, users must choose one of the existing MME services as a starting point. In addition, in order to build the content of the MME over time, users must deposit their data in the point of entry into the MME. To guide users in where to deposit their data, Tables 3 and 4 provide a summary of the data fields that are maintained for each of the participating MME services and the parameters used for matching. Users may wish to deposit data in one system or another depending on the type of genotype and phenotype data associated with cases and how queries are supported.
Table 3.
Types of Data Maintained by each MME Service
| MME Service | Phenotype | Genotype | Candidates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name of condition |
Diagnosis code | Phenotypic terms |
Non-Human Models |
Gene Name | Chromosomal coordinates |
Variants | VCF Files | Flagged Gene Candidates |
Evidence for Gene Candidates |
|
| PhenomeCentral (Canada) | √ | √ | √ | √ | √ | √ | √ | √ | ||
| GeneMatcher (USA) | √ | √ | √ | √ | √ | √ | √ | |||
| DECIPHER (UK) | √ | √ | √ | √ | √ | √ | ||||
Current State of the MME
The success of matching is directly related to the volume of cases that are deposited into the MME services and therefore, to identify all causes of rare disease, we will need to engage the community broadly in encouraging deposition of cases into the system. Building off the birthday paradox, the probability of a match increases with number of patient records that are matchable (Krawitz et al., 2015 (this issue)). As such, even a small number of cases will begin yielding matches as has been demonstrated in the accompanying papers in this issue. After connecting these databases through the MME API, several additional matches have already been made between the Phenome Central and Gene Matcher Systems, including two promising hits undergoing further evaluation (Buske et al., 2015b). Furthermore, implementation of the API is underway in other systems that will collectively bring on thousands of additional cases and model organism data from databases that have already been serving as matchmakers inside their own systems (Gonzalez et al., 2015; Mungall et al., 2015; Lancaster et al., 2015; all in this issue).
Evolving the Matchmaker Exchange
As outlined above, the initial launch of the MME is focusing on the simple matching of unsolved rare disease cases that share a common phenotype and candidate gene. However, additional uses of a federated case-level database containing genotypic and phenotypic data have not escaped the view of the MME. Large, shared datasets have been leveraged throughout the genomics era to identify the genetic basis of common and rare diseases. This has been through both hypothesis-free approaches such as GWASs (Altshuler et al., 2008) or PheWASs (Denny et al., 2010), as well as targeted approaches in Mendelian diseases.
As such, one goal of the MME is to expand the scope of discovery to allow matching in the absence of an identified candidate gene within the genomic dataset. Enabling broader, hypothesis-free approaches to discovery requires MME services to support deeper queries that can return data from entire genomic datasets as opposed to a small number of genes or variants flagged as potentially causal.
A second future goal of the MME is to expand the scope of analysis to genes and genomic variation already implicated in genetic disorders. In this scenario, the goal is to better define the phenotypic spectrum associated with individual genes as well as facilitate the understanding of specific variants identified in known disease genes. Use of sophisticated deep phenotyping approaches, combined with databases like the MME, can better objectively define the phenotypic spectrum of diseases. To support this, solved cases of Mendelian disease must be added and remain in the databases to gradually build larger datasets.
A third goal is to more effectively support the role of patient-initiating matchmaking in the MME. There are already examples of patient's who have played such roles in identifying causes of rare disease (Lambertson et al., 2015) and the MME intends to better support their efforts. Two manuscripts in this special issue describe how patients themselves have taken an interest in matchmaking and are creating their own systems both within and apart from the MME (Kirkpatrick et al., 2015, Lambertson et al., 2015).
A fourth goal of the MME effort is to contribute to the growing array of tools and strategies for broader data sharing and use. The first iteration of the MME enables investigators with unsolved rare disease cases to submit their patient data and thereby find each other and undertake selective data sharing. This balances support for gene discovery with a researcher's desire to protect resource investment in identifying candidate genes. Alternative methods could be used for matchmaking within controlled access and open access environments, some of which would allow researchers to query databases even without patient data in hand (or in situations where submission of patient data is not permitted). Many argue for a far more open environment for data sharing, which would drive scientific discovery in many more ways. For example, a researcher studying a biological pathway may hypothesize that genes in that pathway, when mutated, could cause disorders affecting a certain organ system and wish to validate that hypothesis in the absence of having access to real cases. If that researcher could query MME services for cases with relevant phenotypes and deleterious variants in pathway genes, such a hypothesis could be more quickly validated and form the basis for future studies. Similarly, researchers may wish to perform meta-analyses of large datasets to arrive at generalized conclusions as well as have access to large datasets to train algorithms for pathogenicity detection. To enable these types of investigations, MME systems will need to designate datasets and provide services that allow searching without requiring data deposition of a patient case. Some MME services already have apportioned some or all of their data for open interrogation such as DECIPHER (Chatzimichali et al., 2015, this issue) and the Monarch Initiative (Mungall et al., 2015, this issue), or enable direct searches within private networks as in the case of Cafe Variome (Lancaster et al., 2015 (this issue)). Others services are committed to supporting such activities in the future.
Finally, now that a core federated network has been formed with successful implementation of the MME API v1.0, efforts will turn toward encouraging use of the MME and bringing new MME services into the network. We hope that the MME will grow into a large and vibrant community of commercial, clinical, and academic users who are committed to a federated model of data sharing for the advancement of science and medicine.
Conclusions
In summary, this paper provides an overview of the Matchmaker Exchange, from its founding principles and goals to the steps required to launch it as a robust platform for rare disease discovery. The ensuing papers in this special issue of Human Mutation define many of the individual matchmaker services already connected (Buske, et al., 2015a; Chatzimichali et al., 2015; Sobreira et al., 2015b), or intending to connect to the federated network (Lancaster et al., 2015; Kirkpatrick et al., 2015; Lambertson et al., 2015; Mungall et al., 2015), as well as other core components (Buske et al., 2015b) and concepts (Krawitz et al, 2015; Akle et al., 2015) that support genomic matchmaking. A few case examples of discoveries already made through use of matchmaking approaches are highlighted to add further support for this robust approach to rare disease gene discovery (Au et al., 2015; Jurgens et al., 2015; Loucks et al., 2015). It is our hope that the success of the MME will serve as a model and foundation for innovative data sharing that leverages the increasing role of computational infrastructure to support the scaling of genomics as we collectively advance medicine and improve human health.
Supplementary Material
Acknowledgments
Members of the MME acknowledge the contributions of GA4GH and IRDiRC in advancing this collaborative initiative. H. Rehm, D. Azzariti and J. Krier were supported in part by NIH grant U41HG006834. J. Krier was also supported by NIH T32 GM007748-34 grant U54HG007990. C. Brownstein and I. Holm are supported in part by grant HG007530. I. Holm is also supported by grant HG007690 and grant HD077671. A.J. Brookes is supported in part by the BioSHaRE-EU project (EC FP7, #261433). S.F. Terry is supported in part by PCORI contract PPRN-1306-04899, Robert Wood Johnson Foundation grant 71636 and PXE International. S. Zuchner is supported by NIH (R01NS075764, 5R01NS072248, U54NS065712), MDA and CMTA. A. Hamosh, N. Sobreira and F. Schiettecatte are supported in part by NIH grant 1U54HG006542. K. Boycott, M. Brudno, O. Buske, S. Dumitriu, M. Girdea, and A. Misyura are supported by the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research and the Ontario Genomics Institute, as well as by a NSERC/CIHR Collaborative Health Research Project (CHRP) grant. O. Buske is supported by the Garron Family Cancer Centre and Hospital for Sick Children Foundation Student Scholarship Program. A. Philippakis is supported by a Broad Ignite Award, and an NCI Cloud Pilot award, grant N01CO42400-80. DECIPHER is supported by the Wellcome Trust, grant number WT098051. M. Haendel, C. Mungall, and N. Washington are supported by Monarch NIH #5R24OD011883. Additional support for C. Mungall and N. Washington was received from the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under [Contract No. DE-AC02-05CH11231]. RD-Connect is supported by the European Union Seventh Framework Programme (FP7/2007-2013) grant agreement No. 305444. R. Gibbs is supported in part by grant U54 HG003273. S. Dyke is supported by the Canadian Institutes of Health Research (grants EP1-120608; EP2-120609), the Canada Research Chair in Law and Medicine, and the Public Population Project in Genomics and Society (P3G).
Footnotes
Conflicts
The following authors have a commercial conflict of interest: S. Zuchner is Chair of the Scientific Advisory Board of the non-for-profit charity The Genesis Foundation (501(c)(3); R. Gibbs is the acting C.S.O. of Baylor-Miraca Genetics Laboratories; A. Philippakis is a Venture Partner at Google Ventures.
References
- Akle S, Jordan DM, Cassa CA. Quantifying and mitigating false-positive disease associations in rare disease matchmaking. Hum Mutat. 2015;36 doi: 10.1002/humu.22847. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Daly MJ, Lander ES. Genetic Mapping in Human Disease. Science. 2008 Nov 7;322(5903):881–8. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au PYB, You J, Caluseriu O, Schwartzentruber J, Majewski J, Bernier FP, Ferguson M, Care for Rare Canada Consortium. Valle D, Parboosingh JS, Sobreira S, Innes AM, Kline AD. GeneMatcher aids in the identification of a new malformation syndrome with intellectual disability, unique facial dysmorphisms, and skeletal and connective tissue caused by de novo variants in HNRNPK. Hum Mutat. 2015;36 doi: 10.1002/humu.22837. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein CA, Holm I, Ramoni R, Goldstein DB. Data sharing in the Undiagnosed Disease Network. Hum Mutat. 2015;36 doi: 10.1002/humu.22840. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske OJ, Girdea M, Dumitriu S, Gallinger B, Hartley T, Trang H, Misyura A, Friedman T, Beaulieu C, Bone WP, Links AE, Washington NL, et al. PhenomeCentral: a Portal for Phenotypic and Genotypic Matchmaking of Patients with Rare Genetic Diseases. Hum Mutat. 2015a;36 doi: 10.1002/humu.22851. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske OJ, Schiettecatte F, Hutton B, Dumitriu S, Misyura A, Huang L, Hartley T, Girdea M, Sobreira N, Mungall C, Brudno M. The Matchmaker Exchange API: automating patient matching through the exchange of structured phenotypic and genotypic profiles. Hum Mutat. 2015b;36 doi: 10.1002/humu.22850. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzimichali E, Brent S, Hutton B, Perrett D, Wright CF, Bevan AP, Hurles ME, Firth HV, Swaminathan GJ. Facilitating collaboration in rare genetic disorders through effective matchmaking in DECIPHER. Hum Mutat. 2015;36 doi: 10.1002/humu.22842. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010 May 1;26(9):1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MA, Van Booven D, Hulme W, Ulloa RH, Lebrigio RF, Osterloh J, Logan M, Freeman M, Zuchner S. Whole Genome Sequencing and a New Bioinformatics Platform Allow for Rapid Gene Identification in D. melanogaster EMS Screens. Biology. 2012;1(3):766–77. doi: 10.3390/biology1030766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MA, Lebrigio RFA, Van Booven D, Ulloa RH, Powell E, Speziani F, Tekin M, Schule R, Zuchner S. GEnomes Management Application (GEM.app): A new software tool for large-scale collaborative genome analysis. Hum Mutat. 2013;34(6):842–846. doi: 10.1002/humu.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Falk M, Gai X, Schüle R, Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum Mutat. 2015;36 doi: 10.1002/humu.22836. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens J, Sobreira N, Modaff P, Reiser CA, Seo SH, Seong M, Park SS, Kim OH, Cho T, Pauli RM. Type II collagenopathy due to a novel variant (p.Gly207Arg) manifesting as a phenotype similar to progressive pseudorheumatoid dysplasia and spondyloepiphyseal dysplasia, Stanescu type. Hum Mutat. 2015;36 doi: 10.1002/humu.22839. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz P, Buske O, Zhu Na, Brudno M, Robinson PN. The Genomic Birthday Paradox: How Much is Enough? Hum Mutat. 2015;36 doi: 10.1002/humu.22848. xxx-yyy. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick BE, Riggs ER, Azzariti DR, Rangel Miller V, Ledbetter DH, Miller DT, Rehm H, Martin CL, Faucett WA. GenomeConnect: matchmaking between patients, clinical laboratories and researchers to improve genomic knowledge. Hum Mutat. 2015;36 doi: 10.1002/humu.22838. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster O, Beck T, Atlan D, Swertz M, Dagleish R, Brookes AJ. Cafe Variome: general-purpose software for making genotype-phenotype data discoverable in restricted or open access contexts. Hum Mutat. 2015;36 doi: 10.1002/humu.22841. xxx-yyy. [DOI] [PubMed] [Google Scholar]
- Lambertson K, Damiani S, Might M, Shelton R, Terry S. Participant-Driven Matchmaking in the Genomic Era. Hum Mutat. 2015;36 doi: 10.1002/humu.22852. xxx-yyy. [DOI] [PubMed] [Google Scholar]
- Loucks CM, Parboosingh JS, Shaheen R, Bernier FP, McLeod DR, Seidahmed MZ, Puffenberger EG, Ober C, Hegele RA, Boycott KM, Alkuraya FS, Innes M. Matching two independent cohorts validates DPH1 as a gene responsible for autosomal recessive intellectual disability with short stature, craniofacial and ectodermal anomalies. Hum Mutat. 2015;36 doi: 10.1002/humu.22843. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall C, Washington N, Nguyen Xuan J, Condit C, Smedley D, Köhler S, Groza T, Shefchek K, Hochheiser H, Robinson P, Lewis S, Haendel M. Use of Model Organism and Disease Databases to Support Matchmaking for Human Disease Gene Discovery. Hum Mutat. 2015;36 doi: 10.1002/humu.22857. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Köhler S, Oellrich A, Sanger Mouse Genetics Project. Wang K, Mungall CJ, Lewis SE, Washington N, Bauer S, Seelow D, Krawitz P, Gilissen C, et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24(2):340–8. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New Tools for Mendelian Disease Gene Identification: PhenoDB Variant Analysis Module; and GeneMatcher, a Web-Based Tool for Linking Investigators with an Interest in the Same Gene. Hum Mutat. 2015a;36(4):425–31. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum Mutat. 2015b;36 doi: 10.1002/humu.22844. xxx-yyy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan GJ, Bragin E, Chatzimichali EA, Corpas M, Bevan AP, Wright CF, Carter NP, Hurles ME, Firth HV. DECIPHER: web-based, community resource for clinical interpretation of rare variants in developmental disorders. Hum Mol Genet. 2012;21(R1):R37–R44. doi: 10.1093/hmg/dds362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryka KA, Hao L, Sturcke A, Jin Y, Kimura M, Wang ZY, Ziyabari L, Lee M, Feolo M. The NCBI Handbook [Internet] 2nd edition National Center for Biotechnology Information; Bethesda (MD): 2013. The Database of Genotypes and Phenotypes (dbGaP) and PheGenI. [Google Scholar]
- Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7(11):e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Köhler S, Mackenroth L, Jäger M, Hecht J, Krawitz P, Graul-Neumann L, Doelken S, Ehmke N, Spielmann M, Oien NC, Schweiger MR, et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med. 2014;6(252):252ra123. doi: 10.1126/scitranslmed.3009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Baran J, Cros A, Guberman JM, Haider S, Hsu J, Liang Y, Rivkin E, Wang J, Whitty B, Wong-Erasmus M, Yao L, et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database. 2011;2011:bar026. doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.