Abstract
Tamoxifen is a selective estrogen receptor modulator widely used for the treatment of breast cancer. In addition to its activity as an estrogen receptor agonist/antagonist, tamoxifen also modulates sphingolipid biosynthesis, which has been shown to play an important role in the regulation of neutrophil activity. Here, we find that tamoxifen stimulation enhances several pro-inflammatory pathways in human neutrophils, including chemotaxis, phagocytosis and neutrophil extracellular trap (NET) formation. The enhancement of NET production occurs via a ceramide/PKCζ-mediated pathway, and treatment with synthetic ceramide is sufficient to promote NET formation. Pretreatment of human neutrophils with tamoxifen boosts neutrophil bactericidal capacity against a variety of pathogens in vitro and enhances clearance of the leading human pathogen methicillin-resistant Staphylococcus aureus in vivo. Our results suggest that tamoxifen, and the lipid signaling pathways it modulates, merit further exploration as targets for boosting host innate immune function.
Tamoxifen, a selective estrogen receptor modulator (SERM) taken by hundreds of thousands of patients for the treatment of breast cancer1, is included on the World Health Organization’s list of essential medicines due to its cost effectiveness and safety profile2. In addition to its activity as an estrogen receptor agonist/antagonist3, tamoxifen has many other effects that include modulation of sphingolipid biosynthesis4. Such “off target” effects have been shown to contribute to the therapeutic action of tamoxifen5; however, its impact on some normally functioning cell types, in particular circulating neutrophils, remains poorly understood. Earlier investigations of the impact of tamoxifen on neutrophil function have largely been limited to its inhibition of transendothelial migration, suggesting a primary immunosuppressant activity6, 7. Because sphingolipids are known to be important modulators of immune cell function8, we investigated the effects of tamoxifen on human neutrophils, the primary phagocytic leukocyte of the immune system.
Using in vitro and in vivo approaches, we find that tamoxifen, in addition to enhancing neutrophil chemotaxis and phagocytosis, strongly promotes the formation of DNA-based neutrophil extracellular traps (NETs). NETs, which are coated with antimicrobial peptides, histones and/or granule proteases, are capable of entrapping and killing bacteria and other pathogens9, 10. Tamoxifen enhances killing of pathogens in vitro and enhances clearance of methicillin-resistant Staphylococcus aureus (MRSA) in vivo. We find that such alterations in neutrophil activity are dependent upon intracellular ceramide.
Results
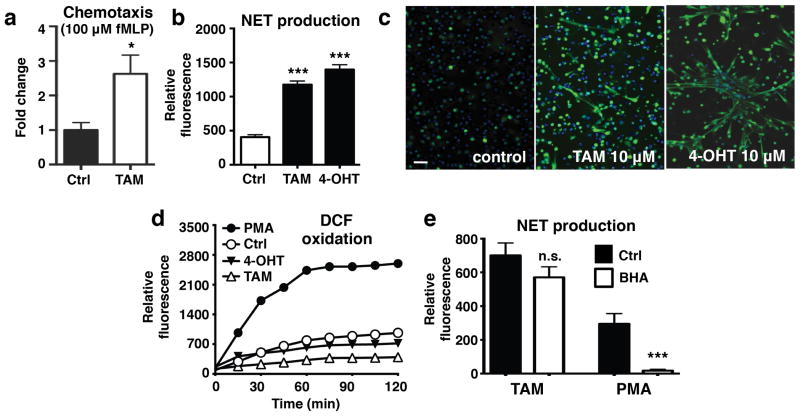
Direct assessment of N-Formyl-Met-Leu-Phe (fMLP)-induced neutrophil chemotaxis indicated that tamoxifen, while not stimulating chemotaxis by itself (Supplementary Fig. 1), significantly enhanced directional cell migration in response to fMLP stimulation (Fig. 1a). These differences in findings may be due to variations in protocol (e.g., previous studies preincubated cells with tamoxifen and removed it prior to assessment of chemotaxis rather than assessing migration in its presence) or the added complexity of in vitro transendothelial migrations models. In addition to these findings, fluorescence-based quantification of extracellular DNA release revealed that tamoxifen (Fig. 1b), its active metabolite 4-hydroxytamoxifen (Fig. 1b), and its primary metabolite N-desmethyltamoxifen (Supplementary Fig. 2), strongly stimulated NET production in freshly isolated human neutrophils. These results were confirmed by immunostaining of stimulated neutrophils using a primary antibody against myeloperoxidase, a NET marker (Fig. 1c)11. Because tamoxifen is generally described to have antagonist activity at the estrogen nuclear receptors ERα and ERβ and agonist activity at the G protein-coupled estrogen receptor GPR30, we attempted to replicate these results using selective agonists/antagonists. In these studies, NET production was not observed in response to treatment with MPP or PHTPP (selective ERα and ERβ antagonists, respectively12, 13), G-1 (a GPR30 agonist14) or fulvestrant (a SERM that mimics the mixed agonist/antagonist characteristics of tamoxifen15, 16) (Supplementary Fig. 3), indicating that tamoxifen-induced NET production is mediated by an estrogen receptor-independent mechanism.
Figure 1. Tamoxifen promotes extracellular trap formation in human neutrophils in vitro.
(a) The effect of 10 μM tamoxifen (TAM; upper well) on chemotaxis in the presence or absence of 100 nM fMLP (lower well) was determined using a transwell system (n = 12). (b) Extracellular DNA was quantified to determine the effect of both TAM and 4-hydroxytamoxifen (4-OHT) on NET release (n = 9). (c) NET production was confirmed by fixing and immunostaining TAM/4-OHT-stimulated cells with DAPI and a primary antibody for myeloperoxidase (blue: DAPI; green; NETs/myeloperoxidase); images representative of three independent experiments shown (scale bar = 50 μm). (d) DCF-based reactive oxygen species (ROS) assays were performed to evaluate the effect of TAM and 4-HT on ROS production (n = 9). (e) Neutrophils were pre-incubated with the reactive oxygen species scavenger BHA prior to addition of either PMA to TAM to determine whether ROS production is required for tamoxifen-induced NET production (n = 9).
To elucidate the pathway underlying the neutrophil activating effects of tamoxifen, we next assessed signaling molecules/effectors typically associated with NET release. Although NET production has been reported to be dependent on reactive oxygen species (ROS) generation17, we found that both tamoxifen and 4-hydroxytamoxifen lowered ROS production in human neutrophils (Fig. 1d), mirroring our previous findings with statins18. This finding was supported by experiments using the reactive oxygen species scavenger butylated hydroxyanisole (BHA), which potently inhibited phorbol myristate acetate (PMA)-induced NET production, but did not significantly reduce tamoxifen-induced NET production (Fig. 1e).
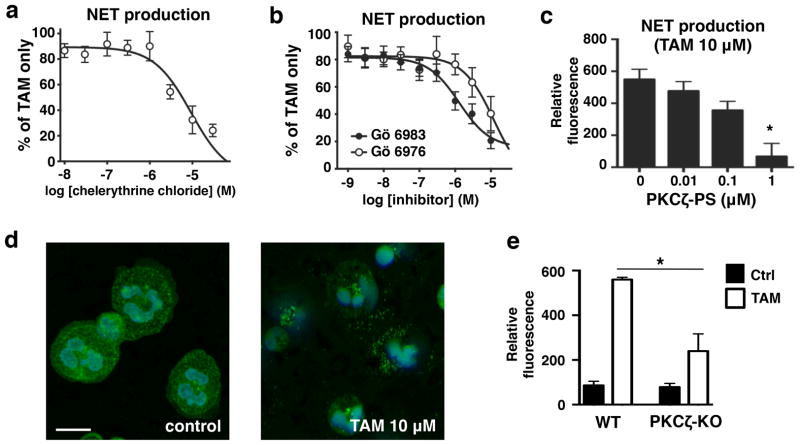
In contrast, pre-incubation with the general PKC antagonist chelerythrine chloride inhibited tamoxifen-induced NET production (Fig. 2a). To determine the specific PKC isoform responsible for tamoxifen-induced NET release, we stimulated neutrophils with tamoxifen in the presence of two broad-spectrum PKC inhibitors with different selectivity profiles, Gö 6973 and Gö 6983, the latter of which is a more potent inhibitor of PKCζ and PKCμ19. Inhibition curves revealed that Gö 6983 exhibited greater inhibition of tamoxifen-induced NET production, pointing to either PKCζ or PKCμ as the primary PKC isoform responsible for this induction (Fig. 2b). Further experiments using a highly selective inhibitor of PKCζ, PKCζ pseudo substrate (PKCζ-PS)20, resulted in near-complete inhibition of tamoxifen-induced NET production, strongly implicating PKCζ as a key mediator of this process (Fig. 2c). Inhibition of PI3K had no effect on tamoxifen-induced NET production (Supplementary Fig. 4), though tamoxifen promoted translocation of neutrophil PKCζ (Fig. 2d). The importance of PKCζ in tamoxifen-induced NET production was confirmed in experiments using bone marrow-derived neutrophils collected from PKCζ knockout animals, which showed significantly reduced NET production following tamoxifen treatment compared to wild-type littermate controls (Fig. 2e).
Figure 2. Tamoxifen-induced extracellular trap production is PKCζ dependent.
(a) To determine the whether PKC is required for TAM-induced NET production, neutrophils were pre-incubated with (a) the general PKC antagonist chelerythrine chloride (n = 8) or (b) the more selective inhibitors Gö 6976 and Gö 6983 (n = 9) prior to treatment with 10 μM TAM. (c) Neutrophils were pre-incubated with PKCζ pseudo-substrate (PKCζ-PS) to specifically evaluate the role of PKCζ in TAM-induced NET production. (d) Immunostaining/confocal microscopy was used to visualize translocation of PKCζ in human neutrophils in response to TAM (10 μM) treatment (blue: DAPI; green; PKCζ); images representative of three independent experiments shown (scale bar = 10 μm). (e) Bone-marrow derived neutrophils were collected from PKCζ knockout mice to determine TAM-induced NET production in comparison to and wild type littermate controls (n = 4–5). Where applicable, results were analyzed by one-way ANOVA and post hoc Newman Keuls test. *P < 0.05, ***P < 0.001 vs. control values.
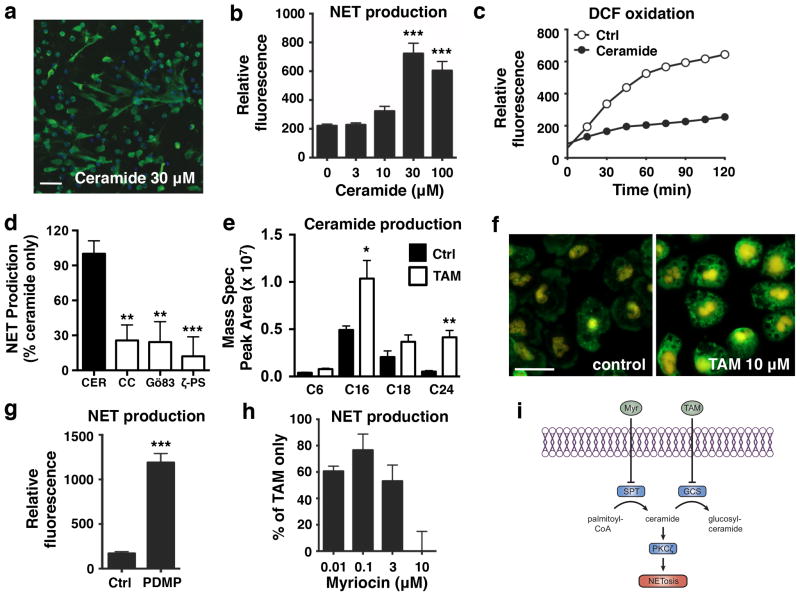
Through its ability to inhibit glucosylceramide synthase, a key enzyme in the sphingolipid synthesis pathway that converts ceramide to glucosylceramide, tamoxifen has been shown in some cell types to promote intracellular accumulation of ceramide, a potent PKCζ activator21. Thus, we next determined if ceramide accumulation could drive tamoxifen-induced NET production. Indeed, like tamoxifen, ceramide promoted NET release in a dose-dependent manner (Fig. 3a, b) while inhibiting ROS generation (Fig. 3c). Ceramide-induced NET production was also PKC/PKCζ dependent (Fig. 3d). Mass spectrometry revealed a statistically significant increase in C16 and C24 ceramide species in neutrophils upon tamoxifen treatment. Structural verification of ceramide species was obtained with high mass accuracy of the parent ion and additionally by monitoring the hallmark 264 Dalton ion fragment released during the fragmentation of ceramides via tandem mass spectrometry (Fig. 3e, Supplementary Fig. 5). Elevated intracellular ceramides were also visually confirmed via immunocytochemistry using an anti-ceramide antibody (Fig. 3f). Further evidence of the role of ceramide in NET production was obtained by treating cells with DL-PDMP, a selective inhibitor of glucosylceramide synthase that mimics the intracellular ceramide-increasing effect of tamoxifen22, which also promoted NET production (Fig. 3g). Sphingosine-1-phosphate, which is generated from ceramide via the cascade initiated by glucosylceramide synthase23, did not significantly promote NET production (Supplementary Fig. 6). To further confirm that tamoxifen-induced NET production is ceramide-dependent, we pre-incubated neutrophils with myriocin, an inhibitor of serine palmitoyltransferase24. Inhibition of this enzyme, which initiates the pathway that generates ceramide from palmitoyl-CoA, prevents tamoxifen-mediated ceramide accumulation25. Although it does not directly inhibit PKCζ (Supplementary Fig. 7), myriocin treatment resulted in complete inhibition of tamoxifen-induced NET production (Fig. 3h) and partially reversed inhibition of PMA-induced ROS production by tamoxifen (Supplementary Fig. 8), illustrating the key role of ceramide accumulation in this pathway.
Figure 3. Tamoxifen-induced NET production is mediated by intracellular ceramide.
(a) Ceramide-induced NET production was visualized by immunostaining neutrophils for myeloperoxidase (blue: DAPI; green; NETs/myeloperoxidase); images representative of three independent experiments shown (scale bar = 50 μm). (b) Quantification of extracellular DNA/NET production in response to ceramide treatment (n = 9). (c) The effect of ceramide (30 μM) on ROS production was determined using DCF-based reactive oxygen species (ROS) assays (n = 9). (d) Neutrophils were pre-incubated with PKC inhibitors (CC = chelerythrine chloride, Gö86 = Gö 6983, ζ-PS = PKCζ pseudo-substrate; 10 μM each; n = 9) to determine the involvement of PKC in ceramide-induced NET production. (e) Mass spectrometry analysis of lipid extracts were performed to quantify intracellular accumulation of several ceramide species in response to TAM treatment (10 μM). (f) Intracellular ceramide levels in control and TAM treated (10 μM) neutrophils were visualized via immunostaining of fixed neutrophils using a ceramide primary antibody (green; ceramide); images representative of three independent experiments shown, with identical exposure and gain settlings used for control and tamoxifen-treated samples (scale bar = 20 μm). (f) The effect of DL-PDMP, which like tamoxifen inhibits glucosylceramide synthase, on NET production was assessed via quantification of extracellular DNA (n = 9). (e) Myriocin, which blocks ceramide synthesis by inhibiting serine palmitoyltransferase, dose-dependently inhibits tamoxifen-induced NET production. (f) Summary of the mechanism of tamoxifen-induced NET production. By inhibiting glucosylceramide synthase, tamoxifen treatment leads to an accumulation of neutrophil intracellular ceramide levels, which promotes NET formation via activation of PKCζ. Where applicable, results were analyzed by one-way ANOVA and post hoc Newman Keuls test. *P < 0.05, ***P < 0.001 vs. control values.
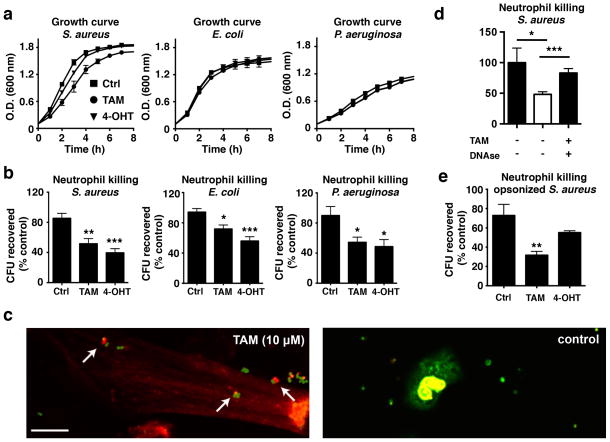
Having established that tamoxifen stimulates neutrophil migration and induces NET production, we examined whether neutrophil bactericidal capacity could be enhanced via tamoxifen treatment. At the concentrations used to stimulate neutrophils, tamoxifen and 4-hydroxytamoxifen had negligible effects on the growth of the USA300 strain of community-associated MRSA, Escherichia coli (K1 serotype) or Pseudomonas aeruginosa (PA01 strain) (Fig. 4a). However, both tamoxifen and 4-hydroxytamoxifen enhanced the killing activity of activated human neutrophils against unopsonized bacteria of all three species (Fig. 4b). Treatment of neutrophils with ceramide mimicked these effects (Supplementary Fig. 9). Live/dead staining of USA300 MRSA exposed to either control or tamoxifen-treated neutrophils revealed dead bacteria within the NETs of stimulated neutrophils, in contrast to mostly live bacteria surrounding the untreated neutrophils producing fewer NETs (Fig. 4c). Addition of DNAse to neutrophils prior to the addition of bacteria significantly enhanced bacterial recovery (Fig. 4d), suggesting that the bactericidal effects of tamoxifen are largely NET driven. Addition of DNase to neutrophil/bacteria culture samples prior to serial dilution and plating indicated that reductions in colony forming units were primarily due to killing, rather than clumping, of bacteria (Supplementary Fig. 10). Several different approaches confirmed that the DNAse used in these experiments was active, being able to both degrade DNA and eliminate NETs (Supplementary Fig. 11). Given that some reports have suggested that the principal activity of NETs is to clump rather than kill bacteria26, 27, our results provide evidence that NETs are heterogeneous in both their mechanism of formation and activity. Both tamoxifen and 4-hydroxytamoxifen also stimulated the uptake of S. aureus-labeled bioparticles (Supplementary Fig. 12), suggesting a further activity to upregulate phagocytic pathways. Additional experiments using USA300 MRSA opsonized by incubation in pooled human serum also revealed enhanced bactericidal activity of neutrophils following a 10 min pre-incubation with tamoxifen (Fig. 4e).
Figure 4. Tamoxifen enhances neutrophil killing.
(a) Bacterial growth curves for USA300 methicillin-resistant Staphylococcus aureus (MRSA), K1 strain Escherichia coli, or PA01 strain Pseudomonas aeruginosa (n = 6) in the presence or absence of TAM or 4-hydroxytamoxifen 4-OHT (10 μM each). (b) Bactericidal assays evaluating the effect of a 4 h TAM or 4-HT pre-treatment on neutrophil killing (represented by a reduction of CFUs) of USA300 MRSA, E. coli and P. aeruginosa (n = 9–12). (c) Live/dead cell analysis of fixed cells; dead bacteria (red; propidium iodide) can be observed in tamoxifen-stimulated NETs (left panel). Live bacteria (green; SYTO 8) can be observed outside of the NETs and surrounding unstimulated cells (right panel). Images shown are representative of three independent experiments (scale bar = 10 μm). (d) Addition of DNAse prior to addition of bacteria to tamoxifen-treated neutrophils significantly reduces killing (n = 9). (e) Neutrophils were incubated with TAM and bacteria concurrently to determine killing of USA300 MRSA opsonized using pooled human serum (n = 8–9). Where applicable, results were analyzed by one-way ANOVA and post hoc Newman Keuls test. *P < 0.05, ***P < 0.001 vs. control values.
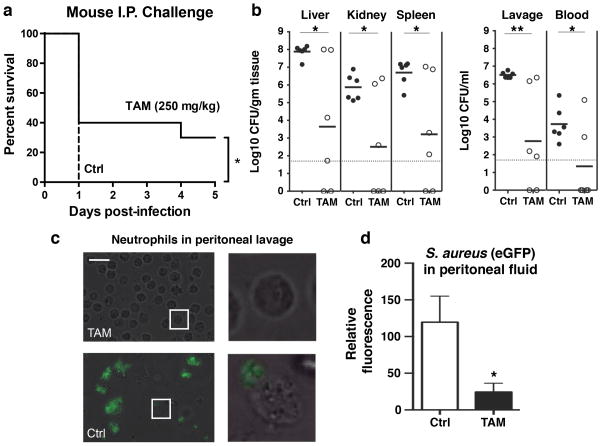
We next tested tamoxifen in a murine model of systemic MRSA infection. One hour following intraperitoneal (IP) treatment with tamoxifen or vehicle control (corn oil), outbred CD-1 mice were infected IP with a lethal dose of USA300 MRSA. Mice were subsequently treated with tamoxifen or vehicle control at 1 h and 8 h post infection. Tamoxifen provided significant protection against mortality (Fig. 5a); whereas all mice in the control group died on day 1, 40% of the tamoxifen-treated mice remained alive at day 3, and 33% surviving to the experimental endpoint (5 days). In subsequent experiments, mice were treated with tamoxifen or vehicle 2 h before and 6 h after intraperitoneal infection with an EGFP-expressing USA300 MRSA. Mice were euthanized 8 h post infection to assess bacterial dissemination; mice in the tamoxifen-treated group exhibited significantly lower bacterial counts in peritoneal lavage fluid as well as in the liver, kidney, spleen, and blood (Fig. 5b), with bacterial counts below the limit of detection in the tissues of several tamoxifen-treated animals. Analysis of peritoneal lavage fluid using fluorescence microscopy revealed very few intact bacteria in samples from tamoxifen-treated mice, in stark contrast to samples collected from vehicle-treated animals, whose neutrophils contained large numbers of intact bacteria (Fig. 5c, d). Of note, neutrophils from these control animals often appeared damaged, consistent with previous findings that S. aureus can promote neutrophil necrosis following phagocytosis28.
Figure 5. Tamoxifen reduces mortality and enhances clearance of USA300 MRSA in an in vivo model of acute infection.
(a) One hour following intraperitoneal (IP) treatment with tamoxifen (TAM; 250 mg/kg) or vehicle control (VEH; corn oil), CD-1 background CD-1 background mice were infected IP with a lethal dose of USA300 MRSA (5 × 108 CFU/mL, 200 μl). Mice were subsequently treated with tamoxifen or vehicle control at 1 h and 8 h post infection and monitored for five days. Tamoxifen significantly protected against mortality. (b) To assess bacterial dissemination, CD-1 background mice were received IP injections tamoxifen (250 mg/kg) or vehicle control 2 h prior to and 6 h following IP injection of enhanced green fluorescent protein (EGFP)-expressing USA300 MRSA (5 × 108 CFU/mL, 200 μl). Bacterial counts were determined in the peritoneal lavage fluid and blood, kidney, liver and spleen tissue samples. (c) Light microscopy revealed that neutrophils in the vehicle-control samples often appeared damaged and contained multiple intact bacteria (scale bar = 25 μm). (d) Quantitative analysis of fixed peritoneal lavage samples confirmed the presence of elevated EGFP-USA300 MRSA levels in vehicle vs. tamoxifen-treated animals. Results were analyzed by the log-rank test (survival curve comparison) or one-way ANOVA and post hoc Newman Keuls test. *P < 0.05, ***P < 0.001 vs. control values.
Discussion
In sum, we have shown that tamoxifen enhances neutrophil bactericidal activity, with increased NET production a likely contributory mechanism. Our evidence suggests that this enhancement occurs in significant part through the modulation of intracellular ceramide levels. Ceramide, an important bioactive sphingolipid and cellular stress signal, has previously been linked to neutrophil apoptosis29, 30, and tamoxifen-induced ceramide accumulation mediates certain estrogen receptor-independent effects of the drug against cancer tissues5. Here, we show that both tamoxifen and synthetic ceramide are potent inducers of NETosis, a novel cell death pathway with an important role in innate immunity31, Interestingly, ceramide can activate both apoptotic and autophagocytic pathways32, the later of which have recently been implicated in NET production33. Shifting the balance between apoptotic and autophagocytic pathways may influence relative levels of NET production; indeed, we found that rapamycin, a potent inducer of autophagy, enhanced ceramide-induced NET production while having little effect on its own (Supplementary Fig. 13).
Further investigation of compounds that modulate the ceramide biosynthesis pathway may reveal more potent enhancers of neutrophil antimicrobial activity. Along these lines, it is interesting to note that the active metabolite of tamoxifen, 4-hydroxytamoxifen, is even more potent at enhancing NET release and bactericidal activity than tamoxifen. Exploring the mechanism underlying tamoxifen/4-hydroxytamoxifen-induced NET formation may facilitate discovery of derivatives compounds specifically optimized to promote targeted enhancement of innate immune responses.
The outcomes of enhanced NET production are likely to be pathogen-specific. Several bacterial species are capable of evading NET capture through various mechanisms (e.g., via the expression of extracellular nucleases capable of cleaving DNA-based structures34, 35). In the context of S. aureus, a focus of this study, tamoxifen may exhibit particularly dramatic effects due to the pathogen’s ability not only to evade killing via opsonophagocytosis36, 37 and oxidative burst38, but also to invade neutrophils and trigger a unique necrolytic pathway. We observed neutrophils in peritoneal lavage samples collected from tamoxifen-treated mice exhibited a significantly reduced bacterial load in comparison to untreated controls.
In addition to their role in innate immunity, excessive or aberrant production of NETs has been implicated in inflammatory diseases such as vasculitis and bronchial asthma39, 40. The cumulative effect of tamoxifen on neutrophil responses hold clinical implications given the large number of patients receiving this drug and related therapeutic agents.
Methods
Materials
Tamoxifen citrate, ceramide, MPP dihydrochloride, G-1, Gö 6983, Gö 6976, PKCζ-PS and DL-PDMP were all obtained from Tocris Bioscience (Bristol, UK). Myriocin was obtained from Cayman Chemical (Ann Arbor, MI). Unless otherwise noted, all other chemicals were obtained from Sigma Aldrich.
Bacterial strains
Methicillin-resistant Staphylococcus aureus (MRSA) strain USA300 (UAMS 1182), Escherichia coli strain K1, and Pseudomonas aeruginosa strain PA01 were used in this study. EGFP-transformed MRSA strain USA300 (LAC) were used for in vivo studies where indicated.
Neutrophil isolation
Human venous blood was collected from healthy volunteers, with heparin added as an anticoagulant. Neutrophils were isolated using Polymorphprep (Axis-Shield, Dundee, Scotland) according to the manufacturer’s protocol. Blood was drawn according to a protocol approved by the local ethics committee. Neutrophils were collected from PKCζ knockout animals (C57BL/6 background) and wild-type littermate controls by extracting bone marrow from harvested tibia. Bone marrow was plated on polystyrene dishes and incubated for 37°C with 5% CO2 for 1 h; subsequently, neutrophils (still in suspension) were collected, counted, and used in experiments.
Transwell chemotaxis assay
Neutrophils, preincubated for 10 min at 24°C in HBSS alone or HBSS with 10 μM tamoxifen, were seeded in 6-mm transwell permeable supports (3-μm pore size; Corning Inc.,) that were placed in 24 well plates; lower chambers contained either HBSS alone, or 100 nM fMLP. Following a 45-min incubation at 37°C, inserts were removed and cells were lysed by addition of Triton-X 100 (0.1% final, 10 min, 24°C). To determine the relative levels of migration to the lower well, the colorimetric elastase substrate N-methoxysuccinyl-Ala-Ala-Pro-Val-p-nitroanilide was added to the lysed cell samples (10 mM final); following a 30 min incubation at 24°C, absorbance at 405 nm was measured using a SpectraMAX Gemini EM fluorescence reader (Molecular Devices, Sunnyvale, CA).
Induction and quantification of neutrophil extracellular traps
To induce extracellular trap production, neutrophils were seeded in 96-well plates at a density of 2 × 105 cells/well in HBSS (final volume: 200 μl). Where applicable, cells were incubated with NETosis inhibitors at 37°C with 5% CO2 for 60 min (myriocin), 45 min (PKCζ-PS) or 30 min (all other inhibitors) prior to addition of NET inducing agents. NET inducers were then added, and cells were incubated for a further 2 h at 37°C with 5% CO2. Where applicable, 25 nM PMA was included as a positive control. Micrococcal nuclease was then added (final concentration: 500 mU/ml) prior to an additional 10 min incubation at 37°C. Micrococcal nuclease activity was stopped via the addition of 5 mM EDTA, and plates were spun at 200 × g for 8 min. Supernatant samples (100 μl) were collected from each well, and extracellular DNA content was quantified using a Quant-IT PicoGreen dsDNA Assay kit (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions (Grand Island, NY).
NET visualization and fluorescence microscopy
To visualize NETs, cells were seeded on 24-well plates at a density of 2 × 105 cells/well. Following stimulation of NET production (2 h, 37°C with 5% CO2 with indicated drugs), cells were fixed by the addition of paraformaldehyde (4% final) for 10 min at 24°C. After blocking for 45 minutes with PBS containing 2% bovine serum albumin (2% PBS-BSA) and 2% goat serum, cells were immunostained via the following incubation steps (each at room temperature): 1 h with rabbit anti-human myeloperoxidase primary antibody (1:300 in 2% PBS-BSA; Dako North America, Inc., Carpinteria, CA; cat # A039829-2), 45 min (protected from light) with Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (1:500 in 2% PBS-BSA); Life Technologies, Carlsbad, CA, cat # A11070), and 10 min with 1 μM Hoechst-33342-trihydrochloride. Three washes with PBS were performed after each staining step. Images were obtained using a Zeiss AxioObserver D1 microscope equipped with an LD A-Plan 20X/0.35 Ph1 objective. Imaging of intracellular ceramide was performed by permeabilizing cells with 0.25% Triton-X 100 following fixation with paraformaldehyde, blocking with PBS containing 2% bovine serum albumin (2% PBS-BSA) and 2% donkey serum for 1 h, followed by incubation for 1 h with mouse anti-ceramide primary antibody (1:300 in 2% PBS-BSA; Sigma Aldrich, St. Louis, MO; cat # C8104-50TST) and 45 min (protected from light) with Alexa Fluor 488 donkey anti-mouse IgG secondary antibody (1:500 in 2% PBS-BSA; Life Technologies, Carlsbad, CA; cat # A21202). Representative images shown were collected the Zeiss AxioObserver D1 microscope and objective described above, with exposure and gain settings kept consistent during collection of control and tamoxifen-treated images. A similar protocol in combination with a PKCζ primary antibody (1:200 in 2% PBS-BSA; Abcam, Cambridge, MA, cat # ab59364) was used to visualize intracellular PKCζ distribution using a Leica TCS SPE confocal microscope equipped with a HC PLAN APO 63X/1.40-0.60 NA oil immersion objective. For live/dead cell analysis, neutrophils were incubated in 8-well LabTek II glass-bottom slides (Nalge Nunc, Penfield, NY) for 2 h with or without tamoxifen, followed by addition of USA300 MRSA (MOI: 5). After 30 min, cells were stained using a LIVE/DEAD BacLight Bacterial Viability Kit according to the manufacturer’s instructions. Cells were fixed with 4% paraformaldehyde and imaged using the Leica TCS SPE confocal microscope and objective described above.
ROS production assays
Neutrophils were incubated in HBSS containing 10 μM 2′,7′-dichlorofluorescein diacetate for 20 min at 37°C with gentle agitation. Neutrophils were then centrifuged at 400 × g for 5 min, washed with HBSS, and centrifuged again using the same settings before being counted and resuspended in HBSS at a concentration of 5 × 106 cells/ml. Samples of the cell suspension were then added to a 96-well plate (5 × 105 cells/well) and mixed at a 1:1 ratio with PMA, ceramide, tamoxifen or 4-hydroxytamoxifen. Fluorescence intensity (485 nm excitation, 530 nm emission) was measured in 15 min intervals using a SpectraMAX Gemini EM fluorescence reader over the course of 2 h (plates were incubated at 37°C, protected from light and with gentle shaking, between reads).
Extraction and quantification of ceramide species
Cellular lipids were extracted from neutrophils using the protocol described by Zhang et al. 41. Samples obtained from these extractions were analyzed using Orbitrap Fusion Tribrid mass spectrometry technology. Briefly, nano ultra high-performance liquid chromatography (HPLC) was performed on a Thermo Fisher EASY-nLC (Thermo Fisher Scientific Inc., Waltham, MA) with an incorporated cooled autosampler compartment calibrated for a 6x8 sample grid. The flow rate was set to 300 nl/min, and an 85 min gradient was run. Mobile phase A consisted of 0.1% formic acid in water and mobile phase B consisted of 0.1% formic acid in acetonitrile. The nano-analytical column was prepared by pulling Polymicro fused silica, 30 cm × 75 μm, (Polymicro Technologies, Phoenix, AZ) and packing with 3 μm UChrom C18 bulk packing material (nanoLCMS Solutions LLC). Analysis was performed on a finely calibrated Thermo Fisher Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA). Positive electrospray ion mode was used in all runs in the targeted selective ion monitoring (TSIM) scan mode. Relative quantification of the structurally confirmed ceramide species was determined by integration of the peak areas obtained from the extracted ion chromatogram using Xcalibur software.
Bacterial growth curves
Glass tubes containing Todd Hewitt broth (THB) plus vehicle (10% HBSS), tamoxifen (10 μM) or 4-hydroxytamoxifen (10 μM) were inoculated with sufficient amounts of overnight bacterial cultures to achieve an optical density (600 nm) of 0.1. Tubes were incubated in a shaking 37°C incubator, and absorbance measurements at 600 nm were taken every hour up to 7 h, and once at 24 h, using a Spectronic 20D+ spectrophotometer (Thermo Scientific, Waltham, MA, USA).
In vitro bactericidal assays
Freshly isolated neutrophils in serum-free RPMI were added to 48-well plates at a density of 4 × 105 cells/well. HBSS, tamoxifen, or 4-hydroxytamoxifen was added to applicable wells, resulting in a final volume of 200 μl, and cells were incubated for 4 h at 37°C with 5% CO2 (identical wells containing no neutrophils were also prepared). Overnight cultures of bacteria were resuspended in RPMI containing 10% 70°C heat-inactivated FBS to achieve a density of 8 × 10^5 CFUs/ml. 50 μl of this bacterial suspension was added to applicable wells of the 48-well plate, resulting in a 2% final concentration of FBS and a multiplicity of infection (MOI) of 0.1. Plates were then centrifuged at 1600 rpm for 5 min prior to a 15-min incubation at 37°C. A total of 25 μl was then collected from each well and serially diluted in a 96-well plate containing sterile water (225 μl/well). Samples (25 μl) from each well were collected and plated were collected and plated on Todd Hewitt agar (THA) plates, which were incubated at 37°C overnight. Colonies were counted the next day to calculate bacterial survival. For bactericidal assays using opsonized bacteria, USA300 MRSA were incubated for 30 min in 80% 56°C heat-inactivated pooled human serum in RPMI before being combined with neutrophils in a 96-well plate (such that the final concentration of serum was 2% and the MOI was 1). Samples were collected, serially diluted in water, and plated on THA plates to quantify CFUs.
Phagocytosis assay
Neutrophils were combined in a 96-well plate (2 × 105 cells/well) with pHrodo Red S. aureus Bioparticles (Life Technologies, Carlsbad, CA) as specified by the manufacturer. Plates were incubated at 37°C, and phagocytosis assessed by measuring fluorescence intensity (560 nm excitation, 585 nm emission) at 15 min intervals using a SpectraMAX Gemini EM fluorescence reader (Molecular Devices, Sunnyvale, CA).
Murine infection studies
For in vivo infection studies, 8-week-old female CD1 mice (Charles River Laboratories, Wilmington, MA, USA) were injected intraperitoneally with tamoxifen (250 mg/kg) or vehicle (corn oil) (n = 6 mice per group). After an hour, mice were infected intraperitoneally with a lethal dose (~1 × 109 CFU) of USA300 MRSA strain (x). The mice received additional tamoxifen or vehicle treatments 1 and 8 h following infection, and survival was monitored over 7 days. Mice that appeared moribund were humanely killed. To determine CFUs in tissues of tamoxifen-treated and control animals, mice were treated with tamoxifen or vehicle 2 h before and 6 h after intraperitoneal infection with EGFP-expressing USA300 MRSA. Mice were euthanized 8 h post infection. Peritoneal lavage and blood samples were collected along with the kidneys livers, and spleens of the infected animals. Following homogenization of organs using a MagNA Lyser instrument (Roche Diagnostics Corporation, Indianapolis, IN), samples were serially diluted in PBS and plated on THA plates to determine CFUs/g of tissue. 200 μl samples of the peritoneal lavages were fixed via addition of 4% paraformaldehyde and imaged using the Zeiss AxioObserver system described above. This in vivo infection study was performed in duplicate, and data from one representative experiment are shown. All studies involving animals were reviewed and approved by the University of California San Diego Animal Care and Use Committee.
Statistical Analysis
All of the statistical analyses described in the figure legends were performed using Prism v6.0. Unless otherwise noted, n values indicate the number of replicates performed in at least three independent experiments.
Supplementary Material
Acknowledgments
Research was supported by NIH grants HD071600, AI057153, and AI052453 (to V.N.) and OD008469 (to J.T.C.).
Footnotes
Conflict of Interest Statement: The authors listed above certify that there is no conflict of interest with any financial organization regarding the material discussed in this manuscript.
Author Contributions
All authors planned and designed experiments. R.C., A.H, J.O., J.D., J.L. and D.G. conducted experiments. All authors interpreted data. R.C. and V.N. drafted and revised the manuscript.
References
- 1.Waters EA, Cronin KA, Graubard BI, Han PK, Freedman AN. Prevalence of tamoxifen use for breast cancer chemoprevention among U.S. women. Cancer Epidemiol Biomarkers Prev. 2010;19:443–446. doi: 10.1158/1055-9965.EPI-09-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. WHO Model List of Essential Medicines: 18th List. 2013 [Google Scholar]
- 3.Komm BS, Mirkin S. An overview of current and emerging SERMs. J Steroid Biochem Mol Biol. 2014;143:207–222. doi: 10.1016/j.jsbmb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Paul R, et al. Both the immunosuppressant SR31747 and the antiestrogen tamoxifen bind to an emopamil-insensitive site of mammalian Delta8-Delta7 sterol isomerase. J Pharmacol Exp Ther. 1998;285:1296–1302. [PubMed] [Google Scholar]
- 5.Mandlekar S, Kong AN. Mechanisms of tamoxifen-induced apoptosis. Apoptosis. 2001;6:469–477. doi: 10.1023/a:1012437607881. [DOI] [PubMed] [Google Scholar]
- 6.Ligeiro de Oliveira AP, Oliveira-Filho RM, da Silva ZL, Borelli P, Tavares de Lima W. Regulation of allergic lung inflammation in rats: interaction between estradiol and corticosterone. Neuroimmunomodulation. 2004;11:20–27. doi: 10.1159/000072965. [DOI] [PubMed] [Google Scholar]
- 7.Moreland JG, Davis AP, Bailey G, Nauseef WM, Lamb FS. Anion channels, including ClC-3, are required for normal neutrophil oxidative function, phagocytosis, and transendothelial migration. J Biol Chem. 2006;281:12277–12288. doi: 10.1074/jbc.M511030200. [DOI] [PubMed] [Google Scholar]
- 8.Baumruker T, Bornancin F, Billich A. The role of sphingosine and ceramide kinases in inflammatory responses. Immunol Lett. 2005;96:175–185. doi: 10.1016/j.imlet.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Brinkmann V, Zychlinsky A. Beneficial suicide: why neutrophils die to make NETs. Nat Rev Microbiol. 2007;5:577–582. doi: 10.1038/nrmicro1710. [DOI] [PubMed] [Google Scholar]
- 10.von Kockritz-Blickwede M, Nizet V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med. 2009;87:775–783. doi: 10.1007/s00109-009-0481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Köckritz-Blickwede M, Chow O, Ghochani M. Nizet V. 7 - Visualization and Functional Evaluation of Phagocyte Extracellular Traps. In: Dieter K, Stefan HEK, editors. Methods in Microbiology. Academic Press; 2010. [Google Scholar]
- 12.Sun J, Huang YR, Harrington WR, Sheng S, Katzenellenbogen JA, Katzenellenbogen BS. Antagonists selective for estrogen receptor alpha. Endocrinology. 2002;143:941–947. doi: 10.1210/endo.143.3.8704. [DOI] [PubMed] [Google Scholar]
- 13.Compton DR, et al. Pyrazolo[1,5-a]pyrimidines: estrogen receptor ligands possessing estrogen receptor beta antagonist activity. J Med Chem. 2004;47:5872–5893. doi: 10.1021/jm049631k. [DOI] [PubMed] [Google Scholar]
- 14.Bologa CG, et al. Virtual and biomolecular screening converge on a selective agonist for GPR30. Nat Chem Biol. 2006;2:207–212. doi: 10.1038/nchembio775. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt JM, et al. De novo design, synthesis, and evaluation of novel nonsteroidal phenanthrene ligands for the estrogen receptor. J Med Chem. 2003;46:1408–1418. doi: 10.1021/jm020536q. [DOI] [PubMed] [Google Scholar]
- 16.Thomas P, Pang Y, Filardo EJ, Dong J. Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology. 2005;146:624–632. doi: 10.1210/en.2004-1064. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs TA, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow OA, et al. Statins enhance formation of phagocyte extracellular traps. Cell Host Microbe. 2010;8:445–454. doi: 10.1016/j.chom.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gschwendt M, Dieterich S, Rennecke J, Kittstein W, Mueller HJ, Johannes FJ. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett. 1996;392:77–80. doi: 10.1016/0014-5793(96)00785-5. [DOI] [PubMed] [Google Scholar]
- 20.Laudanna C, Mochly-Rosen D, Liron T, Constantin G, Butcher EC. Evidence of zeta protein kinase C involvement in polymorphonuclear neutrophil integrin-dependent adhesion and chemotaxis. J Biol Chem. 1998;273:30306–30315. doi: 10.1074/jbc.273.46.30306. [DOI] [PubMed] [Google Scholar]
- 21.Lozano J, et al. Protein kinase C zeta isoform is critical for kappa B-dependent promoter activation by sphingomyelinase. J Biol Chem. 1994;269:19200–19202. [PubMed] [Google Scholar]
- 22.Rani CS, et al. Cell cycle arrest induced by an inhibitor of glucosylceramide synthase. Correlation with cyclin-dependent kinases. J Biol Chem. 1995;270:2859–2867. doi: 10.1074/jbc.270.6.2859. [DOI] [PubMed] [Google Scholar]
- 23.Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell Biochem. 2008;49:413–440. doi: 10.1007/978-1-4020-8831-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wadsworth JM, et al. The Chemical Basis of Serine Palmitoyltransferase Inhibition by Myriocin. J Am Chem Soc. 2013;135:14276–14285. doi: 10.1021/ja4059876. [DOI] [PubMed] [Google Scholar]
- 25.Scarlatti F, et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004;279:18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 26.Menegazzi R, Decleva E, Dri P. Killing by neutrophil extracellular traps: fact or folklore? Blood. 2012;119:1214–1216. doi: 10.1182/blood-2011-07-364604. [DOI] [PubMed] [Google Scholar]
- 27.Malachowa N, Kobayashi SD, Freedman B, Dorward DW, DeLeo FR. Staphylococcus aureus leukotoxin GH promotes formation of neutrophil extracellular traps. J Immunol. 2013;191:6022–6029. doi: 10.4049/jimmunol.1301821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi SD, et al. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J Innate Immun. 2010;2:560–575. doi: 10.1159/000317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seumois G, et al. De novo C16- and C24-ceramide generation contributes to spontaneous neutrophil apoptosis. J Leukoc Biol. 2007;81:1477–1486. doi: 10.1189/jlb.0806529. [DOI] [PubMed] [Google Scholar]
- 30.Scheel-Toellner D, et al. Clustering of death receptors in lipid rafts initiates neutrophil spontaneous apoptosis. Biochem Soc Trans. 2004;32:679–681. doi: 10.1042/BST0320679. [DOI] [PubMed] [Google Scholar]
- 31.Wartha F, Henriques-Normark B. ETosis: a novel cell death pathway. Sci Signal. 2008;1:pe25. doi: 10.1126/stke.121pe25. [DOI] [PubMed] [Google Scholar]
- 32.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 33.Remijsen Q, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011;21:290–304. doi: 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uchiyama S, Andreoni F, Schuepbach RA, Nizet V, Zinkernagel AS. DNase Sda1 allows invasive M1T1 group A Streptococcus to prevent TLR9-dependent recognition. PLoS Pathog. 2012;8:e1002736. doi: 10.1371/journal.ppat.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schilcher K, Andreoni F, Uchiyama S, Ogawa T, Schuepbach RA, Zinkernagel AS. Increased neutrophil extracellular trap-mediated Staphylococcus aureus clearance through inhibition of nuclease activity by clindamycin and immunoglobulin. J Infect Dis. 2014;10:473–82. doi: 10.1093/infdis/jiu091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko Y-P, et al. Phagocytosis Escape by a Staphylococcus aureus protein that connects complement and coagulation proteins at the bacterial surface. PLoS Pathog. 2013;9:e1003816. doi: 10.1371/journal.ppat.1003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laarman AJ, Ruyken M, Malone CL, van Strijp JA, Horswill AR, Rooijakkers SH. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol. 2011;186:6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 38.Liu GY, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessenbrock K, et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med. 2009;15:623–625. doi: 10.1038/nm.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dworski R, Simon HU, Hoskins A, Yousefi S. Eosinophil and neutrophil extracellular DNA traps in human allergic asthmatic airways. J Allergy Clinical Immuno. 2011;127:1260–1266. doi: 10.1016/j.jaci.2010.12.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R, Shen Z, Nauseef WM, Hazen SL. Defects in leukocyte-mediated initiation of lipid peroxidation in plasma as studied in myeloperoxidase-deficient subjects: systematic identification of multiple endogenous diffusible substrates for myeloperoxidase in plasma. Blood. 2002;99:1802–1810. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.