Abstract
Two distinct subsets of CD4+Foxp3+ regulatory T (Treg) cells have been described based on the differential expression of Helios, a transcription factor of the Ikaros family. Efforts to understand the origin and biological roles of these Treg populations in regulating immune responses have, however, been hindered by the lack of reliable surface markers to distinguish and isolate them for subsequent functional studies. Using a single-cell cloning strategy coupled with microarray analysis of different Treg functional subsets in humans, we identify the mRNA and protein expression of TIGIT and FCRL3 as a novel surface marker combination that distinguishes Helios+FOXP3+ from Helios−FOXP3+ memory cells. Unlike conventional markers that are modulated on conventional T cells upon activation, we show that the TIGIT/FCRL3 combination allows reliable identification of Helios+ Treg cells even in highly activated conditions in vitro as well as in PBMCs of autoimmune patients. We also demonstrate that the Helios−FOXP3+ Treg subpopulation harbors a larger proportion of nonsuppressive clones compared with the Helios+ FOXP3+ cell subset, which is highly enriched for suppressive clones. Moreover, we find that Helios− cells are exclusively responsible for the productions of the inflammatory cytokines IFN-γ, IL-2, and IL-17 in FOXP3+ cells ex vivo, highlighting important functional differences between Helios+ and Helios− Treg cells. Thus, we identify novel surface markers for the consistent identification and isolation of Helios+ and Helios− memory Treg cells in health and disease, and we further reveal functional differences between these two populations. These new markers should facilitate further elucidation of the functional roles of Helios-based Treg heterogeneity.
Forkhead box protein 3+ regulatory T (Treg) cells are critical mediators of immunological self-tolerance. Their absence results in severe multiorgan autoimmunity in humans and mice (1, 2). Although the significant contribution of Treg cells in the pathogenesis of autoimmunity has been established based on several animal models (3), investigations on exact pathogenic roles of Treg dysfunction in human autoimmune disorders have resulted in inconclusive findings, mainly due to the lack of specific markers that allow the reliable identification and isolation of a pure Treg population across donors. Most human studies rely on the high expression of CD25 and the low CD127 expression to identify Treg cells (4). However, the expression levels of these two markers are modulated on conventional CD4+ T (Tconv) cells upon activation, making them indistinguishable from Treg cells during immune activation, thereby complicating the interpretation of findings based on these markers. Whereas the expression of FOXP3 can reliably identify human Treg cells in the resting state, its intracellular localization precludes its use for sorting of live cells. Moreover, TCR-mediated activation leads to a substantial upregulation of FOXP3 in a fraction of Tconv cells, thus confounding any ex vivo Treg phenotypic or functional analysis (5, 6). To circumvent these issues and to characterize bona fide Treg cells, we previously used a single-cell cloning approach to dissect the functional heterogeneity within the FOXP3+ population of healthy individuals (7, 8). We observed that the FOXP3+ T cell population, although composed mostly of highly suppressive Treg clones, contains a sizeable subpopulation (~25–30%) of nonsuppressive FOXP3+ clones that are indistinguishable from their functional counterparts in terms of the conventional Treg markers (8).
In the present study, we used the same single-cell cloning strategy to identify suppressive and nonsuppressive FOXP3+ Treg functional subsets in humans. We further performed microarray analysis to identify gene products that potentially discriminate these subsets. By comparing the gene expression profiles of these FOXP3+ Treg subsets, we found suppressive clones to have an increased transcription level of the IKZF2 gene, which encodes the Ikaros family transcription factor, Helios. Helios has been recently proposed as a marker to distinguish thymus-derived Treg cells from peripherally induced ones in mice (9). However, in humans, naive FOXP3+ cells isolated from healthy blood contain a Helios− population, suggesting that not all Helios−FOXP3+ cells are generated in the periphery (10–12). Investigation of the functional relevance of Helios expression in human Treg biology is desired. However, such studies have been hindered by the paucity of surface markers to distinguish them.
Comparing suppressive and nonsuppressive clones, we also found an increased expression of the genes encoding two surface proteins: T cell immunoreceptor with Ig and ITIM domains (TIGIT) and FcR-like 3 (FCRL3). TIGIT is an immunoregulatory molecule expressed on memory and activated T cells (13). Functionally, TIGIT has been reported to render dendritic cells (DCs) tolerogenic through interaction with its ligand (CD155) on DCs and induction of IL-10 production (13). TIGIT has also been shown to act as an intrinsic inhibitor of T cell proliferation, similar to the effect of CTLA-4 signaling (14). Recently, Harrison and colleagues (15) showed that TIGIT is transcriptionally targeted by FOXP3, and a role for TIGIT signaling in enhancing Treg-mediated suppression has recently been suggested (16).
FCRL3 (CD307c) is a member of the FCRL family of classical FcR homologs that is expressed on human B cells, some memory T lymphocytes, as well as NK cells (17, 18). Although the ligand(s) and physiological function of FCRL3 on T cells are yet to be unraveled, the presence of both ITIM and ITAM motifs in its intracellular domain suggests a role for FCRL3 in the maintenance of homeostatasis via regulation of immune responses (19, 20). Indeed, some studies have shown a potential role of FCRL3 in regulating B cell differentiation and proliferation initiated by BCR signaling (20, 21). Interestingly, a functional variant in the FCRL3 promoter has been linked to susceptibility to rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and autoimmune thyroiditis (22), and it was associated with increased expression of FCRL3 on T cells of RA patients (23).
Examining the surface expression of TIGIT and FCRL3 on memory FOXP3+ Treg cells, we found a positive correlation of both markers with Helios expression. We demonstrated that the simultaneous use of TIGIT and FCRL3 surface expression allows the consistent discrimination and isolation of live Helios+ and Helios− memory FOXP3+ populations, each endowed with different functionalities. Notably, we demonstrate in the present study that TIGIT/FCRL3 is a stable marker combination under inflammatory conditions in vitro, in PBMCs of autoimmune patients, as well as in the resting state, thus providing a novel approach for further characterization of Treg heterogeneity in health and disease.
Materials and Methods
Donors and cell isolation
PBMCs were isolated from buffy coats of healthy donors (n = 11; Sanguine BioSciences) using Ficoll-Paque Plus density gradient (GE Healthcare) and cryopreserved. PBMC samples from untreated patients with relapsing-remitting multiple sclerosis (n = 6), ulcerative colitis (n = 6), Crohn’s disease (n = 6), and their age- and sex-matched healthy controls were obtained from the Montreal Neurological Institute as part of the Canadian Institutes for Health Research NET grant in clinical autoimmunity. PBMC samples from patients with active systemic lupus erythematosus (SLE; SLE Disease Activity Index 2000 update score of >6) were obtained from the Montreal General Hospital SLE clinic. All samples were collected in accordance with McGill University’s Research Ethics Board.
Flow cytometry
For ex vivo flow cytometry analysis, PBMCs were thawed and stained with viability dye (eFluor 780; eBioscience). Purified anti-FCRL3 (24) was used where mentioned, followed by staining with F(ab′)2 anti-mouse IgG (eFluor 660 or PE; eBioscience) and extensive washing with PBS. Cells were then stained with fluorochrome-conjugated Abs against CD4 (FITC or V500), CD25 (PE-CF594), CD45RA (Alexa Fluor 700) (BD Biosciences), CD127 (PE), TIGIT (PerCP–eFluor 710), FOXP3 (PE-Cy7), CD62L (FITC), HLA-DR (PE-Cy7), CTLA-4 (allophycocyanin) (eBioscience), and Helios (Pacific Blue; BioLegend).
For cytokine detection, PBMCs were incubated with PMA (25 ng/ml), ionomycin (1 μg/ml) (both from Sigma-Aldrich), and GolgiStop (eBioscience) for 4 h, followed by intracellular cytokine staining with Abs against IFN-γ (FITC), IL-2 (PerCP-Cy5.5), and IL-17 (allophycocyanin) (eBioscience).
Flow cytometry analysis was performed on an LSRFortessa analyzer, and sorting throughout this study was performed on a FACSAria IIu cell sorter (both from BD Biosciences).
Generation of primary CD4+ clones
Primary CD4+ Treg and Tconv clones were generated from healthy PBMCs by single-cell expansion of FACS-sorted CD25high (top 1%) and CD25− cells as described previously (7, 8). The clones were expanded in X-VIVO 15 medium (Lonza) supplemented with 5% FBS (Sigma-Aldrich) and were stimulated with soluble anti-CD3 (30 ng/ml; eBioscience), recombinant human IL-2 (200 U/ml; gift of the Surgery Branch, National Cancer Institute, National Institutes of Health), and irradiated human PBMCs as feeders. Fresh medium and IL-2 were replenished on day 5 and every 2 d thereafter, and clones were passaged as needed. Restimulation was performed on days 11–12 and the clones were further expanded until harvested on days 22–24. Phenotypic and functional assays were performed on the clones in parallel. Clones were generated from seven different healthy subjects (34–66 FOXP3+ clones/donor).
Suppression assays
CFSE-based suppression assays were used throughout this study to measure the suppressive potency of Treg cells as we previously described (8). Briefly, responding allogeneic CD4+CD25− (responder T [Tresp]) cells were sorted by FACS, labeled with CFSE proliferation dye (5 μM; Sigma-Aldrich), and plated at 8000 cells/well in U-bottom 96-well plates (Sarstedt) with irradiated PBMCs as feeders (30,000 cells/well). Treg cells were added at different Treg/Tresp ratios and the assays were stimulated with soluble anti-CD3 (30 ng/ml; eBioscience) for 4 d. Suppression was calculated based on the division index (as calculated by the FlowJo software) of unsuppressed Tresp cells cultured in the absence of Treg cells.
Gene expression analysis
Total mRNA was prepared from individual clones representing three populations: 1) suppressive and 2) nonsuppressive FOXP3+ clones generated from CD4+CD25high cells, and 3) control FOXP3− clones generated from CD4+CD25− cells. The clones were derived from two healthy donors, and each group is represented by two to three pooled clones per donor. Clone selection was based on a thorough analysis of phenotype and function as we described before (8). Cells were pelleted and resuspended in RLT Plus lysis buffer (Qiagen) and immediately frozen at −80°C. mRNA reverse transcription, labeling, and hybridization onto an Illumina HumanHT-12_V3 Expression BeadChip were performed by the Génome Québec Innovation Centre (Montreal, QC, Canada). The microarray data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through Gene Expression Omnibus Series accession no. GSE65650 at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65650.
Total gene expression data were normalized using the robust-spline method, without background correction and with base 2 logarithmic transformation, using the R software. Samples were compared using an F test (ANOVA) with the random variance model–based correction for multiple-hypothesis testing (25) using the R software. To minimize interference from gene expression of the irradiated feeders used to expand the clones, we established a feeder contamination standard curve by expanding the same clones in the presence of titrated numbers of feeder cells and processing the resulting samples on the same chip. For each gene expression value, the corresponding contamination contribution was predicted by regression, and a separate, corrected gene expression value was calculated. A gene was considered differentially expressed in two subsets when its corrected fold change value was ≥1.5 and the ANOVA p value was ≤0.1.
T cell activation assays
To investigate the activation-induced modulation of marker expression, CD4+CD25−TIGIT−FCRL3− cells from healthy PBMCs were FACS sorted, labeled with CFSE, and activated for 4–6 d in vitro with anti-CD3/anti-CD28–coated beads (Miltenyi Biotec) at a ratio of two beads/one cell. Where indicated, irradiated autologous PBMCs were used as feeders. Protein expression levels of FOXP3, Helios, TIGIT, and FCRL3 were measured by flow cytometry over time. In different experiments, whole PBMCs were activated with anti-CD3/anti-CD28–coated beads for 3–5 d followed by flow cytometric analysis of marker expression.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism 5.0 software. One-way ANOVA, followed by Turkey’s range test, and the Student t test were used where indicated. A p value <0.05 was considered significant.
Results
Lack of suppressive function in human FOXP3+ clones is associated with reduced expression of Helios, TIGIT, and FCRL3
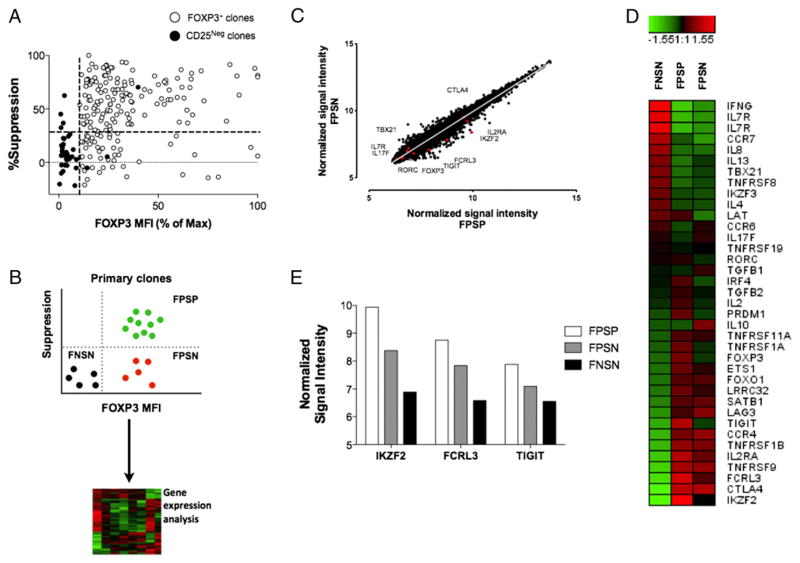
We have recently shown that in ~30% of the FOXP3+ clones derived from healthy individuals, FOXP3 expression does not lead to suppressive function despite the maintenance of a typical Treg phenotype (8) (Fig. 1A). To further understand this functional heterogeneity and to identify markers of the different functional subsets of Treg cells, we compared gene expression profiles of FOXP3+ suppression-positive (FPSP) and FOXP3+ suppression-negative (FPSN) clones, relative to FOXP3− suppression-negative (control) clones (FNSN) (Fig. 1B) (GSE65650, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65650). As expected, we observed that many common Treg cell phenotypic markers are expressed at comparable levels in both FOXP3+ functional subsets (Fig. 1C, 1D). To further compare FPSP and FPSN clones, we analyzed the Treg-specific demethylated region (TSDR) of the FOXP3 locus, which is normally demethylated in Treg cells but methylated in Tconv cells (26, 27). Owing to the female origin of the analyzed clones in this study, a Treg clone is expected to be ~50% methylated in its TSDR region due to X chromosome inactivation (28). Indeed, unlike FNSN clones, which have 77% methylated TSDR, both FPSP and FPSN clones show only ~54% methylation in their TSDR regions (Supplemental Fig. 1), suggesting that both groups are derived from bona fide Treg cells.
FIGURE 1.
Gene expression analysis of suppressive versus nonsuppressive FOXP3+ clones. Illumina BeadChip analysis was used for total mRNA prepared from individual clones representing three populations: 1) suppressive and 2) nonsuppressive FOXP3+ clones generated from CD4+CD25high cells, and 3) control FOXP3− clones generated from CD4+CD25− cells. (A) Correlation of suppressive potency with FOXP3 expression levels in primary FOXP3+ and FOXP3− (CD25−) clones generated from three healthy donors. The dotted lines represent the cutoffs for FOXP3 mean fluorescence intensity (MFI) (x-axis) and percentage suppression (y-axis) determined through the overall FOXP3 MFI and suppressive capacity of the control FOXP3− clones generated from CD4+CD25− cells. (B) Schematic illustrating the process of selection of representative clones. (C) Scatter plot showing normalized gene expression level in suppressive versus nonsuppressive clones in the resting state. (D) Heat map comparing variations in the expression of selected Treg- and Tconv-associated genes relative to the median mRNA levels across the three subsets. (E) Relative mRNA expression levels of TIGIT, FCRL3, and Helios in the three functional categories. Representative clones were derived from two healthy donors, and two to three similar clones/subset/donor were pooled to obtain sufficient mRNA.
However, several genes were found to be differentially expressed between the two FOXP3+ subsets (Fig. 1D, 1E). One such gene, IKZF2, codes for the transcription factor Helios and was found to be significantly more transcribed in FPSP relative to FPSN clones (Fig. 1E). Moreover, in search for potential extracellular markers of Treg functional subsets, we additionally found the genes coding for the surface receptors TIGIT and FCRL3 to be more highly expressed in FPSP clones relative to FPSN. The expression of all three genes was lowest in FNSN clones (Fig. 1E).
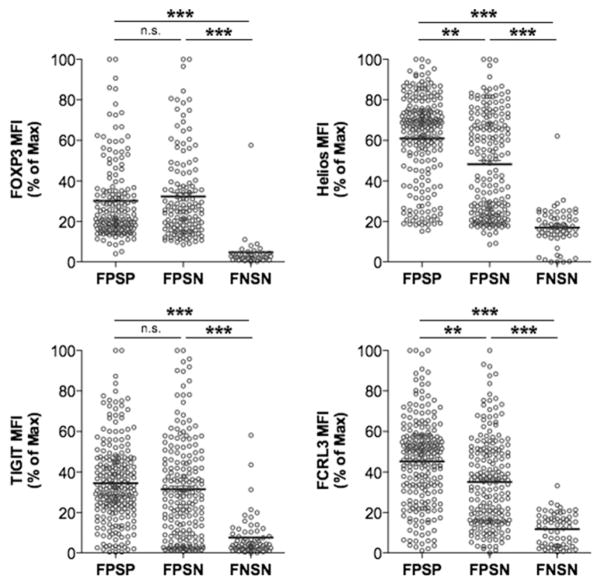
To validate the differences observed in gene transcription, we generated primary clones from healthy individuals and examined the expression of FOXP3, Helios, TIGIT, and FCRL3 proteins in FPSP, FPSN, and FNSN clones. Whereas FOXP3 and TIGIT were expressed equally in FPSP and FPSN clones, both Helios and FCRL3 were significantly lower in FPSN relative to FPSP clones and lowest in FNSN clones (Fig. 2), confirming our observations at the transcription level with regard to Helios and FCRL3, and suggesting a potential role for these markers in the suppressive function of Treg cells.
FIGURE 2.
FPSN clones comprise majorly Helios− clones and display a reduced FCRL3 expression. Primary Treg and Tconv clones were generated by single cell cloning of FACS-sorted CD25high and CD25− cells of four healthy individuals. Marker expression analysis was performed immediately after the harvest on days 22–24, and 4-d CFSE-based suppression assays were carried out using allogeneic CD4+CD25− cells as responders at a 1:1 Treg/Tresp ratio in the presence of irradiated PBMCs and anti-CD3 (30 ng/ml). Suppression was measured relative to the division index of the unsuppressed Tresp-alone control. Shown are the expression levels of FOXP3, Helios, TIGIT, and FCRL3 in FPSP, FPSN, and FNSN clones immediately after harvest. Statistical analysis was done with the one-way ANOVA followed by a Tukey posttest. **p ≤ 0.01, ***p ≤ 0.001. n.s., not significant.
Differential TIGIT and FCRL3 expression delineates Helios+ and Helios− memory Treg cells ex vivo
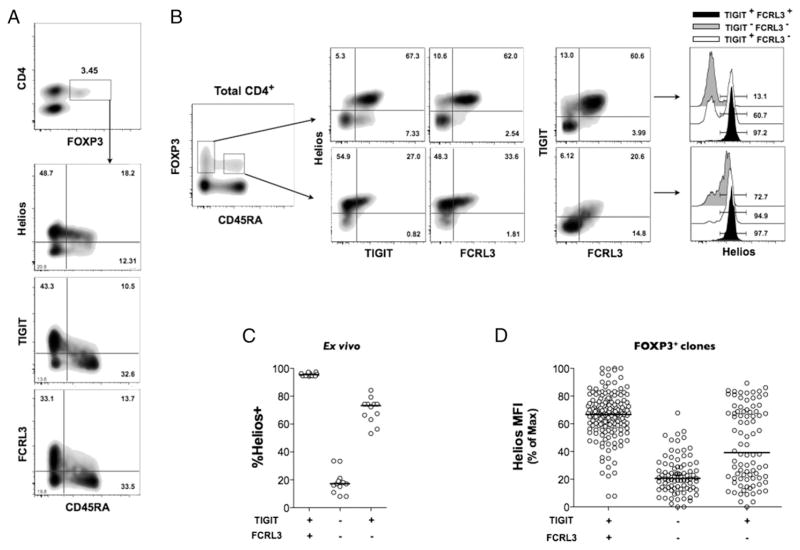
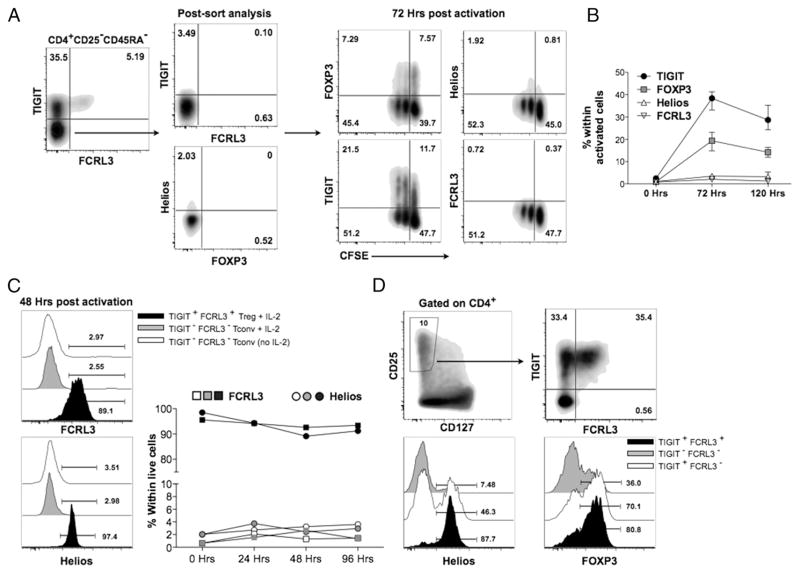
We then examined the protein expression of TIGIT and FCRL3 as well as Helios in CD4+FOXP3+ cells of PBMCs of healthy individuals by flow cytometry. In line with previous studies, Helios protein is expressed in ~70% of CD4+FOXP3+ cells (Fig. 3A) (9, 11, 12). In the same experiments, we measured significant levels of TIGIT and FCRL3 protein expression on ~50% of CD4+FOXP3+ cells, with the vast majority of TIGIT- and FCRL3-expresing cells exhibiting a memory profile (CD45RA−) (Fig. 3A, 3B). Subsequent analyses are therefore restricted to the CD4+CD45RA− population. Interestingly, both TIGIT and FCRL3 are preferentially expressed on the Helios+ population of memory FOXP3+ cells whereby TIGIT+ and FCRL3+ cells comprise ~80 and 90% Helios+ cells, respectively (Fig. 3B). Notably, the combined expression of TIGIT and FCRL3 identifies a memory FOXP3+ population that is even more greatly enriched for Helios+ cells (median, 95.1%; interquartile range [IQR], 1.69%) (Fig. 3B, 3C). Conversely, the lack of both TIGIT and FCRL3 expression on memory FOXP3+ cells denotes a population that is predominantly negative for Helios (median, 17.1%; IQR, 5.48%). The ability to discriminate between Helios subsets was precisely reproducible in primary FOXP3+ clones derived from healthy individuals (Fig. 3D), demonstrating the reliability of TIGIT and FCRL3 as a novel surface marker combination to stratify Treg populations according to Helios expression in vitro and ex vivo.
FIGURE 3.
The combined expression of TIGIT and FCRL3 discriminates between Helios+ and Helios− subsets in memory FOXP3+ cells. (A–C) PBMCs from healthy subjects were analyzed ex vivo by flow cytometry. (A) Expression of TIGIT, FCRL3, and Helios on naive and memory CD4+FOXP3+ populations. (B) Representative plots showing the correlation of TIGIT and FCRL3 expression with Helios expression in naive versus memory CD4+ FOXP3+ cells. (C) Combined analysis of 11 healthy individuals showing the application of TIGIT and FCRL3 in the identification Helios+ and Helios− subsets within memory FOXP3+ cells. (D) Applicability of TIGIT/FCRL3 combination in discriminating Helios subsets in FOXP3+ clones generated from seven healthy donors (n = 299 clones).
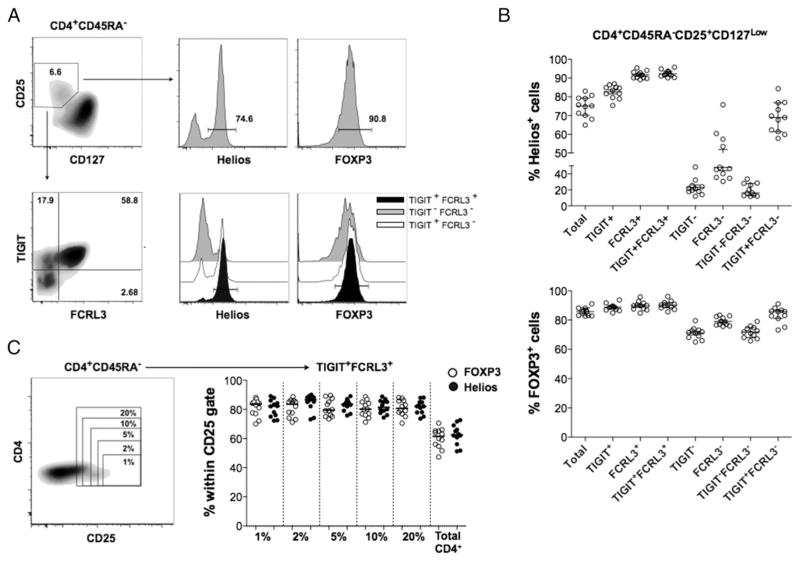
To sort human live Treg cells, most previous studies relied on the high cell surface expression of CD25 (1–2% of CD4+ cells) and the concomitant low expression of CD127. Using this gating strategy, we were able to obtain a population that is enriched for FOXP3+ cells (median, 85.8%; IQR, 4.35%) from memory CD4+ cells of healthy PBMC. As expected, the defined CD4+CD25+CD127low cells contained both Helios+ (median, 75.2%; IQR, 8.55%) and Helios− cells with a similar ratio as that in CD4+FOXP3+ cells (Fig. 4A, 4B). By further defining TIGIT+FCRL3+ cells, we consistently obtained a highly enriched Helios+ population (median, 92.2%; IQR, 2.25%) from CD4+CD25+CD127low cells. In contrast, a majorly Helios− population (median, 16.2%; IQR, 13.7%) was identified in the TIGIT−FCRL3− subpopulation (Fig. 4A, 4B). These results demonstrate that the combined use of TIGIT and FCRL3 achieves the nearly exclusive isolation of live Helios+ and Helios− Treg subsets without sacrificing cell viability. Furthermore, we assessed the enrichment in FOXP3- and Helios-expressing cells that can be attained by gating on CD4+CD45RA−CD25+ TIGIT+FCRL3+ cells with varying stringencies of the CD25 gate. Interestingly, the same high level of enrichment in FOXP3+Helios+ cells can be achieved with a CD25 gate encompassing as little as 1% and as much as 20% of memory CD4+ cells (Fig. 4C), indicating that TIGIT and FCRL3 can be used to sort memory Helios+ Treg cells with enhanced consistency and recovery when compared with the conventional gating strategies that rely on subjective CD25 gating.
FIGURE 4.
TIGIT/FCRL3 combination provides a reliable surface marker for the isolation of Helios+ and Helios− memory Treg cells. PBMCs from 11 healthy subjects were analyzed ex vivo by flow cytometry. (A) Representative FACS plots showing the application of the TIGIT/FCRL3 marker combination in distinguishing Helios subsets within memory CD4+CD25+CD127low cells. (B) Identification of Helios subsets by TIGIT/FCRL3 surface markers is precisely reproducible in healthy samples with a wide range of Helios expression. Shown is the frequency of Helios+ (top) and FOXP3+ (bottom) cells in Treg populations gated using the conventional markers (CD25+CD127low; referred to as Total) compared with further gating using different combinations of TIGIT and FCRL3. (C) The TIGIT/FCRL3 combination allows the identification of consistently enriched FOXP3+Helios+ populations with less stringent gating on CD25. Shown is the frequency of FOXP3+ and Helios+ cells in TIGIT+FCRL3+ cells obtained from variably stringent CD25+ gates on memory CD4+ cells.
Finally, we compared the expression of a panel of Treg-associated surface molecules on TIGIT+FCRL3+ and TIGIT−FCRL3− Treg cells of healthy individuals. Although minor differences in the expression of markers such as CD62L, HLA-DR, and CTLA-4 can be seen (Supplemental Fig. 2), none of the examined molecules has the ability to define TIGIT+FCRL3+ or TIGIT−FCRL3− Treg cells.
TIGIT and FCRL3 reliably identify Helios+ Treg cells in activated conditions in vitro
A major drawback of the currently used Treg markers is their modulation on Tconv cells following cell activation, which precludes their reliable use to distinguish Treg and Tconv cells in inflammatory conditions. Thus, we next assessed whether TIGIT and FCRL3 expression could be induced on Tconv following T cell activation. We isolated CD4+CD45RA−CD25− (Tconv) cells and stimulated them with anti-CD3/anti-CD28–coated beads in vitro. By 72 h postactivation, both TIGIT and FOXP3 were upregulated on 40 and 20% of Tconv cells, respectively. However, FCRL3 and Helios were not upregulated at all on activated Tconv cells even after 5 d in culture (Fig. 5A, 5B). To address the possibility that APCs may be required for the upregulation of Helios and FCLR3 on Tconv cells, we FACS-sorted CD4+CD25−TIGIT−FCLR3− cells and activated them in vitro with anti-CD3/anti-CD28–coated beads in the presence of irradiated autologous feeders with or without IL-2. Neither Helios nor FCLR3 was upregulated over the 4-d culture period (Fig. 5C). Additionally, we examined the use of TIGIT and FCRL3 on whole PBMC cultures stimulated with anti-CD3/anti-CD28–coated beads. At the peak of activation on day 3, CD4+CD25+CD127lowTIGIT+FCRL3+ cells were still majorly FOXP3+ (>80%) and highly enriched in Helios+ cells (>83%) (Fig. 5C). Moreover, we did not observe a significant change in the frequency of FCRL3+ or Helios+ cells in the CD4+ population upon activation of healthy PBMCs (Supplemental Fig. 3), further indicating that both Helios and FCRL3 are not upregulated in Tconv cells upon activation. Finally, we examined the applicability of the TIGIT/FCRL3 combination for flow cytometric analysis of PBMCs derived from patients with various active autoimmune disorders, including untreated relapsing-remitting multiple sclerosis, ulcerative colitis, Crohn’s disease, and active SLE. Similar to our observations in healthy subjects, the memory CD4+CD25+CD127low cell population could be segregated into Helios+ and Helios− subpopulations by further gating using TIGIT and FCRL3 (Supplemental Fig. 4A). Collectively, these data indicate the stability of this marker combination in inflammatory conditions, and they provide a reliable approach for the consistent isolation of Treg cells.
FIGURE 5.
The TIGIT/FCRL3 marker expression can reliably identify Helios+ and Helios− Treg cell subsets in inflammatory contexts. (A) FACS-sorted TIGIT−FCRL3− memory CD4+CD25− cells from PBMCs of a healthy donor were labeled with CFSE and activated in vitro with anti-CD3/anti-CD28–coated beads at a ratio of two beads/one cell for 5 d. Shown are representative FACS plots and (B) the kinetics of activation-induced marker upregulation in three separate experiments on three healthy individuals. (C) Total CD4+CD25−TIGIT−FCRL3− cells from PBMCs activated in vitro with anti-CD3/anti-CD28–coated beads at a ratio of two beads/one cell with or without recombinant human IL-2 in the presence of irradiated autologous feeders for 4 d. Shown is the expression of FCRL3 and Helios on the activated CD4+CD25−TIGIT−FCRL3− cells compared with FACS-sorted CD4+CD25+CD127lowTIGIT+FCRL3+ cells plated in parallel. (D) Whole PBMCs from a healthy individual were stimulated with anti-CD3/anti-CD28–coated beads at a ratio of two beads/one cell for 72 h. Representative FACS plots from similar experiments on three healthy donors are shown.
Nonsuppressive Treg cells are enriched within the Helios− subset
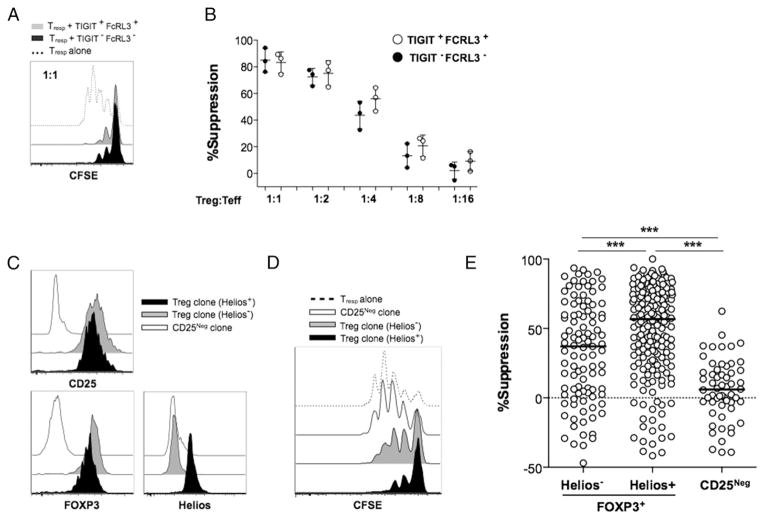
The assessment of the role of Helios expression in the suppressive function of Treg cells has been hindered by the inability to discriminate Helios+ and Helios− Treg cells ex vivo. We exploited the use of our novel combination of surface markers to isolate Helios+ and Helios− cells directly ex vivo for functional assessment. To this end, we cocultured sorted CD4+CD45RA−CD25+ CD127lowTIGIT+FCRL3+ (Helios+) or CD4+CD45RA−CD25+ CD127lowTIGIT−FCRL3− (Helios−) cells from healthy PBMCs with CFSE-labeled CD4+CD25− responding T cells in the presence of soluble anti-CD3 and irradiated feeder PBMCs. Suppression of proliferation of responding T cells was then measured in a 4-d assay. We show that both Helios+ and Helios− FOXP3+ cell subsets efficiently suppressed the proliferation of responder T cells, and no significant difference was observed between these subsets (Fig. 6A, 6B). We then assessed whether FOXP3+ Treg cells that differentially express Helios displayed equivalent suppressive activities with prolonged T cell activation and expansion. To this end, we generated individual FOXP3+ clones from healthy PBMCs and assessed suppressive function relative to Helios expression (Fig. 6). Our results show that the Helios− population contains a significantly higher proportion of nonsuppressive clones compared with its Helios+ counterpart, although both populations harbored highly suppressive clones (Fig. 6D, 6E).
FIGURE 6.
An increased frequency of nonsuppressive clones is found within the FOXP3+Helios− subset. The suppressive potency of Helios+ and Helios− Treg cells was compared ex vivo (A and B) and in primary clones (C–E). For ex vivo suppression assays, CD4+CD25+CD127lowTIGIT+FCRL3+ and CD4+CD25+CD127lowTIGIT−FCRL3− cells were FACS sorted and tested for the capacity to suppress the proliferation of CFSE-labeled CD4+CD25− Tresp cells stimulated with soluble anti-CD3 and irradiated PBMCs for 4 d. (A) Representative CFSE dilution histograms showing suppression of Tresp cells. (B) Combined suppression analysis from three different experiments using cells from three different healthy individuals. (C–E) Primary Treg and Tconv clones were generated by single-cell cloning of FACS-sorted CD25high and CD25− cells of seven healthy individuals. Marker expression was performed immediately after the harvest on days 22–24, and 4-d CFSE-based suppression assays were carried out using allogeneic CD4+CD25− cells as responders at a 1:1 Treg/Tresp ratio in the presence of irradiated PBMCs and anti-CD3 (30 ng/ml). (C) The expression of CD25, FOXP3, and Helios in representative clones at harvest. (D and E) Suppressive potency of FOXP3+Helios+ (n = 196 clones), FOXP3+Helios− (n = 103 clones), and FOXP3− (n = 59 clones). Suppression was measured relative to the division index of the unsuppressed Tresp-alone control. Statistical analysis was done with the one-way ANOVA followed by a Tukey posttest. ***p ≤ 0.001.
Overall, our results show that differential TIGIT and FCRL3 expression on human FOXP3+ Treg cells can efficiently be used to isolate Helios-expressing subsets. Moreover, our data also indicate a greater level of functional heterogeneity within the Helios− subset of FOXP3+ T cells, as this subset manifests compromised suppressive activity after prolonged cellular expansion.
Helios+ and Helios− CD4+FOXP3+ cells differ in cytokine production capacity
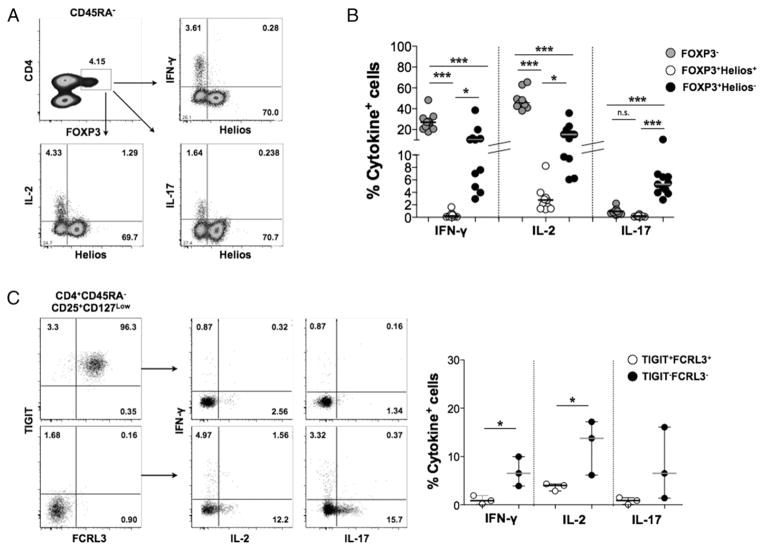
The stable expression of FOXP3 in Treg cells results in the transcriptional repression of a number of cytokines such as IFN-γ, IL-17, and IL-2 (29, 30). However, the production of these cytokines has been observed in a small population of memory FOXP3+ cells (31, 32). We analyzed cytokine production in healthy PBMCs ex vivo and found that, compared with FOXP3− cells, FOXP3+ cells are indeed poor producers of IFN-γ, IL-17, and IL-2 in response to stimulation with PMA and ionomycin (Fig. 7B). However, consistent with previous reports (10, 33), we detected a subpopulation of inflammatory cytokine-producing cells within the FOXP3+ population, and these cells were exclusively restricted to the Helios− subpopulation (Fig. 7A, 7B). Similarly, increased levels of inflammatory cytokines were measured in FACS-sorted TIGIT−FCRL3−, but not TIGIT+FCRL3+, memory Treg cells (CD4+CD45RA+CD25+ CD127low) (Fig. 7C). Moreover, analysis of IFN-γ, IL-17, and IL-2 production in CD4+CD45RA+CD25+CD127low Treg cells of patients of several autoimmune diseases further confirms the restriction of cytokine production to the TIGIT−FCRL3− fraction of Treg cells (Supplemental Fig. 4B). These data highlight important differences between Helios+ and Helios− cells and emphasize the need for further characterization of their functional relevance.
FIGURE 7.
Inflammatory cytokine production in FOXP3+ cells is restricted to the Helios− fraction. (A) PBMCs were incubated ex vivo with PMA (25 ng/ml), ionomycin (1 μg/ml), and GolgiStop for 4 h, followed by intracellular cytokine staining. Shown are representative flow cytometry plots (A) and the frequency of cytokine-producing cells in the indicated subsets (B) analyzed from 11 different healthy samples. (C) Cytokine production in healthy FACS-sorted TIGIT+FCRL3+ versus TIGIT−FCRL3− Treg cells (CD4+CD45RA−CD25+CD127low). Statistical analysis was done with the one-way ANOVA followed by a Tukey posttest (B) or with the Student t test (C). *p ≤ 0.05, ***p ≤ 0.001. n.s., not significant.
Discussion
The characterization of the relative function of Helios-defined Treg subsets has been hindered by the lack of reliable surface markers to delineate and isolate them. In this study, we identify the coexpression of TIGIT and FCRL3 as a novel marker combination that allows for a reliable stratification and isolation of human memory FOXP3+ Treg cells according to Helios expression. Importantly, we demonstrate the reliability of this marker combination under immune-activating conditions in vitro and in the periphery of autoimmune patients presenting with various active systemic and organ-specific autoimmune diseases. Moreover, our combined use of TIGIT and FCRL3 has enabled us to highlight significant functional differences between Helios+ and Helios− Treg cells, emphasizing the need for further investigation of their respective roles in regulating immune responses.
The modulation of the currently used Treg markers on Tconv upon activation makes it very challenging to identify bona fide Treg cells in inflammatory conditions. An important observation in this study is that FCRL3 is selectively expressed by FOXP3+ Treg cells and is not upregulated by Tconv cells upon TCR-mediated stimulation. This is in line with previous observations that reported the lack of FCRL3 upregulation in Treg cells induced from Tconv cells upon activation in the presence of TGF-β (24, 34). Thus, the TIGIT/FCRL3 combination provides an excellent surface marker for Helios+ Treg cells in inflammatory conditions. Such marker stability will render comparative functional analysis of Treg function in health and disease more informative. Additionally, our observations concerning the upregulation of TIGIT and FOXP3, but not FCRL3 or Helios, on activated Tconv cells indicate that differential expression of these markers can also be used to identify and exclude recently activated Tconv cells, which are expected to exhibit the phenotype TIGIT+FCRL3−FOXP3+Helios− in CD25+CD127lowcells.
The contribution of Helios expression to the suppressive function of Treg cells is poorly defined. Whereas Helios-deficient mice show normal Treg development (9, 35), small interfering RNA–mediated knockdown of Helios expression in human CD4+CD25+ cells resulted in reduced suppressive function (36). Using our novel marker combination, we find that Helios+ and Helios− cells exert equal levels of suppression when assessed directly ex vivo. This result is in accordance with recent observations by Raffin et al. (33) that both Helios+ cells and Helios− cells sorted based on the combined expression of CCR7 and IL-1RI are highly suppressive, although their study reported that Helios− cells are more suppressive than Helios+ cells. Note that the suppression assays in that study used monocytes as feeders and were stimulated with PHA, whereas the assays reported in the present study used irradiated PBMCs and were stimulated with soluble anti-CD3. Interestingly, our analysis of the suppressive capacity of FOXP3+ clones showed that, unlike Helios+ clones that are highly enriched in suppressive clones, Helios− clones constitute a significantly more heterogeneous mixture of suppressive and nonsuppressive clones. Such functional heterogeneity in the Helios− population may have been masked in our ex vivo suppression analysis due to the polyclonal nature of the suppressor population and the functional complementation by the highly suppressive cells within the Helios− population. Alternatively, the wider functional distribution of Helios− cells could also be attributed to the possibility that Helios− cells may be more prone to modulating their suppressive function than Helios+ cells. This could be a result of prolonged TCR-mediated expansion or, in the presence of certain inflammatory signals, a potential example of what was recently proposed by Raffin et al. (33) and involves IL-1β signaling, which causes Helios− Treg cells to downregulate their suppressive function. Such functional diversity within the Helios− population may suggest a higher plasticity potential in this Treg population in response to the changing microenvironment.
The biological significance of effector cytokine production by FOXP3+ Treg cells is unclear. Whereas some studies on mouse models suggest that upregulation of inflammatory cytokines such as IL-17 and IFN-γ occurs in unstable Treg cells, and precedes loss of FOXP3 and adoption of a Tconv phenotype (37, 38), other studies suggest that some FOXP3+ Treg cells acquire a transient ability to produce IFN-γ and IL-17 to allow efficient homing to Th1 and Th17 inflammation sites, respectively, without loss of suppressive function or downregulation of FOXP3 (8, 32, 39, 40). We show that the expression of Helios in FOXP3+ cells correlates with a strong repression of effector cytokine production. These findings are in agreement with, and extend, recently reported observations that inflammatory cytokine production in human FOXP3+ cells is restricted to the Helios− population (10, 12, 33). These results further highlight some potentially important functional differences between Helios+ and Helios− Treg cells, and they warrant more in-depth analysis of the two subsets.
The degree of demethylation in the TSDR region of the FOXP3 promoter is now used as a measure of stability of FOXP3 expression and suppressive function (26, 27). A number of studies have analyzed this epigenetic parameter in Helios subsets of human Treg cells. Supporting our result, Kim et al. (10) reported partial TSDR demethylation (55%) in total FOXP3+Helios− cells compared with complete demethylation in FOXP3+Helios+ cells, suggesting heterogeneity within the Helios− subset. Note, however, that Helios−FOXP3+ cells are not restricted to the memory fraction of Treg cells, but are also present in naive cells in peripheral blood. More recently, Himmel et al. (12) analyzed TSDR demethylation in naive (CD45RA+) FOXP3+ cells and observed comparable demethylation patterns in Helios+ and Helios− populations, suggesting that the methylation observed in Helios− cells in earlier studies is specific to the memory fraction. Indeed, McClymont et al. (41) reported predominant TSDR methylation in IFN-γ–producing FOXP3+ cells, which are majorly restricted to the memory Helios− fraction, suggesting that the Helios− population is heterogeneous and encompasses cells with enhanced functional plasticity to allow homeostatic adjustments to the needs of the immune response. Owing to the lack of reliable markers for Helios subsets of Treg cells, all of these epigenetic analyses were performed on fixed cells, and thus further delineation of the functional significance of Helios expression in human Treg cells was not previously possible.
The functional role of TIGIT in Treg biology has only started to be unraveled. A recent study by Joller et al. (16) found that TIGIT signaling enhanced Treg-mediated suppression of Th1 and Th17, but not of Th2, responses. Such selectivity is achieved through the production of fibrinogen-like protein 2, which prevents the suppression of Th2 responses. Other reported TIGIT-mediated regulatory functions include its active tolerogenization of DCs through binding to CD155 and induction of IL-10 secretion (13), as well as its intrinsic inhibition of T cell activation and proliferation (14).
Two independent studies have previously examined the association between the expression of FCRL3 on Treg cells and their suppressive function. Whereas Swainson et al. (34) reported that FCRL3 expression identified Treg populations with a reduced suppressive potency, Nagata et al. (24) observed a comparable suppressive capacity in the FCRL3+ and FCRL3− fractions of CD25+CD127low Treg cells. Further investigation is warranted to elucidate the potential molecular link between the expression of these markers and Helios expression, as well as to determine the precise function of FCRL3 and TIGIT in Treg cells.
In conclusion, we identified TIGIT and FCRL3 expression as a novel surface marker combination that allows the isolation of live Helios+ and Helios− memory Treg cells. Our data further revealed phenotypic and functional differences between human Helios+ and Helios− Treg cells. The novel surface markers presented in the present study will facilitate in-depth investigations into the functional heterogeneity of these Treg subsets and may provide important clues to allow directed manipulation of specific Treg populations.
Supplementary Material
Acknowledgments
This work was supported by Canadian Institutes for Health Research Grant MOP67211 (to C.A.P.), as well as by Canadian Institutes for Health Research Grant MOP84041 from the New Emerging Team in “Clinical Autoimmunity: Immune Regulation and Biomarker Development in Pediatric and Adult Onset Autoimmune Diseases” (to C.A.P. and A.B.-O.). C.A.P. holds a Canada Research Chair. S.N. was supported by National Institutes of Health Centers of Biomedical Research Excellence Grant P20 GM103548. K.B.D. is the recipient of research fellowships from the Canadian Institutes for Health Research neuroinflammation training program and the Research Institute of the McGill University Health Centre.
We thank Marie-Helene Lacombe (McGill University Health Center Immunophenotyping Platform) for cell sorting. We also thank Helen Mason and Mara Kornete for valuable technical assistance.
Abbreviations used in this article
- DC
dendritic cell
- FCRL3
FcR-like 3
- FNSN
FOXP3− suppression-negative (control)
- FPSN
FOXP3+ suppression-negative
- FPSP
FOXP3+ suppression-positive
- IQR
interquartile range
- RA
rheumatoid arthritis
- SLE
systemic lupus erythematosus
- Tconv
conventional T
- TIGIT
T cell immunoreceptor with Ig and ITIM domains
- Treg
regulatory T
- Tresp
responder T
- TSDR
regulatory T–specific demethylated region
Footnotes
The microarray data presented in this article have been submitted to the National Center for Biotechnology Information’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65650) under accession number GSE65650.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 2.d’Hennezel E, Bin Dhuban K, Torgerson T, Piccirillo CA. The immunogenetics of immune dysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J Med Genet. 2012;49:291–302. doi: 10.1136/jmedgenet-2012-100759. [DOI] [PubMed] [Google Scholar]
- 3.Buckner JH. Mechanisms of impaired regulation by CD4+CD25+ FOXP3+ regulatory T cells in human autoimmune diseases. Nat Rev Immunol. 2010;10:849–859. doi: 10.1038/nri2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S, Gottlieb PA, Kapranov P, Gingeras TR, Fazekas de St Groth B, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203:1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gavin MA, Torgerson TR, Houston E, DeRoos P, Ho WY, Stray-Pedersen A, Ocheltree EL, Greenberg PD, Ochs HD, Rudensky AY. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci USA. 2006;103:6659–6664. doi: 10.1073/pnas.0509484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allan SE, Crome SQ, Crellin NK, Passerini L, Steiner TS, Bacchetta R, Roncarolo MG, Levings MK. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int Immunol. 2007;19:345–354. doi: 10.1093/intimm/dxm014. [DOI] [PubMed] [Google Scholar]
- 7.d’Hennezel E, Piccirillo CA. Analysis of human FOXP3+ Treg cells phenotype and function. Methods Mol Biol. 2011;707:199–218. doi: 10.1007/978-1-61737-979-6_13. [DOI] [PubMed] [Google Scholar]
- 8.d’Hennezel E, Yurchenko E, Sgouroudis E, Hay V, Piccirillo CA. Single-cell analysis of the human T regulatory population uncovers functional heterogeneity and instability within FOXP3+ cells. J Immunol. 2011;186:6788–6797. doi: 10.4049/jimmunol.1100269. [DOI] [PubMed] [Google Scholar]
- 9.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YC, Bhairavabhotla R, Yoon J, Golding A, Thornton AM, Tran DQ, Shevach EM. Oligodeoxynucleotides stabilize Helios-expressing Foxp3+ human T regulatory cells during in vitro expansion. Blood. 2012;119:2810–2818. doi: 10.1182/blood-2011-09-377895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayyoub M, Raffin C, Valmori D. Comment on “Helios+ and Helios− cells coexist within the natural FOXP3+ T regulatory cell subset in humans”. J Immunol. 2013;190:4439–4440. doi: 10.4049/jimmunol.1390018. [DOI] [PubMed] [Google Scholar]
- 12.Himmel ME, MacDonald KG, Garcia RV, Steiner TS, Levings MK. Helios+ and Helios− cells coexist within the natural FOXP3+ T regulatory cell subset in humans. J Immunol. 2013;190:2001–2008. doi: 10.4049/jimmunol.1201379. [DOI] [PubMed] [Google Scholar]
- 13.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 14.Lozano E, Dominguez-Villar M, Kuchroo V, Hafler DA. The TIGIT/CD226 axis regulates human T cell function. J Immunol. 2012;188:3869–3875. doi: 10.4049/jimmunol.1103627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Maksimovic J, Naselli G, Qian J, Chopin M, Blewitt ME, Oshlack A, Harrison LC. Genome-wide DNA methylation analysis identifies hypomethylated genes regulated by FOXP3 in human regulatory T cells. Blood. 2013;122:2823–2836. doi: 10.1182/blood-2013-02-481788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V, et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40:569–581. doi: 10.1016/j.immuni.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrhardt GR, Leu CM, Zhang S, Aksu G, Jackson T, Haga C, Hsu JT, Schreeder DM, Davis RS, Cooper MD. Fc receptor-like proteins (FCRL): immunomodulators of B cell function. Adv Exp Med Biol. 2007;596:155–162. doi: 10.1007/0-387-46530-8_14. [DOI] [PubMed] [Google Scholar]
- 18.Polson AG, Zheng B, Elkins K, Chang W, Du C, Dowd P, Yen L, Tan C, Hongo JA, Koeppen H, Ebens A. Expression pattern of the human FcRH/IRTA receptors in normal tissue and in B-chronic lymphocytic leukemia. Int Immunol. 2006;18:1363–1373. doi: 10.1093/intimm/dxl069. [DOI] [PubMed] [Google Scholar]
- 19.Davis RS, Wang YH, Kubagawa H, Cooper MD. Identification of a family of Fc receptor homologs with preferential B cell expression. Proc Natl Acad Sci USA. 2001;98:9772–9777. doi: 10.1073/pnas.171308498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kochi Y, Myouzen K, Yamada R, Suzuki A, Kurosaki T, Nakamura Y, Yamamoto K. FCRL3, an autoimmune susceptibility gene, has inhibitory potential on B-cell receptor-mediated signaling. J Immunol. 2009;183:5502–5510. doi: 10.4049/jimmunol.0901982. [DOI] [PubMed] [Google Scholar]
- 21.Li FJ, Schreeder DM, Li R, Wu J, Davis RS. FCRL3 promotes TLR9-induced B-cell activation and suppresses plasma cell differentiation. Eur J Immunol. 2013;43:2980–2992. doi: 10.1002/eji.201243068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, Bae SC, Tokuhiro S, Chang X, Sekine A, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37:478–485. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajpai UD, Swainson LA, Mold JE, Graf JD, Imboden JB, McCune JM. A functional variant in FCRL3 is associated with higher Fc receptor-like 3 expression on T cell subsets and rheumatoid arthritis disease activity. Arthritis Rheum. 2012;64:2451–2459. doi: 10.1002/art.34457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagata S, Ise T, Pastan I. Fc receptor-like 3 protein expressed on IL-2 nonresponsive subset of human regulatory T cells. J Immunol. 2009;182:7518–7526. doi: 10.4049/jimmunol.0802230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 26.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 28.Golding A, Hasni S, Illei G, Shevach EM. The percentage of FoxP3+Helios+ Treg cells correlates positively with disease activity in systemic lupus erythematosus. Arthritis Rheum. 2013;65:2898–2906. doi: 10.1002/art.38119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bettelli E, Dastrange M, Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-κB to repress cytokine gene expression and effector functions of T helper cells. Proc Natl Acad Sci USA. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichiyama K, Yoshida H, Wakabayashi Y, Chinen T, Saeki K, Nakaya M, Takaesu G, Hori S, Yoshimura A, Kobayashi T. Foxp3 inhibits RORγt-mediated IL-17A mRNA transcription through direct interaction with RORγt. J Biol Chem. 2008;283:17003–17008. doi: 10.1074/jbc.M801286200. [DOI] [PubMed] [Google Scholar]
- 31.Ayyoub M, Deknuydt F, Raimbaud I, Dousset C, Leveque L, Bioley G, Valmori D. Human memory FOXP3+ Tregs secrete IL-17 ex vivo and constitutively express the TH17 lineage-specific transcription factor RORγt. Proc Natl Acad Sci USA. 2009;106:8635–8640. doi: 10.1073/pnas.0900621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putnam AL, Brusko TM, Lee MR, Liu W, Szot GL, Ghosh T, Atkinson MA, Bluestone JA. Expansion of human regulatory T-cells from patients with type 1 diabetes. Diabetes. 2009;58:652–662. doi: 10.2337/db08-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raffin C, Pignon P, Celse C, Debien E, Valmori D, Ayyoub M. Human memory Helios− FOXP3+ regulatory T cells (Tregs) encompass induced Tregs that express Aiolos and respond to IL-1β by downregulating their suppressor functions. J Immunol. 2013;191:4619–4627. doi: 10.4049/jimmunol.1301378. [DOI] [PubMed] [Google Scholar]
- 34.Swainson LA, Mold JE, Bajpai UD, McCune JM. Expression of the autoimmune susceptibility gene FcRL3 on human regulatory T cells is associated with dysfunction and high levels of programmed cell death-1. J Immunol. 2010;184:3639–3647. doi: 10.4049/jimmunol.0903943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Q, Dierich A, Oulad-Abdelghani M, Chan S, Kastner P. Helios deficiency has minimal impact on T cell development and function. J Immunol. 2009;183:2303–2311. doi: 10.4049/jimmunol.0901407. [DOI] [PubMed] [Google Scholar]
- 36.Getnet D, Grosso JF, Goldberg MV, Harris TJ, Yen HR, Bruno TC, Durham NM, Hipkiss EL, Pyle KJ, Wada S, et al. A role for the transcription factor Helios in human CD4+CD25+ regulatory T cells. Mol Immunol. 2010;47:1595–1600. doi: 10.1016/j.molimm.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci USA. 2009;106:1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yurchenko E, Shio MT, Huang TC, Da Silva Martins M, Szyf M, Levings MK, Olivier M, Piccirillo CA. Inflammation-driven reprogramming of CD4+ Foxp3+ regulatory T cells into pathogenic Th1/Th17 T effectors is abrogated by mTOR inhibition in vivo. PLoS ONE. 2012;7:e35572. doi: 10.1371/journal.pone.0035572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, Hulme MA, Hoffmüller U, Baron U, Olek S, Bluestone JA, Brusko TM. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol. 2011;186:3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.