Abstract
Background & Aims
Chronic inflammation promotes development and progression of colorectal cancer (CRC). We explored the distribution of the corticotropin-releasing-hormone (CRH) family of receptors and ligands in CRC and their contribution in tumor growth and oncogenic epithelial-to-mesenchymal transition (EMT).
Methods
The mRNA expression of CRH-family members was analyzed in CRC (n = 56) and control (n = 46) samples, seven CRC cell lines, and normal NCM460 cells. Immunohistochemical detection of CRHR2 was performed in 20 CRC and five normal tissues. Cell proliferation, migration, and invasion were compared between urocortin-2 (Ucn2)-stimulated parental and CRHR2-overexpressing (CRHR2+) cells in the absence or presence of interleukin-6 (IL-6). CRHR2/Ucn2-targeted effects on tumor growth and EMT were validated in SW620-xenograft mouse models.
Results
CRC tissues and cell lines showed decreased mRNA and protein CRHR2 expression compared with controls and NCM460 cells, respectively. The opposite trend was shown for Ucn2. CRHR2/Ucn2 signaling inhibited cell proliferation, migration, invasion, and colony formation in CRC-CRHR2+ cells. In vivo, SW620-CRHR2+ xenografts showed decreased growth, reduced expression of EMT-inducers, and elevated levels of EMT-suppressors. IL-1b, IL-6, and IL-6R mRNAs were diminished in CRC-CRHR2+ cells, while CRHR2/Ucn2 signaling inhibited IL-6-mediated Stat3 activation, invasion, migration, and expression of downstream targets acting as cell cycle– and EMT-inducers. Expression of cell cycle– and EMT-suppressors was augmented in IL-6/Ucn2-stimulated CRHR2+ cells. In patients, CRHR2 mRNA expression was inversely correlated with IL-6R and vimentin levels and metastasis occurrence, while positively associated with E-cadherin expression and overall survival.
Conclusions
CRHR2 down-regulation in CRC supports tumor expansion and spread through maintaining persistent inflammation and constitutive Stat3 activation. CRHR2low CRC phenotypes are associated with higher risk for distant metastases and poor clinical outcomes.
Keywords: Colorectal Cancer, Inflammation, Metastasis, Neuropeptides
Abbreviations used in this paper: Ast2B, astressin-2B; BSA, bovine serum albumin; CRC, colorectal cancer; CRH, corticotropin-releasing hormone; CRHR, corticotropin-releasing hormone receptor; EMT, epithelial-to-mesenchymal transition; EV, empty virus; FBS, fetal bovine serum; GI, gastrointestinal; IBD, inflammatory bowel disease; IHC, immunohistochemistry; IL, interleukin; JAK, Janus kinase; LUMC, Leiden University Medical Center; PBS, phosphate-buffered saline; PCNA, proliferating cell nuclear antigen; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; RKIP, Raf-1 kinase inhibitory protein; siRNA, small interfering RNA; STAT3, signal transducer and activator of transcription 3; TBS, Tris-buffered saline; TGFβ1, transforming growth factor β1; TMA, tissue microarray; VEGF, vascular endothelial growth factor; Ucn, urocortin
Summary.
Chronic colitis is associated with increased risk for colorectal cancer. CRHR2/Ucn2 has an inhibitory effect on signaling of tumor growth and metastatic potential through regulation of colonic inflammation, and may have a therapeutic and prognostic impact on colorectal cancer.
Chronic inflammation is a driving force for the growth of many human malignancies. Inflammatory diseases such as inflammatory bowel disease (IBD) are associated with increased risk for colorectal cancer (CRC).1 The corticotropin-releasing hormone (CRH) family of receptors, CRHR1 and CRHR2 and their ligands CRH, urocortin-1 (Ucn1), -2, and -3 are well-characterized with an established role in the regulation of intestinal inflammation.2 However, their net effects on CRC development and progression remain unclear, especially under conditions of chronic inflammation.
Although CRH family members are best known as principal neuroendocrine regulators of the stress response in the central nervous system, they are also peripherally expressed and act as autocrine or paracrine pro- or anti-inflammatory factors in several diseases, including IBD.3, 4 The distribution of these peptides and receptors has been well described in the intestine;4, 5 but current knowledge of their expression in gastrointestinal tumors is limited. Expression of CRH-family members has been documented for only a few cancers and reported to follow tissue-specific expression patterns. CRHR2 is preferentially expressed in endocrine tumors, whereas CRHR1 is predominant in tumors of the nervous system.6 CRH and/or Ucn overexpression has been reported in reproductive malignancies and breast cancer tumors.7, 8, 9 Low or absent CRHR1 and CRHR2 expression was found in exocrine ductal pancreatic carcinomas, prostate cancer, and non-small cell lung cancer.6
In addition to expression discrepancies, the functional role of the CRH-system in cancer remains controversial. CRHR2 activation by CRH showed inhibition of apoptosis in gynecologic cancers and induction of tumor cell proliferation, migration, and invasion.10, 11 In contrast, CRHR1/CRH promoted apoptosis in breast cancer and endometrial adenocarcinoma, and Ucn and Ucn2 enhanced malignant cell growth in the human stomach via CRHR2.12, 13, 14 In hepatocellular carcinoma, Ucn inhibited tumor growth via thwarting angiogenesis, while neovascularization and prostate cancer development are associated with CRHR2 down-regulation.15, 16 Different molecular mechanisms triggering CRHR1- and CRHR2-driven effects on tumors might contribute to these inconsistencies, but further clarification is needed. In addition, endogenous ligands might complicate tumorigenesis, favoring progression or suppression depending on the cancer type and relative distribution of CRHRs.
To our knowledge, it has not been clearly elucidated whether CRH family members are differentially expressed in CRC and/or impact CRC pathophysiology, especially in response to colonic inflammation. Contrary to an initial study showing lost CRHR2 expression in 10 CRC samples,6 a recent study reported elevated levels of CRHR2 and Ucn3 in 30 CRC patient samples that were correlated with decreased cell adhesion and enhanced cell motility.17 Thus, studies with larger cohorts are needed to elucidate the functional role of CRH receptors and their peptides on CRC. The present study was designed to 1) determine the distribution of CRH-related components in CRC cell lines and human tissues relative to their normal counterparts, 2) delineate the functional impact of any detected expression differences in CRH molecules on tumor growth and metastatic potential, and 3) explore the underlying molecular mechanisms associated with chronic inflammatory responses.
Materials and Methods
Cell Lines and Culture Conditions
Seven human CRC cell lines (SW480, SW620, DLD1, SW403, HT29, HCT116, Caco2) and one immortalized human colonic epithelial cell line (NCM460) (all from American Type Culture Collection [ATCC], Manassas, VA) were used in the study. The cell lines were cultured in the following media: SW480, SW620, and DLD1 in RPMI 1640 (cat. no. 11875-093; Invitrogen, Grand Island, NY); SW403 in Ham’s F-12 medium (cat. no. 11765-054; Invitrogen); HT29 and HCT116 in McCoy’s 5A (cat. no. 30-2007; ATCC); CaCo2 in minimal essential medium (11095-080; Invitrogen), and NCM460 in M3:D (cat no. M3DEF; InCell Corporation, San Antonio, TX). All media were supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 10 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, Carlsbad, CA). Cells were incubated at 37°C with 5% CO2. The CRHR2-transduced cells were cultured as above with 2 μg/mL of puromycin (Sigma-Aldrich, Natick, MA). The institutional biosafety committee approved all procedures involving human cells.
Human Samples for mRNA Analysis
Fifty-six CRC samples were obtained from surgical resections from patients with CRC diagnosed with established criteria (Table 1). The control group included samples from disease-free donors and noninvolved colonic tissues obtained from ulcerative colitis patients (n = 46). Each tissue was immediately snap frozen for RNA extraction as described herein. The CRC tissue samples were provided by the Department of Gastroenterology and Hepatology of Leiden University Medical Center (LUMC) and were confirmed by histopathologic evaluation. The samples were obtained according to the instructions and guidelines of the LUMC medical ethics committee, in accordance with the Helsinki Declaration, as regulated and approved by the LUMC MDL biobanking protocol CuraRata. The control samples were provided by the Center for Neurobiology of Stress in the Division of Digestive Diseases at the University of California–Los Angeles (UCLA) or by the Department of Gastroenterology and Hepatology at LUMC. All experiments were approved by the institutional review boards of all the institutions.
Table 1.
Colorectal Cancer Patient Clinicopathologic Characteristics Used for the mRNA Analysis of Corticotropin-Releasing Hormone Family Members
| Characteristicsa | Cases (n = 56) | CRHR2 |
Ucn2 |
||
|---|---|---|---|---|---|
| Mean ± SEM | P Value | Mean ± SEM | P Value | ||
| Age, median (y) | 69 (42–90) | ||||
| Sex | 33 | ||||
| M | 23 | ||||
| F | |||||
| Localization | |||||
| Left colon | 39 | ||||
| Right colon | 13 | ||||
| Dukes classification | NS | NS | |||
| A | 1 | 1.28 ± 0.0 | 3 ± 0.0 | ||
| B1 + B2 | 17 | 0.8 ± 0.52 | 45 ± 13.7 | ||
| C1 + C2 | 29 | 0.67 ± 0.25 | 54.8 ± 17 | ||
| D | 9 | 0.68 ± 0.53 | 47.8 ± 27.5 | ||
| TNM classification | NS | NS | |||
| Low | 18 | 0.83 ± 0.5 | 42.65 ± 13.2 | ||
| High | 38 | 0.67 ± 0.22 | 53.16 ± 14.4 | ||
| Differentiation | NS | NS | |||
| Well | 7 | 0 ± 0.0 | 58.41 ± 33 | ||
| Moderate | 18 | 0.79 ± 0.44 | 24.67 ± 6.4 | ||
| Poor | 31 | 0.84 ± 0.29 | 62.42 ± 17 | ||
| Metastasisb | .047 | NS | |||
| NM | 21 | 1.37 ± 0.47 | 51.69 ± 15 | ||
| DM | 18 | 0.36 ± 0.21 | 46.5 ± 13.3 | ||
| Body mass indexb | .048a | NS | |||
| Underweight (<18.5) | 3 | 0 ± 0.0 | 16.8 ± 11 | ||
| Normal (18.5–24.9) | 23 | 1.07 ± 0.41 | 41.91 ± 9.2 | ||
| Overweight (25–29.9) | 13 | 0.06 ± 0.06 | 69.87 ± 31.6 | ||
| Obese (>30) | 3 | 1.21 ± 0.92 | 145.3 ± 111 | ||
Note: CRHR2, corticotropin-releasing hormone receptor 2; DM, distant metastasis; NM, no metastasis; NS, not statistically significant (P > .05); SEM, standard error of the mean; TNM, tumors/nodes/metastases; Ucn2, urocortin-2.
Normal weight versus overweight.
Information was not available for all cases.
Human Samples for Tissue Microarrays
The study cohort consisted of samples from 20 randomly selected CRC patients who underwent surgery for tumor removal. The control group included five samples from adjacent morphologically normal tissue. The study was approved by the ethics committee of the Hospital Infantil de México Federico Gomez (Mexico City, Mexico; HIM/2007/061). Both CRC and normal samples were collected between 2002 and 2007 from the Department of Pathology of Hospital General Regional No. 25, IMSS and Speciality Hospital CMN La Raza, IMSS (Mexico City, Mexico). All CRC cases were classified according to the Dukes or TNM (tumors/nodes/metastases) staging. The sample collection and use was approved by the ethics committee of the Hospital Infantil de México Federico Gomez. Tissue microarrays (TMAs) were constructed as follows: formalin-fixed, paraffin-embedded archival tumor specimens were prepared at the Immunology and Infection Research Unit, National Medical Center La Raza and at the Oncology Disease Research Unit, Hospital Infantil de México Federico Gomez. At least three core tissue biopsies (each 0.6 mm in diameter) were taken from morphologically representative regions of each colorectal tumor and precisely arrayed according to the microarray technique with a semiautomatic ATA-100 Chemicon system (advanced tissue array; Chemicon, Temecula, CA), as previously described elsewhere.18 Tissues were arrayed into five TMA blocks. For staining, sections (4 μm) were transferred to glass slides using an adhesive slide system (PSA-CS 4; Instrumedics, St. Louis, MO) to support cohesion of the array elements.
Cell Treatments
Cells were treated with the CRHR2-specific agonist urocortin-2 (Ucn2, H-5852; Bachem, Torrance, CA), the CRHR2 specific antagonist astressin-2B (Ast2B, cat. no. 2391; Tocris Bioscience, Bristol, United Kingdom) and interleukin-6 (IL-6, CYT-213; ProSpec-Tany Technogene, East Brunswick, NJ) used either as single agents or in combination at the indicated concentrations of each experimental condition. All cell treatments were performed in media containing 0.1% FBS. Before treatment, the cells remained in serum-deprived media for overnight incubation.
Gene Expression Analysis
Total RNA was extracted from tissues and cell lines using the RNeasy Plus Universal Mini kit (cat. no. 73404; Qiagen, Valencia, CA) and RNeasy Plus Mini Kit (cat. no. 74134; Qiagen), respectively. cDNA was synthesized using the quantitative reverse-transcription kit (cat. no. 205311; Qiagen). Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed using TaqMan universal PCR master mix (4352042; Applied Biosystems, Foster City, CA) and on-demand gene-specific primers (Applied Biosystems). Primer sets included the following: CRHR1 (Hs00366363_a1), CRHR2 (Hs00266401_m1), CRH (Hs01921237_s1), Ucn1 (Hs01849155_s1), Ucn2 (Hs00264218_s1), Ucn3 (Hs00846499_s1), IL-6 (Hs00985639_m1), IL-6R (Hs01075666_m1), IL-1β (Hs01555410_m1), Vim (Hs00958116_m1), CDH1 (Hs01023894_m1), and 18S (Hs99999901_s1). The reactions were run in an ABI 7500 Fast RT-PCR system (Applied Biosystems) and were analyzed using SDS software (version 1.3; Applied Biosystems). Relative quantification was achieved by normalizing to gene expression of 18S.
Transient Transfections
SW620 and SW480 cells cultured in 24/well plates at 50–60% confluence were transfected with 30 pmol/well of small interfering RNA (siRNA) against Ucn2 (cat. no. Sc-106676; Santa Cruz Biotechnology, Santa Cruz, CA) or scramble control siRNA (cat. no. Sc-37007; Santa Cruz Biotechnology) using Lipofectamine RNAiMAX (cat. no. 13778075; Invitrogen). No cell toxicity was detected owing to the transfection agent. The RNA was extracted 72 hours after transfection for qRT-PCR.
For plasmid transfections, SW480-EV and SW480-CRHR2+ cells cultured at 70% confluence were transfected with 2 μg of pCDNA3.1-SnailS6A or pCDNA3.1, respectively, using Lipofectamine 2000 (cat. no. 11668019; Invitrogen). Mutated SnailS6A protein is more resistant to ubiquitination-dependant proteosome degradation. Twenty-four hours later, cells were transferred to 96-transwells and incubated for 48 hours with 15 ng/mL of recombinant human transforming growth factor β1 (TGFβ1, cat. no. PHG9204; Invitrogen) ± 0.1 μM Ucn2 for determination of cell migration. All transfections were performed in triplicate.
Lentiviral Transductions
Construction of CRHR2-expressing lentivirus and stable cell transduction were performed as described previously elsewhere.19 Briefly, the cells were infected with MCS-IRES-Strawberry-hPGK-Puro lentivirus particles containing a cytomegalovirus promoter driving expression of human CRHR2 (A0634; GeneCopoeia, Rockville, MD) or scramble negative control (EV, empty virus). Selection of transduced clones was performed in culture medium containing 10 μg/mL of puromycin. Only the highly fluorescent cell clones were used in the experiments.
Anchorage-Independent Growth Assay
Triplicates of 5 × 105 CRC cells were mixed 4:1 (v/v) with 2% agarose in growth medium (final, 0.4% agarose). Cell mixtures were plated on a layer of medium containing 0.5% agarose and fed every 6–7 days (0.4% agarose); colonies were counted after 15 days. Where appropriate, 0.1 μM Ucn2 was added every 2 days.
Western Blot Analysis
Western blot analysis in protein lysates derived by cell lines was performed as previously described elsewhere.20 Briefly, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was conducted using 8–12% gradient Tris-glycine gels (Invitrogen) and loaded with 30 μg of total protein lysates. Membranes were blocked in Tris-buffered saline (TBS) + 5% nonfat milk, and primary and secondary antibodies were incubated overnight at 4°C or for 2 hours at room temperature, respectively, in TBS + 0.1% Tween-20 + 5% nonfat milk; phospo-signal transducer and activator of transcription 3 (pStat3) was incubated in TBS + 0.1% Tween-20 + 5% bovine serum albumin (BSA). Primaries used were β-actin ms (1:1000, cat. no. 130065; Santa Cruz Biotechnology), pStat3 (Tyr705) rb (1:1000, cat. no. 9145S; Cell Signaling Technology, Beverly, MA), b-catenin rb (1/3000, cat. no. ab6302; Abcam, Cambridge, MA), vimentin ms (1/2000, ab8069; Abcam), E-cadherin rb (1/500, ab53033; Abcam), ZEB1 rb (1/500, cat. no. SAB3500514; Sigma-Aldrich, Saint Louis, MO), and CRHR2 (1/1000, cat. no. ABN433; Millipore, Temecula, CA). Horseradish peroxidase–tagged IgG secondary antibodies were anti-mouse (cat. no. sc2005; Santa Cruz Biotechnology) or anti-rabbit (cat. no. sc2004; Santa Cruz Biotechnology) used at 1/2000 dilution. Chemiluminescence was detected with enhanced reagent (cat. no. 34080; ThermoScientific, Rockford, IL) using an Eastman Kodak 440 Imaging System (Kodak, Rochester, NY). Beta-actin was used as a loading control.
Immunohistochemistry
Tumors resected via SW620-bearing mice were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) and embedded in paraffin blocks. We cut 2-μm sections, placed them on slides, and used them for Mayer’s H&E staining or for immunohistochemical (IHC) analysis, as previously described elsewhere.21 Slices cut at 4 μm for the TMAs were placed on slides, and they were either H&E stained for histopathologic examination or used for subsequent IHC analysis. In all cases, antigen retrieval was performed after tissue deparaffinization and hydration by immersing the slides in a solution of 0.01% sodium citrate (pH 6.0) for 5 minutes in boiling water. Endogenous peroxidase activity was inhibited by immersing the slides in 3% H2O2-methanol, and background-unspecific binding was decreased by incubating the slides in 2% BSA (Sigma-Aldrich, St. Louis, MO) in PBS for 60 minutes. Primary antibodies were incubated at room temperature overnight. The primaries used were Raf-1 kinase inhibitory protein (RKIP) ms (1:500, cat. no. sc-365973; Santa Cruz Biotechnology), pStat3(Tyr705) rb (1:250, cat. no. 9145S; Cell Signaling Technology), N-cadherin rb (1/500, cat. no. ab18203; Abcam), vimentin ms (1/250, cat. no. ab8069; Abcam), vascular endothelial growth factor (VEGF) rb (1/200, cat. no. ab46154; Abcam), Snail rb (1/100, cat. no. ab135708; Abcam), proliferating cell nuclear antigen (PCNA) ms (1:4000, cat. no. 2586S; Cell Signaling Technology), and CRHR2 rb (1/750, cat. no. ABN433; Millipore). To decrease variability, all samples were processed at the same time in a single experiment, using a single batch of antibody diluted in PBS-BSA.
After washing, the slides were incubated with a biotinylated secondary antibody (Universal LSAB kit; Dako Corporation, Carpinteria, CA) for 30 minutes at room temperature, followed by incubation with a streptavidin–horseradish peroxidase conjugate (Universal LSAB kit) for 30 minutes at room temperature and then with 3,3′-diaminobenzidine tetra-hydrochloride (liquid DAB; Dako Corporation). The reaction was stopped by adding distilled water, and the slides were counterstained with H&E. The tissue was washed in tap water for 5 minutes, dehydrated using an ethanol series (70%, 90%, and 100%) and xylene and mounted with E-2 mounting medium (Shandon Laboratory, Pittsburgh, PA). The slides were examined by light microscopy (Olympus BX-40; Olympus Corporation, Tokyo, Japan), and then expression intensity (brown color) in the total section area (mm3) was evaluated by digital image analysis as described herein.
Analysis of Immunohistochemical Digital Images
Immunohistochemically stained sections were digitized at 40× magnification using an Aperio Scanscope CS (Aperio, Vista, CA). The Aperio Scanscope CS obtains 40× magnification images with a spatial resolution of 0.45 μm/pixels. Once the areas were annotated, they were sent for automated image analysis using Spectrum software (Aperio), and an algorithm was developed to quantify the total expression of the target protein. The output from the algorithm returns a number of quantitative measurements such as intensity, concentration, and percentage of positive staining present. The quantitative scales of intensity and percentage were categorized to four and five classes, respectively, after the cutoff values were determined. The intensity of staining was categorized as 0 (no staining), 2+ (moderate), and 3+ (strong). The final IHC score was calculated from a combination of the intensity and percentage scores.
In Vitro Migration and Invasion Assays
The invasive and migratory potentials of SW620 and HCT116 cells were assessed using Cultrex 96-transwell assays containing inserts with 8-micron polyethylene terephthalate membranes and according to the manufacturer’s instructions (3465-096-K; 3455-096-K; Trevigen, Gaithersburg, MD). Briefly, to analyze invasion, the inserts were first coated with 50 μL of 0.5X basement membrane extract solution and incubated overnight at 37°C. Subsequently, 5 × 104 EV or CRHR2+ cells, previously deprived of serum (0.1%), were seeded into the upper chambers in serum-free medium supplemented with 0.1 μM Ucn2, alone or in combination with 1 μM Ast2B in the presence or absence of 20 ng/mL IL-6. Medium supplemented with 15% FBS was applied to the lower chambers as a chemoattractant to induce invasion/migration.
The chambers were incubated at 37°C in a 5% CO2 incubator for 48 hours. At the end of incubation, the inserts were extensively washed, and the cells that invaded were labeled with 5 μg/mL of calcein acetomethylester diluted in cell dissociation solution at 37°C for 1 hour. Fluorescence was measured at 485 nm in a Wallac 1420 Victor2 multilabel plate reader (Perkin-Elmer, Shelton, CT). In the migration assay, the same procedure of invasion assay was followed, except that inserts were not coated with basement membrane extract. For detecting IL-6-induced invasion and migration of DLD1, the cells were pretreated with 20 ng/mL IL-6 for 72 hours as previously described elsewhere.22 Migration and invasion were monitored using 24-well transwell chambers.
Monitoring Invasion and Migration of DLD1 Cells
The invasion and migration assays of IL-6-pretreated DLD1 cells were performed using Matrigel Invasion Chambers (8-micron pore size membrane) (cat. no. 354480; Corning, Bedford, MA) and transwell permeable supports (8-micron polycarbonate membrane) (cat. no. 3422; Costar, Corning, NY), respectively. Briefly, serum-deprived cells were collected with Accutase (cat. no. A11105-01; Life Technologies, Grand Island, NY) and seeded at a concentration of 5 × 104 cells in each upper chamber in serum-free medium supplemented with 0.1 μM Ucn2 alone or in combination with 1 μM Ast2B. As a chemoattractant, 20% FBS was placed in the lower chamber. Control inserts were included in all assays (cat. no. 354578; Corning).
After 72 hours, the nonmotile/invasive cells in the chambers were wiped away using a cotton swab, and the invaded/migrated cells were fixed and stained with 0.5% crystal violet solution containing 20% methanol. After multiple washes, the membranes were cut, air dried, mounted on slides, and observed under a microscope. For each sample, six random optical fields at 10× magnification were analyzed to determine the mean number of invading/migrating cells. Photographs were taken with an inverted Imager Z.1 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) equipped with an AxioCam HRc camera (Carl Zeiss Microscopy) and analyzed with the accompanying AxioVision software.
cAMP Determination
Ucn2 dose-response studies related to cAMP production were performed in parental and CRHR2+ CRC cells using the Direct cAMP Enzyme Imunoassay Kit (cat. no. 901-066; Assay Designs, Ann Arbor, MI). Cells were stimulated with Ucn2 (10−9 to 10−6 M) for 30 minutes. After removal of media, cells were lysed for 20 minutes in 1 mL 0.1M HCl. Cell lysates were centrifuged at 600g at room temperature for 5 minutes and cAMP levels determined in 50 μL supernatant samples using triplicate wells and duplicate assays for each treatment.
Ucn2 Protein Determination
Ucn2 protein secreted by different CRC and NCM460 cell lines was quantified by an enzyme-linked immunosorbent assay kit (cat. no. MBS2023605; MyBioSource, San Diego, CA) according to the manufacturer’s instructions with slight modifications. Briefly, serum-free conditioned media were collected from cells seeded in 35-mm wells. We incubated 100 μL of undiluted cell culture supernatants in precoated wells in enzyme-linked immunosorbent assay plate at 4°C overnight. Subsequent procedures for target protein detection were performed according to the manufacturer’s instructions. The concentration of Ucn2 secreted to the culture medium was measured at optical density at 450 nm and normalized by the total amount of protein in cell lysates harvested by each well.
Cell Proliferation Assay
Cell proliferation was quantified by a colorimetric XTT-based assay (cat. no. 11465015001; Roche Applied Science, Indianapolis, IN). Briefly, 4 × 103 EV or CRHR2+ cells were plated in 96-well plates and allowed to adhere for 24 hours. Then cells were treated with 0.1 μM Ucn2 alone or in combination with 1 μM Ast2B in presence or absence of 20 ng/mL IL-6 for 72 hours. At the end of incubation, 50 μL of XTT labeling mixture was added to each well, and the cells incubated at 37°C for 4 hours. Cell growth was determined by measuring absorbance at 570 nm in a Wallac 1420 Victor2 multilabel plate reader (Perkin-Elmer).
xCELLigence Real-Time Cell Analysis Technology
To access NCM460 proliferation and migration under various conditions, we used a label-free real-time cell analysis platform (xCELLigence; Roche Applied Science). Briefly, for cell proliferation 100 μL of complete culture medium at room temperature was added into each well of E-plate 96. After this the E-plate 96 was connected to the system and checked in the cell-culture incubator for proper electrical contacts, and the background impedance was measured during 24 seconds. Meanwhile, NCM460-EV and NCM460-CRHR2 cells were resuspended in complete culture medium and adjusted to 10,000 cells/mL (previously determined optimal seeding concentration). We added 100 μL of each cell suspension to the 100 μL-medium-containing wells on E-plate 96. After 30 minutes of incubation at room temperature, E-plate 96 was placed into the cell culture incubator. Approximately 24 hours after seeding, when the cells were in the log growth phase (cell index >0.5), the 100 μL of medium was removed from each well and replaced with 100 mL of complete medium containing Ucn2, Ast2B, or the combination. Controls received medium only. Cell proliferation was monitored every 15 minutes for a period of up to 72 hours via the incorporated sensor electrode arrays of the E-Plate 96. The electrical impedance was measured by the real-time cell analysis–integrated software of the xCELLigence system as a dimensionless parameter termed “CI.”
The rate of NCM460-EV and NCM460-CRHR2 cell migration was also monitored in real-time with the xCELLigence system (CIM-plate 16). At 24 hours before we conducted the experiment, the cells were serum starved. We added 100 μL of complete culture medium into the lower chamber and 50 μL of serum-free medium into the upper chamber. The plate was inserted into the platform and left for incubation for no more than 1 hour. Meanwhile, dilutions of 1 × 106 cells/mL plus the appropriate factors (Ucn2, Ast2B, or the combination) were prepared in serum-free medium. We added 100 μL of each cell suspension in each well of the upper chamber and left it to equilibrate at room temperature for 30 minutes. The impedance value of each well was automatically monitored by the xCELLigence system for a duration of 50 hours and expressed as a “CI” value.
Superarray Analysis
The EV and CRHR2+ SW620 and SW480 cells were treated with 20 ng/mL IL-6 and 0.1 μM Ucn2 for 18 hours. The RNA was isolated as described earlier, and the expression profiles of 84 genes related to cell cycle and 84 genes related to epithelial-to-mesenchymal transition (EMT) were determined using 96-well format RT2 Profiler PCR arrays (PAHS-020Z; PAHS-090Z; SABiosciences/Qiagen, Frederick, MD) including six housekeeping genes and three RNAs as internal controls. The qRT-PCRs were run on an ABI 7500 Fast qPCR instrument (Applied Biosystems) with SDS software using RT2 SYBR Green qPCR Mastermix (cat. no. 330522; SABiosciences/Qiagen). Data analysis was performed using the 2−ΔCt method described on the manufacturer’s Web site (www.SABiosciences.com/pcrarraydataanalysis.php). Heat maps of the relative gene expression were generated using the online Hierarchical Clustering Explorer (HCE) software.
Detection of Apoptosis by Terminal Deoxynucleotidyl Transferase–Mediated Digoxigenin-Deoxyuridine Nick-End Labeling Assay
Apoptosis in tissue sections was assessed using In Situ Cell Death Detection Kit, POD (11684817910; Roche Applied Science) according to the manufacturer’s instructions.
In Vivo Mouse Xenograft Model
Eight-week-old athymic nude mice were used for monitoring SW620-CRHR2+ and SW620-EV growth in vivo under various treatment protocols. Briefly, 1 × 106 SW620-EV or SW620-CRHR2+ cells diluted in 100 μL PBS were injected subcutaneously into the right hind limb under sevoflurane anesthesia. When animals developed palpable tumors (50–100 mm3), mice from each tumor group were randomly assigned to the following subgroups: 1) Ucn2-treated (15 μg/kg; n = 5) intratumorally every 2 days for 2 weeks; 2) Ucn2+Ast2B-treated (15 μg/kg + 30 μg/kg; n = 5) intratumorally with every 2 days for 2 weeks; and 3) vehicle control (0.1% acetic acid in PBS; n = 5). Tumor volume was measured with a caliper weekly and calculated with the following formula: A (length) × B (width) × C (height) × 0.5236. Treatment efficacy was evaluated by change in tumor volume. After treatment, the mice were euthanized, and the tumors were harvested for histologic studies. All animal experiments were done in accordance with institutional guidelines for animal welfare.
Processing of Digital Images
Adobe Photoshop CS6 software (Adobe Systems, Mountain View, CA) was used as a digital tool to 1) adjust contrast, brightness, or color, in digital images after uniform application to the entire image, 2) crop unneeded fractions from gel images, and 3) add scale bars to microscope-derived IHC digital images based on the formula: Actual Pixel size = CCD Pixel × Binning/Lens Magnification × C mount × Objective Magnification.
Statistical Analysis
Statistical and Kaplan-Meier survival analyses were performed with GraphPad Prism Software (version 5; GraphPad Software, La Jolla, CA). Statistical differences between two groups were determined by Student t test. Differences between multiple groups were determined using analysis of variance (ANOVA). P ≤ .05 was considered statistically significant. Data are expressed as the mean ± standard error of the mean (SEM).
Results
Diminished Corticotropin-Releasing Hormone Receptor 2 and Elevated Urocortin-2 Expression in Colorectal Cancer Tissues and Cell Lines
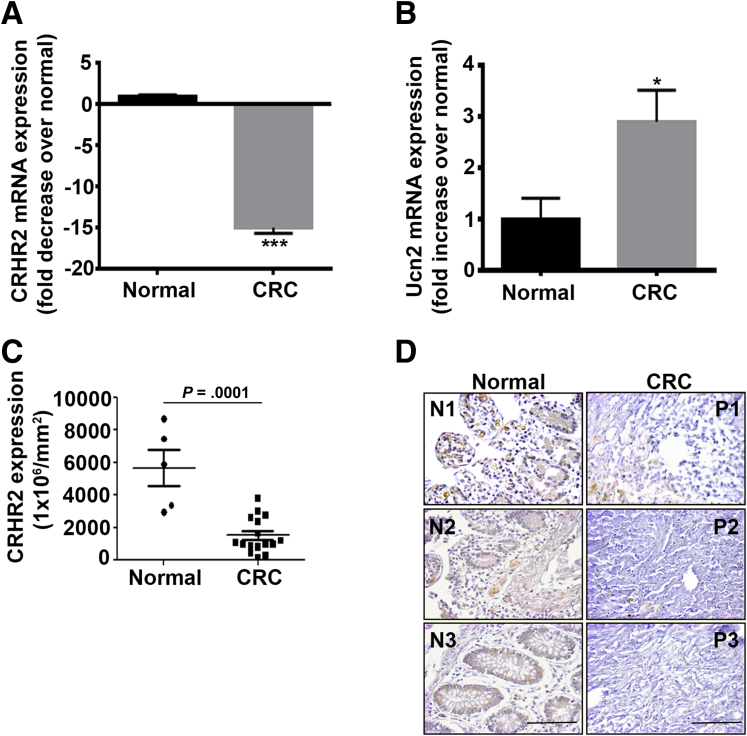
We explored the expression of CRH family members in CRC human tissues (n = 56) (Table 1) and normal controls (N = 46). We found statistically significantly decreased CRHR2 mRNA levels (P < .0001) (Figure 1A) and elevated Ucn2 expression (P = .016) in CRC tissues compared with normal controls (Figure 1B). None of the other CRH family members showed any statistically significant differences in expression between CRC and control tissue groups (Table 2). CRH was not detectable in any group. The reduced CRHR2 expression in CRC was also confirmed by IHC at the protein level in 20 CRC samples compared with five normal controls (Figure 1C). Representative CRHR2 staining in CRC and normal tissues is shown in Figure 1D.
Figure 1.
Corticotropin-releasing hormone receptor 2 (CRHR2) and urocortin-2 (Ucn2) are differentially expressed in colorectal cancer (CRC) patients. (A) Statistically significant inhibition (***P < .001) of CRHR2 mRNA levels in CRC tissues (n = 56) compared with normal controls (n = 46). (B) Statistically significant overexpression (*P = .016) of Ucn2 mRNA in CRC tissues (n = 56) compared with normal controls (n = 46). (C) CRHR2 protein down-regulation in CRC tissues (n = 20) over normal (n = 5) (P = .0001). (D) Representative CRHR2 staining in CRC and normal tissue arrays. Original magnification ×20, scale bar: 50 μm (scale bar proportional to the original microscope-derived image).
Table 2.
mRNA Expression of Corticotropin-Releasing Hormone Peptides and Receptors in Normal and Colorectal Cancer Human Tissues
| Peptide | CRC (n = 56) | Normal (n = 46) | P Value |
|---|---|---|---|
| CRHR1 | 0.28 ± 0.11 | 0.42 ± 0.23 | NS |
| CRHR2 | 0.72 ± 0.21 | 10.55 ± 2 | <.0001 |
| Ucn1 | 20.69 ± 2.9 | 23.72 ± 3 | NS |
| Ucn2 | 49.74 ± 10.6 | 17.2 ± 7 | .0161 |
| Ucn3 | 26.4 ± 4.5 | 37.56 ± 3.4 | NS |
Note: Values are mean ± standard error of the mean. CRHR, corticotropin-releasing hormone receptor; NS, not statistically significant (P > .05); Ucn, urocortin.
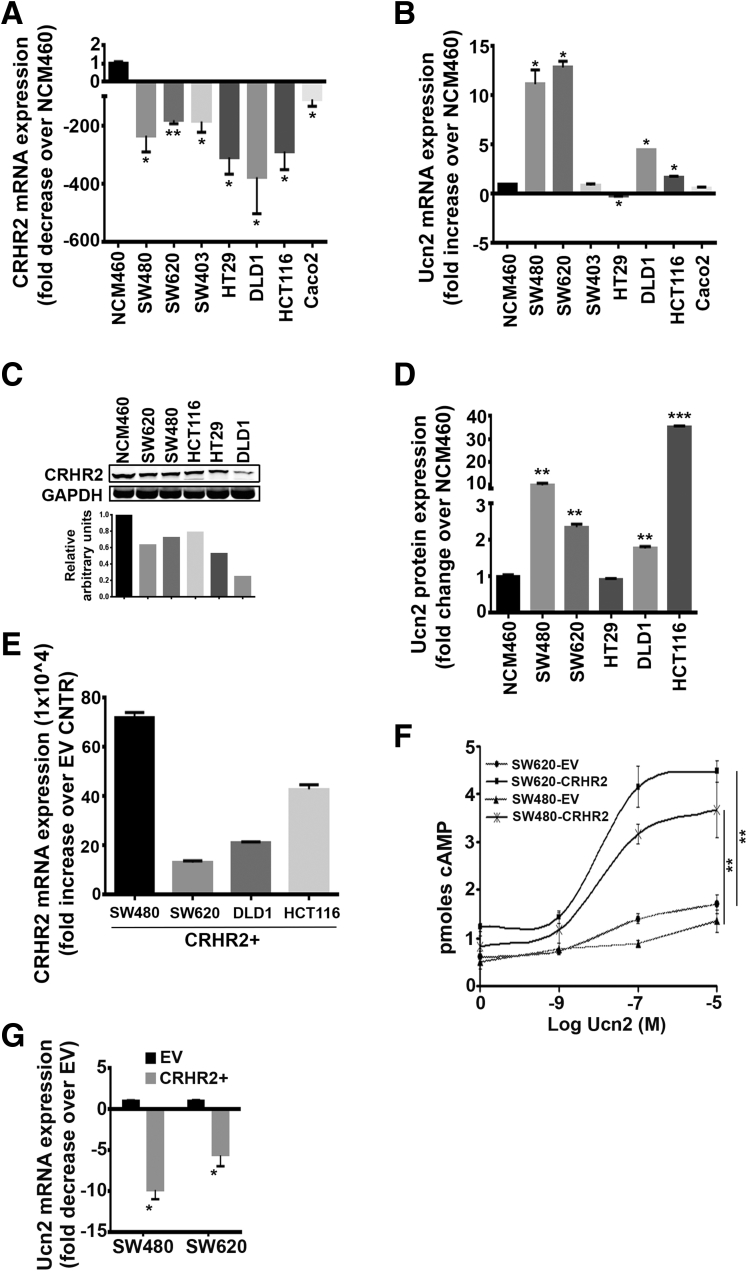
We performed the same analysis in human CRC cell lines (N = 7) and the immortalized colonic epithelial cell line NCM460. Compared with NCM460, all tested cell lines had statistically significantly lower CRHR2 mRNA expression (Figure 2A) (fold decrease range: 50–350), and Ucn2 mRNA levels were found to be statistically significantly up-regulated in four out of seven CRC cell lines tested (Figure 2B) (fold increase range: 2–13). CRHR2 protein levels were also monitored in these cell lines by Western blot analysis, confirming the down-regulated expression in most CRC cell lines tested compared with NCM460 (Figure 2C).
Figure 2.
Corticotropin-releasing hormone receptor 2 (CRHR2) and urocortin-2 (Ucn2) are differentially expressed in colorectal cancer (CRC) cell lines. (A) Statistically significant inhibition of CRHR2 mRNA expression in seven CRC cell lines (SW480, SW620, SW403, HT29, DLD1, HCT116, and CaCo2) compared with an immortalized colonic epithelial cell line NCM460. *P < .05; **P < .01. (B) Ucn2 is statistically significantly overexpressed in the CRC cell lines compared with NCM460. *P < .05. (C) Representative Western blot showing CRHR2 down-regulation at the protein level in CRC cell lines over NCM460. Densitometric analysis corresponds to the presented Western blot. Lanes of bands shown have been cropped from the original whole gel image. (D) Fold change of Ucn2 protein expression in CRC cell culture supernatants compared with NCM460. **P < .01; ***P < .001. (E) CRC cells with profiles of CRHR2 down-regulation and Ucn2 up-regulation versus NCM460 were lentivirally transduced to overexpress CRHR2 (CRHR2+ cells). Induced expression was validated by quantitative reverse-transcription polymerase chain reaction. EV stands for parental cells transduced with an empty (CRHR2−) virus. (F) Statistically significant elevation of cAMP production in CRHR2+ versus EV cells as response to increasing Ucn2 concentrations. **P < .01. (G) CRHR2 induction lowers Ucn2 baseline mRNA levels in SW480 and SW620 cells. *P < .05.
Concomitantly, Ucn2 overexpression in CRC cell lines versus NCM460 cells was further validated at the protein level in cell culture supernatants (Figure 2D). With the exception of HCT116, no statistically significant differences were observed in CRHR1 mRNA expression between the CRC cell lines and NCM460 (Table 3), whereas the Ucn1 and Ucn3 expression profiles followed a cell line-dependant increase or decrease compared with NCM460 (Table 4). These findings show statistically significant differences in CRHR2 and Ucn2 expression between malignant and normal colonic cells that might reflect dysregulated CRHR2/Ucn2 signaling in CRC.
Table 3.
mRNA Expression of Corticotropin-Releasing Hormone Receptors in NCM460 and Colorectal Cancer Cell Lines (N = 7)
| Cell Line | CRHR1 |
CRHR2 |
||
|---|---|---|---|---|
| Mean ± SEM | P Value | Mean ± SEM | P Value | |
| NCM460 | 0.07 ± 0.06 | 1.95 ± 0.19 | ||
| SW480 | 0.06 ± 0.03 | NS | 0.01 ± 0.00 | <.0001 |
| SW620 | 2.03 ± 0.4 | NS | 0.01 ± 0.00 | .01 |
| SW403 | 0.32 ± 0.25 | NS | 0.14 ± 0.12 | .001 |
| HT29 | 2.79 ± 2.7 | NS | 0.007 ± 0.001 | <.0001 |
| DLD1 | 0.17 ± 0.02 | NS | 0.007 ± 0.002 | <.0001 |
| HCT116 | 3.22 ± 0.1 | .001 | 0.008 ± 0.001 | <.0001 |
| CaCo2 | 0.07 ± 0.06 | NS | 0.38 ± 0.36 | .048 |
Note: P values are NCM460 versus colorectal cancer cell lines. CRHR, corticotropin-releasing hormone receptor; NS, not statistically significant (P > .05); SEM, standard error of the mean.
Table 4.
mRNA Expression of Corticotropin-Releasing Hormone Analogs in NCM460 and Colorectal Cancer Cell Lines (n = 7)
| Cell Line | Ucn1 |
Ucn2 |
Ucn3 |
|||
|---|---|---|---|---|---|---|
| Mean ± SEM | P Value | Mean ± SEM | P Value | Mean ± SEM | P Value | |
| NCM460 | 30.24 ± 1.2 | — | 0.98 ± 0.01 | — | 0.43 ± 0.009 | — |
| SW480 | 3.43 ± 0.07 | .002 | 10.97 ± 1.4 | .02 | 0.0 ± 0.0 | <.001 |
| SW620 | 169.7 ± 3.4 | <.001 | 12.64 ± 0.6 | .003 | 20.26 ± 5.03 | .058 |
| SW403 | 65.55 ± 6.9 | .035 | 0.84 ± 0.17 | NS | 0.35 ± 0.05 | NS |
| HT29 | 18.52 ± 0.32 | .01 | 0.19 ± 0.01 | <.001 | 1.9 ± 0.13 | .009 |
| DLD1 | 25.74 ± 1.6 | NS | 4.46 ± 0.001 | <.001 | 5.24 ± 0.65 | .018 |
| HCT116 | 19.37 ± 0.63 | .015 | 1.64 ± 0.14 | .04 | 0.0 ± 0.0 | <.001 |
| CaCo2 | 1.5 ± 0.16 | .002 | 0.54 ± 0.15 | NS | 0.1 ± 0.1 | NS |
Note: P values are NCM460 versus colorectal cancer cell lines. CRHR, corticotropin-releasing hormone receptor; NS, not statistically significant (P > .05); SEM, standard error of the mean.
Corticotropin-Releasing Hormone Receptor 2/Urocortin-2 Signaling Inhibits Colorectal Cancer Proliferation, Migration, Invasion, and Colony Formation
To test the impact of CRHR2/Ucn2 imbalance in CRC pathophysiology, including tumor growth and EMT occurrence, we stably transduced SW480, SW620, DLD1, and HCT116 (cell lines with diminished CRHR2 and elevated Ucn2 expressions versus NCM460 cells) with CRHR2-expressing lentivirus. Validation of CRHR2 overexpression (CRHR2+) over parental (EV) cells was performed using qRT-PCR (Figure 2E); confirmation of receptor activity upon stimulation with its selective agonist Ucn2 was shown by increased cAMP production (Figure 2F). Interestingly, Ucn2 mRNA levels in SW480 and SW620 cells were statistically significantly decreased after CRHR2 induction, suggesting that Ucn2 and CRHR2 expressions might share overlapping regulatory mechanisms (Figure 2G); however, this is beyond the focus of our present study.
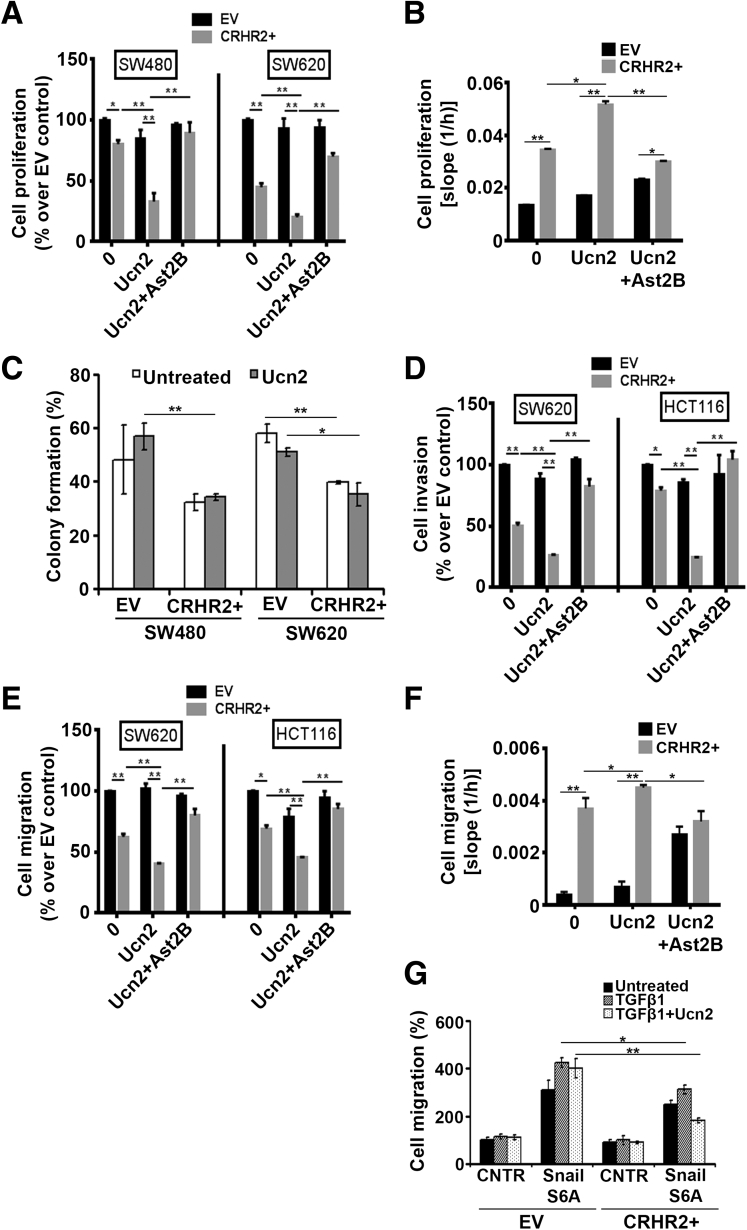
We initially compared the proliferation responses of SW480 and SW620 cells transduced with CRHR2 with those of the corresponding EV cells before and after stimulation with Ucn2 (Figure 3A). Both CRHR2+ cell lines had statistically significantly decreased baseline proliferation compared with EV cells, suggesting that endogenous Ucn2 levels are effective in stimulating CRHR2 signaling to mediate its antiproliferative effects. This inhibition was more pronounced in the presence of exogenous Ucn2. The observed Ucn2-mediated inhibition of CRHR2+ proliferation was reversed by Ast2B, a selective CRHR2 antagonist, demonstrating that this effect is CRHR2 specific. No statistically significant changes were observed in the proliferation of parental cells after treatment with 0.1 μM Ucn2.
Figure 3.
Corticotropin-releasing hormone receptor 2 (CRHR2) induction in colorectal cancer (CRC) cells inhibits tumor growth and oncogenic epithelial-to-mesenchymal transition (EMT) progression. (A) CRHR2 induction in SW480 and SW620 inhibits specifically baseline and urocortin-2 (Ucn2)-mediated cell proliferation. Cells were treated with 0.1 μM Ucn2 ± 1 μM astressin-2B (Ast2B) for 72 hours. (B) NCM460-CRHR2-overexpressing cells have increased and CRHR2-specific baseline and Ucn2-mediated proliferation rates compared with parental NCM460 cells. *P < .05; **P < .01. (C) SW480-CRHR2+ and SW620-CRHR2+ cells are less efficient in forming colonies than corresponding parental EV (empty virus) cells. Cells were treated with 0.1 μM Ucn2 every 2 days for 15 days. (D) SW620- and HCT116-overexpressing CRHR2 showed reduced baseline and Ucn2-mediated migration and (E) invasion potential over parental cells. Both responses were reversed by Ast2B, thus suggesting CRHR2 specificity. In both assays, cells in upper chambers were treated with 0.1 μM Ucn2 ± 1 μM Ast2B for 48 hours. *P ≤ .05; **P ≤ .01. (F) CRHR2 overexpression enhances specifically the migratory properties of NCM460 cells. *P < .05; **P < .01. (G) CRHR2 induction reduces Snail/transforming growth factor β1 (TGFβ1)-mediated migration of SW480 cells. SW620-EV and SW620-CRHR2+ cells were transfected with a plasmid vector expressing SnailS6A and incubated with or without 15 ng/mL TGFβ1 and/or 0.1 μM. Migration was assessed 48 hours after transfection. *P ≤ .05; **P ≤ .01.
In contrast, treatment of NCM460 with Ucn2 did not statistically significantly increase the baseline cell proliferation; however, CRHR2 overexpression in those cells alone or combined with Ucn2 treatment increased specifically the proliferative response, suggesting that CRHR2/Ucn2 signaling might interfere differently with cell cycle regulators in normal and malignant colonic cells (Figure 3B). Concomitantly with the proliferation findings, the baseline colony formation was decreased in CRHR2+ SW460 and SW480 cells (Figure 3C). However, CRHR2+ cell treatment with Ucn2 did not further reduce colony formation in both cell lines.
We next tested the migratory and invasive properties of SW620 and HCT116 after CRHR2 induction. Similarly to cell proliferation, both invasion (Figure 3D) and migration (Figure 3E) of these cells were negatively affected by CRHR2 overexpression, reaching the lowest level after treatment with Ucn2. Although Ucn2 also mediated inhibition of the invasion and migration of HCT116 parental cells, this response was not CRHR2 specific because cell treatment with Ast2B was able to reverse the inhibitory effect only in CRHR2+ cells. Migration of parental NCM460 was not affected by Ucn2; however, cell treatment with the CRHR2 antagonist Ast2B (with or without Ucn2) enhanced both the baseline and Ucn2-mediated migration, thus suggesting that CRHR2 levels might specifically affect NCM460 migration but not exclusively through Ucn2 ligation (Figure 3F). In contrast, NCM460 overexpressing CRHR2 had higher migratory potential than parental NCM460 that was further enhanced by Ucn2 and reversed by Ast2B (Figure 3F). These results further support the notion previously addressed that CRHR2/Ucn2 signaling might interfere differently with regulators of cell growth and migration in normal and malignant CRC cells.
Furthermore, to explore the contribution of CRHR2/Ucn2 signaling in regulating EMT induction in nonmetastatic CRC cells, we monitored the migration rates of SW480-CRHR2+ and SW480-EV cells transfected with Snail in the presence of TGFβ1 with or without Ucn2 treatment (Figure 3G). Transfection of both clones with the degradation-resistant SnailS6A mutant increased the migration rates after all treatments. As expected, cell migration was further enhanced by exposure to TGFβ1. However, compared with the SnailS6A-expressing EV cells, SnailS6A-expressing CRHR2+ cells had lower responses to TGFβ1 and a further reduction in the presence of Ucn2. In addition, Ucn2 inhibited only the TGFβ1-mediated migration of SnailS6A-expressing CRHR2+ cells.
These observations suggest that CRHR2/Ucn2 signaling interferes negatively with both the metastatic potential of CRC cells that have already undergone EMT and the potency of EMT progression in epithelial CRC cells under optimal conditions. Our overall findings imply CRHR2/Ucn2 signaling as a putative negative regulator of tumor growth and oncogenic EMT in CRC.
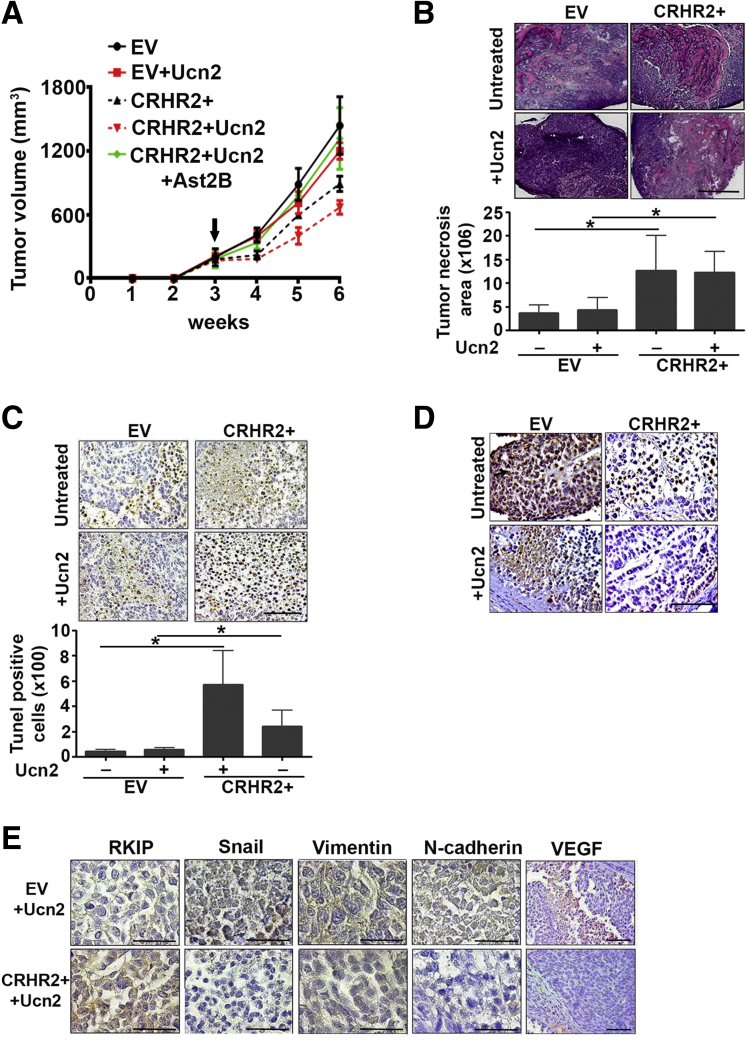
Corticotropin-Releasing Hormone Receptor 2/Urocortin-2 Signaling Inhibits Colorectal Cancer Growth and Epithelial-to-Mesenchymal Transition in Vivo
The impact of CRHR2 overexpression on CRC proliferation and EMT rates was validated in vivo in mice bearing SW620-CRHR2+ or SW620-EV xenografts. Tumor volume was assessed under different conditions (with or without Ucn2 and/or Ast2B) (Figure 4A). CRHR2+ tumors were smaller and volume was further reduced after Ucn2 administration. Ast2B reversed this effect, suggesting that inhibition of tumor growth was CRHR2 dependent. We also found increased necrosis and apoptosis as well as reduced PCNA expression in CRHR2+ tumors, as assessed by H&E (Figure 4B), terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling (Figure 4C), and IHC (Figure 4D) stainings, respectively.
Figure 4.
Corticotropin-releasing hormone receptor 2 (CRHR2)/urocortin-2 (Ucn2) signaling inhibits specifically colorectal cancer (CRC) growth and expression of epithelial-to-mesenchymal transition (EMT) markers in vivo. (A) Reduced tumor volume in mice bearing SW620-CRHR2+ xenografts with or without intratumoral Ucn2 administration. Black arrow indicates treatment initiation. Statistically significant differences in tumor volume were observed at weeks 4, 5, and 6 between the following groups: EV versus CRHR2+ (P < .003), EV/Ucn2 versus CRHR2+/Ucn2 (P < .001), and CRHR2+/Ucn2 versus CRHR2+/Ucn2/Ast2B (P < .004). (B) Increased necrosis in resected SW620-CRHR2+ xenograft specimens versus EV (empty virus) control, as assessed by H&E staining. Scale bar: 2 mm. *P < .05. (C) Increased tumor apoptosis in SW620-CRHR2+ specimens, as assessed by terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling assay. Scale bar: 50 μm. *P < .05. (D) Low proliferating cell nuclear antigen (PCNA) expression in SW620-CRHR2+ specimens indicates reduced tumor proliferation. Scale bar: 50 μm. Black arrow indicates the specific PCNA staining. (E) Induced expression of the metastasis-suppressor Raf-1 kinase inhibitory protein (RKIP) and inhibition of the EMT-inducers vimentin, N-cadherin, and Snail as well as the angiogenesis promoting VEGF in SW620-CRHR2+ specimens derived from Ucn2-treated mice. Scale bars: 50 μm (scale bars are proportional to the original microscope-derived images).
The magnitude of these changes was increased with Ucn2 administration. EMT-inducing gene products such as vimentin, N-cadherin, and Snail were down-regulated in CRHR2+ tumors derived by Ucn2-treated mice accompanied by increased expression of the EMT-suppressor RKIP (Figure 4E). Ucn2-treated CRHR2+ tumor specimens had also decreased VEGF expression, suggesting that angiogenesis could be a possible target of CRHR2/Ucn2 signaling (Figure 4E). These observations validate our in vitro findings on the role of CRHR2 in suppression of CRC proliferation and EMT, at least at the molecular level.
Corticotropin-Releasing Hormone Receptor 2 Induction Inhibits Proinflammatory Cytokine Expression and Interleukin-6-Mediated pSTAT3 Activation in Colorectal Cancer
Several molecular events in chronic inflammation contribute to multistage CRC carcinogenesis and progression in the inflamed colon, including up-regulated proinflammatory cytokines such as IL-1b, IL-6, IL-8, and tumor necrosis factor α (TNFα) expressed by colonic and infiltrating immune cells.23 Given the involvement of the CRH-family in regulating colonic inflammation,3, 4 we hypothesized that the effects of CRHR2/Ucn2 signaling in CRC proliferation, migration, and invasion might be driven by CRHR2-dependent changes in the inflammatory profile of CRHR2+ CRC cells. As such, we compared the expression of proinflammatory cytokines between parental (EV) and CRHR2+ cells.
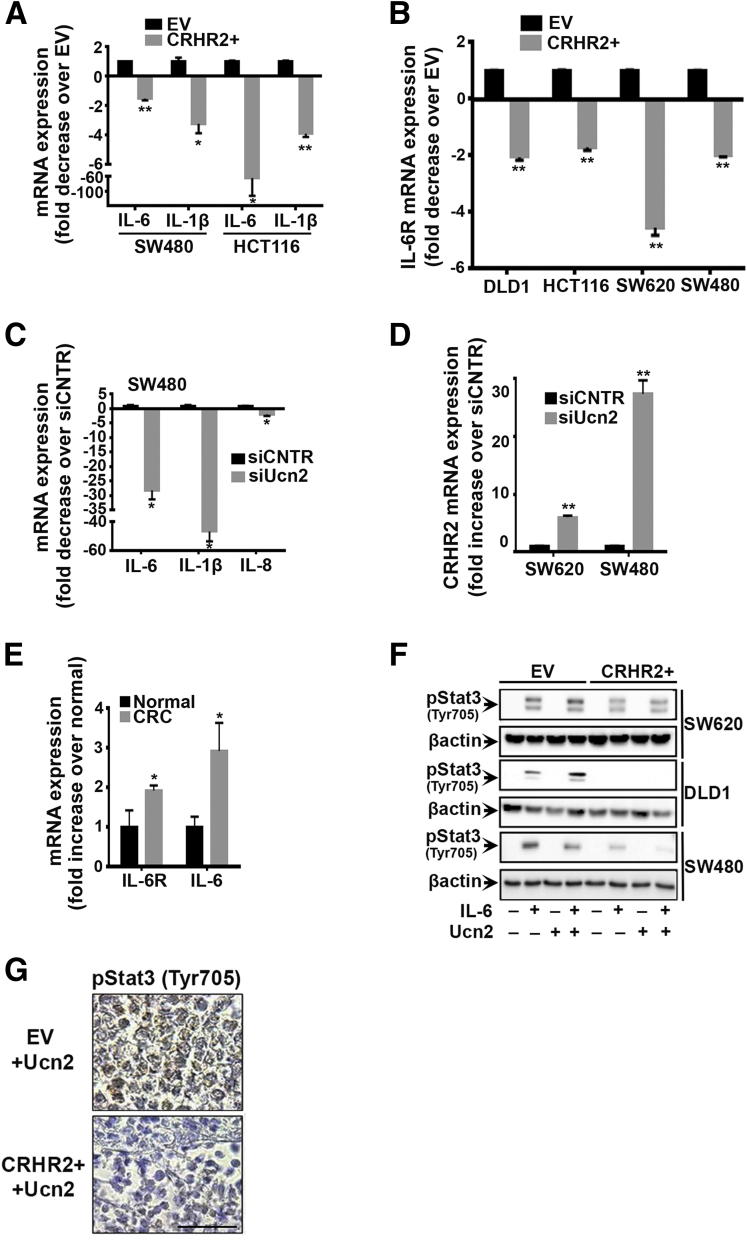
Among the cytokines screened in CRHR2+ cells we detected reduced levels of IL-1b and IL-6 mRNA (Figure 5A), as well as inhibition of IL-6R mRNA expression (Figure 5B). Notably, inhibition of the proinflammatory cytokines IL-6, IL-1b, and IL-8 was also observed in CRC cells after Ucn2 silencing by siRNA (Figure 5C), in parallel with CRHR2 up-regulation (Figure 5D). IL-6 and IL-6R mRNAs were also found elevated in CRC samples compared to healthy controls (Figure 5E). Our overall in vitro findings suggest CRHR2 as a negative mediator of proinflammatory signals produced by the CRC cells, which might contribute to their expansion and progression. In addition, excess production of Ucn2 by CRC cells might contribute to sustaining colonic inflammation, possibly through targeting other cells in the tumor microenvironment.
Figure 5.
Corticotropin-releasing hormone receptor 2 (CRHR2) suppresses inflammatory responses and interleukin-6 (IL-6)-mediated signal transducer and activator of transcription 3 (Stat3) activation in colorectal cancer (CRC). (A) Decreased IL-6 and IL-1b mRNAs in CRHR2+ versus EV (empty virus) CRC cells. *P < .05; **P < .01. (B) Inhibition of IL-6R mRNA expression in CRHR2+ cells compared with EV. **P < .01. (C) Urocortin-2 (Ucn2) inhibition by small interfering RNA diminishes IL-6, IL-1b, and IL-8 mRNA expression in SW480 cells (*P < .05) and (D) up-regulates CRHR2 mRNA expression in SW620 and SW480 cells (**P < .01). (E) CRC specimens express higher IL-6 and IL-6R mRNA levels than controls. *P < .05. (F) Inhibition of IL-6-induced Stat3 (Tyr705) phosphorylation in CRHR2+ cells after stimulation with 20 ng/mL IL-6 and/or 0.1 μM Ucn2 for 30 minutes, as assessed by Western blot. Data shown are from a representative experiment. Lanes of bands shown have been cropped by the original whole gel images. (G) Validation of pStat3 (Tyr705) inhibition in SW620-CRHR2+ resected tumors from mice treated with Ucn2. Scale bar: 50 μm (scale bar proportional to the original microscope-derived image).
Inflammation targets several main regulators of cell growth, invasion, and angiogenesis, including Stat3.24 In CRC in vivo models, Stat3 down-regulation decreases tumorigenicity and cell growth.18, 24 We explored the effects of CRHR2-induction on IL-6-mediated Stat3 activation in CRC because Stat3 is a major downstream target of IL-6/IL-6R signaling and is constitutively active in almost all cancers including CRC.18
SW620-, DLD1- and SW480-CRHR2+ clones had decreased Stat3 (Tyr705) phosphorylation after stimulation with IL-6 (Figure 5F). Inhibition was more pronounced in some clones in the presence of exogenous Ucn2, suggesting that endogenous Ucn2 can also stimulate CRHR2 signaling that leads to efficient pStat3 (Tyr705) down-regulation. The failure to detect pStat3 (Tyr705) baseline levels is likely due to the isolation of cancer cells from the tumor microenvironment as it has been previously reported elsewhere.18 These findings suggest that CRHR2 signaling may negatively regulate IL-6-mediated Stat3 activation in CRC. Inhibition of pStat3 (Tyr705) was also validated in vivo in SW620-CRHR2+ resected xenografts treated with Ucn2, as shown by IHC (Figure 5G).
Corticotropin-Releasing Hormone Receptor 2/Urocortin-2 Signaling Inhibits Interleukin-6–Mediated Colorectal Cancer Growth and Oncogenic Epithelial-to-Mesenchymal Transition in Part Through Regulation of Stat3-Targeted Genes
In addition to proliferation, inflammation in malignant tumors positively impacts tumor metastasis through regulation of oncogenic EMT.25 Given the CRHR2-mediated Stat3 inhibition, we explored the role of CRHR2/Ucn2 signaling in CRC proliferative and EMT responses in the presence of IL-6.
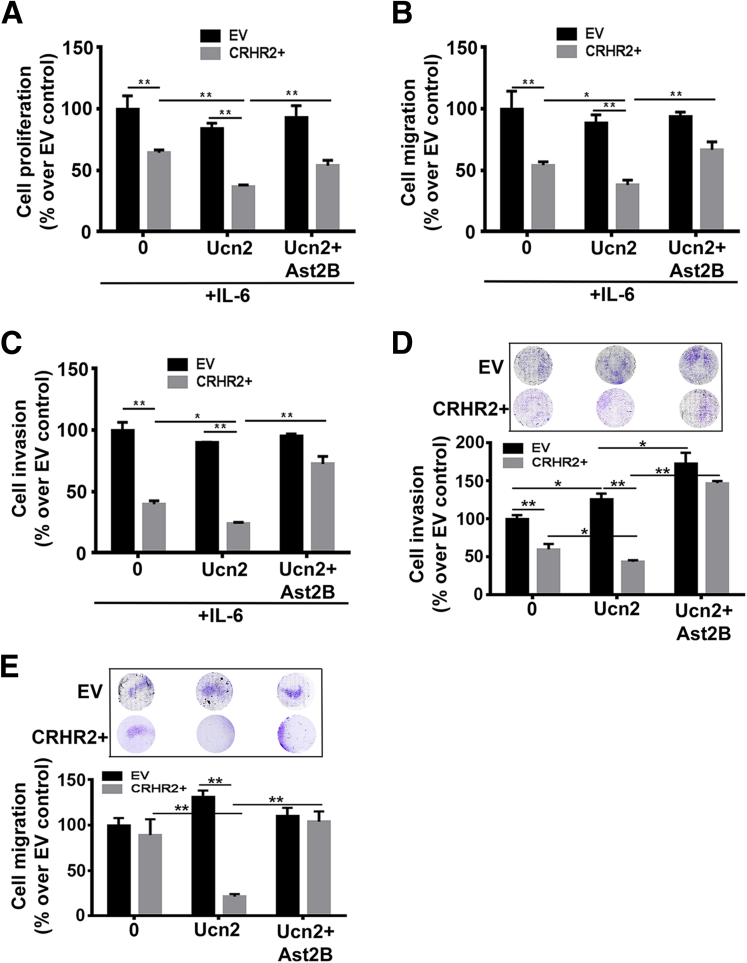
Although IL-6 enhanced the baseline proliferation of both SW480-CRHR2+ and SW480-EV cells (data not shown), CRHR2+ growth was significantly slower than EV (Figure 6A). This inhibition was more pronounced in the presence of exogenous Ucn2 and reversed by Ast2B only in CRHR2+ cells, suggesting a CRHR2-specific effect. Proliferation of EV cells did not statistically significantly change in presence of Ucn2 alone or in combination with Ast2B. Concomitantly, CRHR2-driven and CRHR2-specific inhibition was also observed in IL-6-induced migration (Figure 6B) and invasion (Figure 6C) of SW620-CRHR2+ cells in the absence or presence of Ucn2 treatment. Similarly, CRHR2 induction and stimulation with Ucn2 decreased the invasion and migration of the nonmetastatic cell line DLD1 after pretreatment with IL-6 for 72 hours (Figure 6D and E, respectively). Surprisingly, Ucn2 increased specifically the invasion of IL-6 pretreated DLD1-EV cells but not their migration. These findings suggest the involvement of the CRHR2/Ucn2 signaling in the regulation of IL-6-induced tumor proliferation and EMT onset and/or progression in CRC.
Figure 6.
Corticotropin-releasing hormone receptor 2 (CRHR2)/urocortin-2 (Ucn2) signaling inhibits interleukin-6 (IL-6)-mediated colorectal cancer (CRC) growth, invasion, and migration. (A) CRHR2-dependant inhibition of IL-6-mediated cell proliferation of SW480-CRHR2+ cells. (B, C) CRHR2 specific inhibition of (B) migration and (C) invasion of SW620-CRHR2+ cells. *P ≤ .05; **P ≤ .01. (D) CRHR2 induction inhibits specifically the baseline and Ucn2-mediated invasion of the nonmetastatic cell line DLD1. *P < .05; **P < .01. (E) CRHR2-mediated inhibition of Ucn2-mediated migration of DLD1-CRHR2+ cells. **P < .01. In all assays CRHR2+ and EV CRC cells were treated with 20 ng/mL IL-6 alone or in combination with 0.1 μM Ucn2 ± 1 μM Ast2B for 72 hours for proliferation assessment or 48 hours for migration and invasion monitoring. DLD1 cells were pretreated with 20 ng/mL IL-6 for 72 hours before migration and invasion settings.
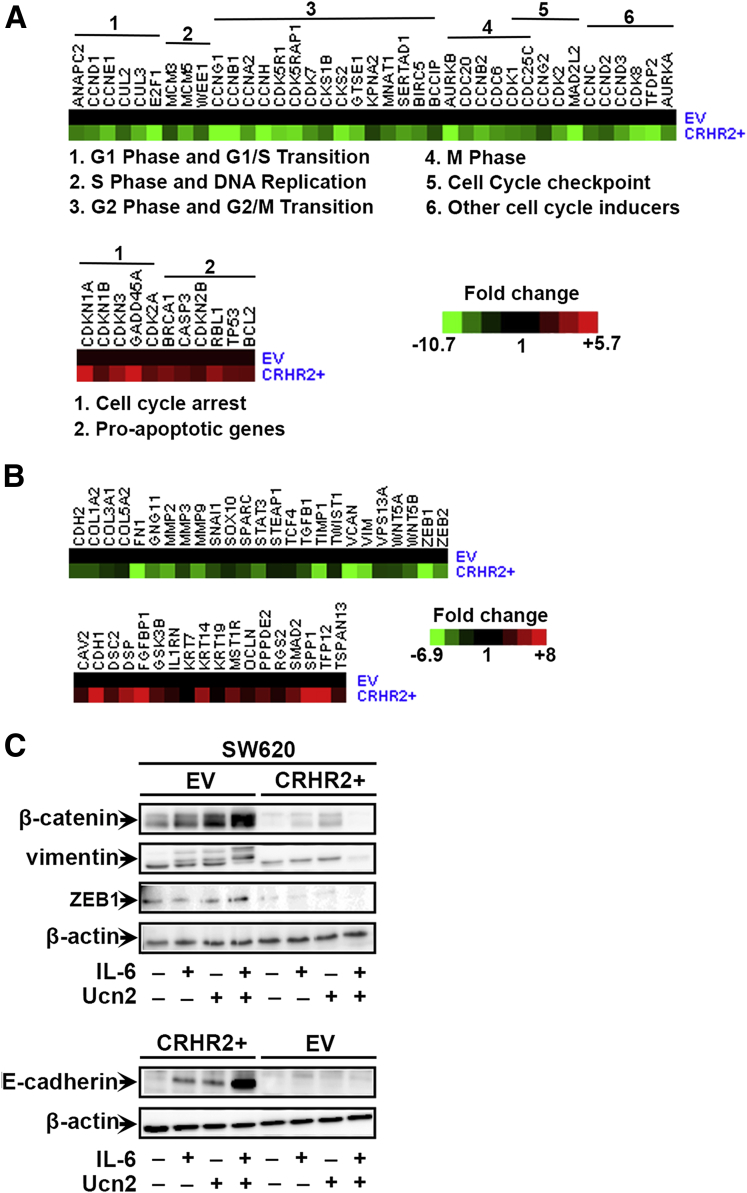
To identify cell cycle– and EMT-relevant gene targets of CRHR2/Ucn2 signaling in CRC under inflammatory conditions at a molecular level, we performed cell cycle– and EMT-specific gene arrays using cDNAs from SW480 and SW620 parental and CRHR2 clones, respectively, treated with Ucn2 and IL-6. The SW480-CRHR2+ cells had lower expression of several positive cell cycle regulators involved in G1 phase and G1/S transition, S phase and DNA replication, G2 phase and G2/M transition, as well as M phase progression (Figure 7A, upper panel). Contrarily, induced expression of genes involved in the regulation of cell cycle checkpoint and arrest such as cyclin inhibitors as well as proapoptotic genes Bcl2, Casp3, and TP53 was found in CRHR2+ cells (Figure 7A, lower panel). Notably, a number of dysregulated genes in CRHR2+ cells, including members of the cyclin superfamily and others, are regulated by Stat3. This suggests that CRHR2/Ucn2 signaling might mediate inhibitory effects on CRC growth through inhibition of Stat3 under inflammatory conditions.
Figure 7.
Corticotropin-releasing hormone receptor 2 (CRHR2)/urocortin-2 (Ucn2) signaling inhibits interleukin-6 (IL-6)-mediated colorectal cancer (CRC) growth and oncogenic epithelial-to-mesenchymal transition (EMT) through regulation of cell cycle- and EMT-related genes. (A) Heat map representing cell cycle–related gene array data derived by SW480-EV and SW480-CRHR2+ cells treated with 20 ng/mL IL-6 and 0.1 μM Ucn2 for 18 hours. Upper and lower panels: Down-regulation of cell cycle-promoting genes and up-regulation of cell cycle-suppressors in SW480-CRHR2+ cells, respectively. (B) Heat map representing EMT-related gene array data derived by SW620-EV and SW620-CRHR2+ cells treated with 20 ng/mL IL-6 and 0.1 μM Ucn2 for 18 hours. Upper and lower panels: Down-regulation of EMT-promoting genes and up-regulation of EMT-suppressors in SW620-CRHR2+ cells, respectively. (C) Validation of EMT inhibition at protein level in SW620-CRHR2+ protein extracts derived after cell treatment with 20 ng/mL IL-6 ± 0.1 μM Ucn2. Lanes of bands shown have been cropped by the original whole gel images.
In an EMT-specific gene array analysis, SW620-CRHR2+ cells treated with IL-6 and Ucn2 had lower expression of EMT-promoting genes, including genes involved in cell migration and motility such as MMPs, CDH2 (N-cadherin), Vim (Vimentin), TGFβ1 and members of the Snail family of transcription factors (Snail, Twist, ZEB1, and ZEB2) (Figure 7B, upper panel). In contrast, CRHR2+ cells had higher levels of critical EMT-suppressor genes including CDH1 (E-cadherin) and GSK3β (inhibitor of Snail and b-catenin) (Figure 7B, lower panel). Along with cell cycle markers, MMPs, Snail, Twist1, CDH1, ZEBs, and other EMT-regulatory genes with altered expression in CRHR2+ clones are also downstream targets of Stat3.
The dysregulated expression of some of these gene products was validated at the protein level by Western blot analysis (Figure 7C). SW60-CRHR2+ cells had decreased expression of ZEB as well as vimentin and β-catenin in all treatments. In contrast, expression of E-cadherin was up-regulated in CRHR2+ cells. Our molecular findings overall suggest that CRHR2/Ucn2 signaling might mediate its inhibitory effects on IL-6-triggered proliferation and oncogenic EMT, at least in part, through Stat3 down-regulation.
Low Corticotropin-Releasing Hormone Receptor 2 Expression in Colorectal Cancer Specimens Is Correlated With Increased Interleukin-6R and Vimentin Levels, Distant Metastasis, and Poor Overall Survival
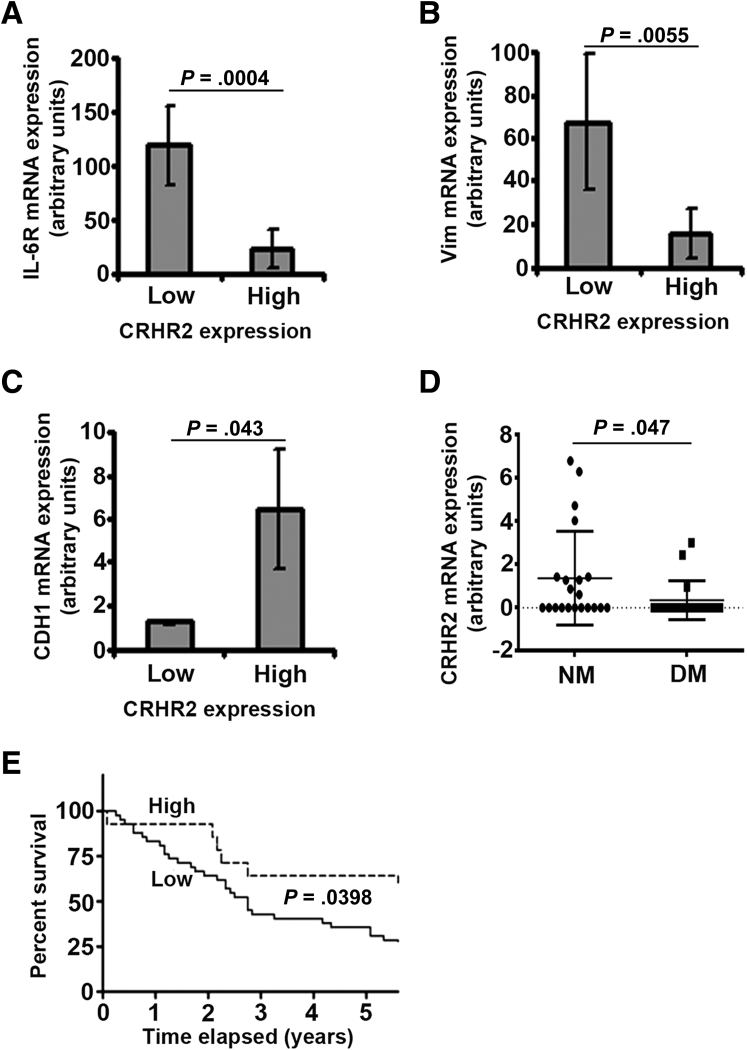
CRHR2, IL-6R, CDH1, and Vim mRNA expression was assessed by qRT-PCR in all 56 human CRC samples and the mean ± SEM value was calculated for each gene independently of any clinicopathologic parameter. Among the 56 CRC samples tested, 42 had undetectable CRHR2 mRNA expression (cycle threshold above 40 cycles that was translated as 0 expression), 3 low [set as CRHR2 levels < mean ± SEM (0.7215 + 0.22 = 0.9415)] and 11 samples high (set as expression > mean ± SEM). Analysis of CRHR2, E-cadherin, IL-6R, and vimentin mRNA levels in CRC revealed that samples with low CRHR2 expression (n = 3) had statistically significantly higher IL-6R (Figure 8A) and vimentin (Figure 8B) expression levels than those with high CRHR2 (n = 11). In contrast, CRC samples with high CRHR2 mRNA levels had significant evidence of more E-cadherin mRNA expression (Figure 8C). Analysis between CRHR2 positive (N = 14) and negative samples (those with undetectable CRHR2 mRNA levels, n = 42) revealed no statistically significant association between the mRNA expression of CRHR2 and any of the tested genes (data not shown).
Figure 8.
Low corticotropin-releasing hormone receptor 2 (CRHR2) mRNA expression in human colorectal cancer (CRC) samples inversely correlates with (A) interleukin-6 (IL-6R) and (B) vimentin mRNA levels. (C) In contrast, a positive correlation was established between E-cadherin (CDH1) and high CRHR2 mRNA expression. Low and high CRHR2 expressions were set as detectable CRHR2 mRNA levels < mean ± standard error of the mean (SEM) or > mean ± SEM, respectively, calculated by the total CRC samples (n = 56). Among the 56 CRC samples tested, 3 were found with low CRHR2 expression, 11 with high expression, and the remaining 42 with undetectable expression. (D) Decreased expression of CRHR2 in CRC samples positively correlates with occurrence of distant metastasis. DM, distant metastasis (n = 18); NM, no metastasis (n = 21). (E) CRC patients with high tumoral CRHR2 mRNA expression (n = 11) have an increased 5-year survival rate compared with patients with low CRHR2 expression (n = 3).
Based on the available clinicopathologic data (Table 1), decreased CRHR2 expression in CRC patients also had evidence of higher occurrence of distant metastasis (Figure 8D) and of poor prognosis for 5-year survival after treatment initiation (Figure 8E). A marginally significant positive association (P = .05) was also observed between CRHR2 expression and normal weight patients as compared with the overweight patients. No associations were established between CRHR2 expression and tumor grade (based on Dukes or TNM classification) or tumor differentiation neither at the mRNA (Table 1) or protein levels (data not shown). Ucn2 expression in the same patients did not reveal any associations with any of the studied clinicopathologic characteristics (Table 1). These findings suggest that CRHR2 levels might serve as a putative prognostic indicator for tumor metastatic activity and overall patient outcome.
Discussion
This study provides clear evidence for dysregulated expression of CRHR2 and its specific agonist Ucn2 in CRC that impacts critically tumor survival and expansion, at least, through sustaining inflammatory signals and Stat3 activation in the colon. We introduce CRHR2 distribution and signaling through Ucn2 as a novel negative modulator of CRC growth in vitro and in vivo as well as a suppressor of oncogenic EMT that associates with less risk for distant metastasis and better overall patient survival. Our current findings introduce possible links among inflammation, the CRH axis, and CRC, but future studies on an inflammation-associated animal model and CRC patients with IBD history are necessary to further support a role of the CRH system in inflammation-associated CRC.
It is well-accepted that many CRC cases are linked to the duration and extension of mucosal inflammation. This is sustained by inflammatory cells and the tumor cells themselves, which modulate the progression of colon carcinogenesis by producing tumor-supportive cytokines such as IL-6 or by stimulating synthesis of molecules with mitogenic effects.26 We show that CRHR2 loss in the malignant colon promotes and/or sustains proinflammatory signals mediated in part by IL-6 and its receptor IL-6R. This finding suggests that CRHR2high CRC tumors may be less responsive to IL-6 that might be present in high levels within the tumor microenvironment. This was further validated in human CRC tissues, where an inverse correlation between CRHR2high and IL-6Rhigh was established.
Concomitantly, Ucn2 up-regulation found on CRC also contributed positively in maintaining or promoting high IL-6 levels. Although activation of CRHR1 typically has been shown to favor proinflammatory responses that contribute to the development of colitis-associated cancer in mouse models27 and CRHR2 to drive mainly anti-inflammatory responses,28, 29 these effects appear to depend on receptor/ligand distribution on colonic mucosa as well as inflammation status (acute or chronic).30, 31 Thus, the concentration of Ucn available intracellularly or in the microenvironment might play a critical role in CRHR2-driven pro/anti-inflammatory responses.
In addition, the reported anti-inflammatory actions of Ucn, regulated via CRHR2, have been evidenced only at low peptide concentrations.32 A rat model of chemically induced colitis illustrated a proinflammatory action for elevated Ucn2, accompanied by a reduction in CRHR2.5 In other tissues, such as smooth muscles, Ucn2 is also reported to support topical inflammation through induction of IL-6.33 This suggests that Ucn2-mediated inflammation might be critically involved in CRC and associated with IBD-related colon cancer. CRHR2 induction in CRC cell lines resulted in decreased mRNA expression of endogenous Ucn2, while silencing of endogenous Ucn2 in parental CRC cell lines up-regulated CRHR2 expression. Although it is likely that endogenous Ucn2 up-regulation might account for the down-regulation of the respective receptor in CRC cells or vice versa,5 this is an area worth investigating in the future.
Most inflammation-related autocrine-paracrine events converging in tumor cells typically result in Stat3 activation, which mediates a transcriptional response favoring tumor survival, proliferation, and angiogenesis.24 Functionally, the most important Stat3 activators are IL-6 and IL-10. Recently, Stat3 has been proposed as a molecular link between intestinal inflammation and tumorigenesis.34 Accumulated evidence indicates that abnormalities in the Janus kinase (JAK)/Stat pathway are involved in CRC oncogenesis,35 with aberrant and persistent Stat3 activation being a frequent observation in human CRC, often associated with poor outcome.18, 36 We demonstrate for the first time that Stat3 is a downstream target of CRHR2/Ucn2 signaling in CRC, with CRHR2 induction suppressing IL-6-mediated Stat3 (Tyr703) phosphorylation in vitro and in vivo. As such, Stat3 inhibition in CRHR2+ CRC cells might confer decreased tumor cell proliferation and diminished tumor growth observed in mice bearing CRHR2+ CRC tumors.
Stat3 activation promotes cell cycle G1/S phase transition in gastric, colon, and squamous cell carcinomas by stimulating transcription of CCNB1, CDK1, and CCND1 along with repression of the cell cycle inhibitor p21 (CDKN1A).35, 36, 37, 38, 39, 40, 41, 42 Consistent with CRHR2-mediated Stat3 inhibition are our findings from the cell cycle gene arrays showing CCNB1, CCND1, and CDK1 down-regulation, induction of CDKN1A, and dysregulated expression of genes involved in G1 phase regulation and G1/S transition, including ANAPC2, CCNE1, CDKN1B, CUL2, CUL3, and E2F1 in CRHR2+ CRC cells. In addition, Stat3-mediated induction of survivin (BIRC5)43 and suppression of p53 (TP53)44 were eliminated in CRHR2+ cells, indicating that CRHR2 also interferes negatively with Stat3-mediated apoptosis suppression. The antiproliferative effects of CRHR2/Ucn2 signaling were further validated in a xenograft model with CRHR2+ tumors to show CRHR2+-specific growth inhibition due to increased necrosis and apoptosis.
As with the majority of cancers, the progression of CRC to invasive and metastatic disease may involve localized occurrences of oncogenic EMT. Key EMT-inducers in CRC include Ras, Wnt, PI3K/Akt, and TGFβ.45 Recently, the Stat3 pathway was found to arbitrate the EMT process directly in CRC cells through induction of cell motility, migration and invasion resulting from increased ZEB1, vimentin, and N-cadherin expression and suppression of E-cadherin.46 To our knowledge, this is the first report demonstrating negative effects of CRHR2/Ucn2 signaling on oncogenic EMT. Previous studies have reported inhibitory effects of CRHR2/CRH and CRHR2/Ucn in EMT properties of TGFβ-treated breast cancer and hepatoma cell lines, respectively.47, 48 We show that CRHR2/Ucn2 signaling triggers inhibition of baseline and IL-6-mediated tumor migration and invasion accompanied by inhibition of mesenchymal markers at the mRNA and protein levels (in vitro and in vivo) and induction of EMT-suppressors including E-cadherin.
The fact that the dysregulated EMT-related gene signature in IL-6- treated CRHR2+ cells contained several Stat3-targeted EMT gene regulators suggests that CRHR2/Ucn2 signaling might mediate its suppressing effects on EMT in CRC at least via modulation of the IL-6/Stat3 pathway and its downstream targets. This is supported by previous reports on other cancers showing dependence of EGF-induced EMT by IL-6R and the JAK2/Stat3 pathway in ovarian cancer49 and IL-6-induced head and neck oncogenic EMT via the JAK/Stat3/Snail pathway.50
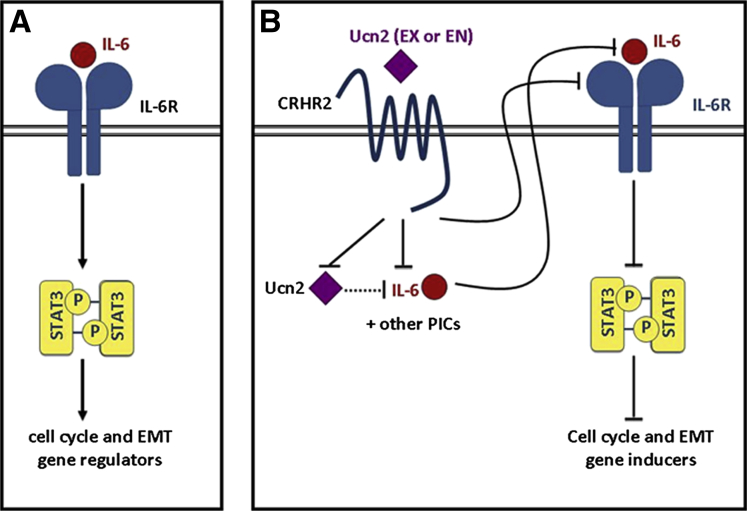
We also found decreased VEGF expression in CRHR2+ xenografts in our in vivo model. VEGF is a prominent transcriptional target for Stat3,51 thus suggesting that CRHR2/Ucn2 might partially affect angiogenesis in CRC through Stat3 suppression. Overall, our findings point to Stat3 as one of the underlying molecular targets of CRHR2 signaling that mediates CRHR2/Ucn2 effects on CRC survival and expansion (Figure 9).
Figure 9.
Schematic representation of the corticotropin-releasing hormone receptor 2 (CRHR2)/urocortin-2 (Ucn2) targeted pathways and downstream events taking place after ectopic expression of CRHR2 in colorectal cancer (CRC) cells. (A) Constitutive activation of STAT3 in CRC by interleukin-6 (IL-6)/IL-6R signaling promotes expression of Stat3-targeted genes that regulate cell cycle and epithelial-to-mesenchymal transition (EMT). (B) CRHR2/Ucn2 signaling inhibits the expression of tumor-produced proinflammatory cytokines (PICs) such as IL-1b or IL-6 either via Ucn2 down-regulation or through a Ucn2-independent manner. CRHR2/Ucn2 also decreases the expression of IL-6R on the surface of CRC cells, thus resulting in attenuation of autocrine- or exocrine-mediated IL6R/IL6 signaling, which in turn leads to significant inhibition of Stat3 phosphorylation and its targeted genes. As constitutively active Stat3 is one of the major regulators of cell cycle and EMT in CRC cells, CRHR2/Ucn2-mediated Stat3 inhibition might facilitate at least in part the decreased proliferative, migratory, and invasive responses shown in CRHR2+ CRC cells. End, endogenous; Ex, exogenous.
Our findings demonstrating CRHR2/Ucn2-mediated inhibition of CRC migratory potential are partially opposed to recently reported findings that show CRHR2/Ucn3 signaling to favor CRC migration through negative regulation of cellular adhesion.17 We also failed to detect overexpression of CRHR2 and Ucn3 in CRC tissues and many CRC cell lines, or to establish positive correlations with high-grade and poorly differentiated tumors as observed by Ducarouge et al.17 These differing results can be also explained by the larger number of tissue samples (both CRC and normal) we used in our study. In addition, the in vivo validation of our in vitro findings strongly supports our proposed role of CRHR2/Ucn2 signaling on negative regulation of CRC growth and EMT.
Finally, we demonstrated inverse correlations among CRHR2, vimentin, and IL-6R expression as well as positive CRHR2 associations with E-cadherin in clinical CRC samples. CRHR2 underexpression or loss in CRC was further associated with increased incidence of distant metastases and poor survival of CRC patients, suggesting a potential clinical relevance of CRHR2 decrease/loss on CRC outcome.
Given the high incidence of CRC cases worldwide and increased lethality due to distant metastases, identifying novel regulatory pathways involved in CRC growth and metastasis is paramount. We present novel findings on the role of the CRH system in CRC pathophysiology and molecular evidence to link CRHR2/Ucn2 signaling with CRC growth and aggressiveness through modulation of inflammatory responses in the colon. We propose that evaluation of CRHR2/Ucn2 expression in CRC may benefit early diagnosis of aggressive tumors and serve as a novel therapeutic target for prevention and treatment of advanced stages of CRC.
Acknowledgments
The authors thank the UCLA Vector Core facility (supported by grants JCCC/P130/P30 CA016042 and CURE/P30 DK041301) for providing us with the lentivirus constructs.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was funded by National Institutes of Health grants T32 HD007512–15 (to J.A.R.), PO-1 DK33506 (to C.P.), P50 DK64539 (to C.P., L.C.), the Blinder Research Foundation for Crohn’s Disease (to I.K.M.L.), the Charles H. Hood Foundation (to D.I.), and the Cure DDRC P30 DK41301/CTSI UL1TR000124 PFS (to S.B.).
References
- 1.Terzic J., Grivennikov S., Karin E. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 2.Gay J., Kokkotou E., O’Brien M. Corticotropin-releasing hormone deficiency is associated with reduced local inflammation in a mouse model of experimental colitis. Endocrinology. 2008;149:3403–3409. doi: 10.1210/en.2007-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paschos K.A., Kolios G., Chatzaki E. The corticotropin-releasing factor system in inflammatory bowel disease: prospects for new therapeutic approaches. Drug Discov Today. 2009;14:713–720. doi: 10.1016/j.drudis.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Moss A.C., Anton P., Savidge T. Urocortin II mediates pro-inflammatory effects in human colonocytes via corticotropin-releasing hormone receptor 2alpha. Gut. 2007;56:1210–1217. doi: 10.1136/gut.2006.110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang J., Hoy J.J., Idumalla P.S. Urocortin 2 expression in the rat gastrointestinal tract under basal conditions and in chemical colitis. Peptides. 2007;28:1453–1460. doi: 10.1016/j.peptides.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reubi J.C., Waser B., Vale W. Expression of CRF1 and CRF2 receptors in human cancers. J Clin Endocrinol Metab. 2003;88:3312–3320. doi: 10.1210/jc.2002-021853. [DOI] [PubMed] [Google Scholar]
- 7.Suda T., Tomori N., Yajima F. Characterization of immunoreactive corticotropin and corticotropin-releasing factor in human adrenal and ovarian tumours. Acta Endocrinol (Copenh) 1986;111:546–552. doi: 10.1530/acta.0.1110546. [DOI] [PubMed] [Google Scholar]
- 8.Miceli F., Ranelletti F.O., Martinelli E. Expression and subcellular localization of CRH and its receptors in human endometrial cancer. Mol Cell Endocrinol. 2009;305:6–11. doi: 10.1016/j.mce.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Ciocca D.R., Puy L.A., Fasoli L.C. Corticotropin-releasing hormone, luteinizing hormone-releasing hormone, growth hormone-releasing hormone, and somatostatin-like immunoreactivities in biopsies from breast cancer patients. Breast Cancer Res Treat. 1990;15:175–184. doi: 10.1007/BF01806354. [DOI] [PubMed] [Google Scholar]
- 10.Minas V., Rolaki A., Kalantaridou S.N. Intratumoral CRH modulates immuno-escape of ovarian cancer cells through FasL regulation. Br J Cancer. 2007;97:637–645. doi: 10.1038/sj.bjc.6603918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo Y.H., Choi Y.J., Kim H.O. Corticotropin-releasing hormone enhances the invasiveness and migration of Ishikawa cells, possibly by increasing matrix metalloproteinase-2 and matrix metalloproteinase-9. J Int Med Res. 2011;39:2067–2075. doi: 10.1177/147323001103900602. [DOI] [PubMed] [Google Scholar]
- 12.Graziani G., Tentori L., Portarena I. CRH inhibits cell growth of human endometrial adenocarcinoma cells via CRH-receptor 1-mediated activation of cAMP-PKA pathway. Endocrinology. 2002;143:807–813. doi: 10.1210/endo.143.3.8694. [DOI] [PubMed] [Google Scholar]
- 13.Graziani G., Tentori L., Muzi A. Evidence that corticotropin-releasing hormone inhibits cell growth of human breast cancer cells via the activation of CRH-R1 receptor subtype. Mol Cell Endocrinol. 2007;264:44–49. doi: 10.1016/j.mce.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Chatzaki E., Lambropoulou M., Constantinidis T.C. Corticotropin-releasing factor (CRF) receptor type 2 in the human stomach: protective biological role by inhibition of apoptosis. J Cell Physiol. 2006;209:905–911. doi: 10.1002/jcp.20792. [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Xu Y., Xu Y. Urocortin’s inhibition of tumor growth and angiogenesis in hepatocellular carcinoma via corticotrophin-releasing factor receptor 2. Cancer Invest. 2008;26:359–368. doi: 10.1080/07357900701788106. [DOI] [PubMed] [Google Scholar]
- 16.Tezval H., Jurk S., Atschekzei F. The involvement of altered corticotropin releasing factor receptor 2 expression in prostate cancer due to alteration of anti-angiogenic signaling pathways. Prostate. 2009;69:443–448. doi: 10.1002/pros.20892. [DOI] [PubMed] [Google Scholar]
- 17.Ducarouge B., Pelissier-Rota M., Laine M. CRF2 signaling is a novel regulator of cellular adhesion and migration in colorectal cancer cells. PLoS One. 2013;8:e79335. doi: 10.1371/journal.pone.0079335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez-Cueto A., Hernandez-Cueto D., Antonio-Andres G. Death receptor 5 expression is inversely correlated with prostate cancer progression. Mol Med Rep. 2014;10:2279–2286. doi: 10.3892/mmr.2014.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corvinus F.M., Orth C., Moriggl R. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7:545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baritaki S., Chapman A., Yeung K. Inhibition of epithelial to mesenchymal transition in metastatic prostate cancer cells by the novel proteasome inhibitor, NPI-0052: pivotal roles of Snail repression and RKIP induction. Oncogene. 2009;28:3573–3585. doi: 10.1038/onc.2009.214. [DOI] [PubMed] [Google Scholar]
- 21.Baritaki S., Huerta-Yepez S., Sahakyan A. Mechanisms of nitric oxide-mediated inhibition of EMT in cancer: inhibition of the metastasis-inducer Snail and induction of the metastasis-suppressor RKIP. Cell Cycle. 2010;9:4931–4940. doi: 10.4161/cc.9.24.14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rokavec M., Oner M.G., Li H. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka T. Animal models of carcinogenesis in inflamed colorectum: potential use in chemoprevention study. Curr Drug Targets. 2012;13:1689–1697. doi: 10.2174/138945012804545452. [DOI] [PubMed] [Google Scholar]
- 24.Klampfer L. The role of signal transducers and activators of transcription in colon cancer. Front Biosci. 2008;13:2888–2899. doi: 10.2741/2893. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Novoa J.M., Nieto M.A. Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 2009;1:303–314. doi: 10.1002/emmm.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller M.M., Fusenig N.E. Friends or foes—bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y., Fang X., Yuan J. The role of corticotropin-releasing hormone receptor 1 in the development of colitis-associated cancer in mouse model. Endocr Relat Cancer. 2014;21:639–651. doi: 10.1530/ERC-14-0239. [DOI] [PubMed] [Google Scholar]
- 28.Dermitzaki E., Tsatsanis C., Minas V. Corticotropin-releasing factor (CRF) and the urocortins differentially regulate catecholamine secretion in human and rat adrenals, in a CRF receptor type-specific manner. Endocrinology. 2007;148:1524–1538. doi: 10.1210/en.2006-0967. [DOI] [PubMed] [Google Scholar]
- 29.Im E., Rhee S.H., Park Y.S. Corticotropin-releasing hormone family of peptides regulates intestinal angiogenesis. Gastroenterology. 2010;138:2457–2467. doi: 10.1053/j.gastro.2010.02.055. e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kokkotou E., Torres D., Moss A.C. Corticotropin-releasing hormone receptor 2-deficient mice have reduced intestinal inflammatory responses. J Immunol. 2006;177:3355–3361. doi: 10.4049/jimmunol.177.5.3355. [DOI] [PubMed] [Google Scholar]
- 31.Wlk M., Wang C.C., Venihaki M. Corticotropin-releasing hormone antagonists possess anti-inflammatory effects in the mouse ileum. Gastroenterology. 2002;123:505–515. doi: 10.1053/gast.2002.34783. [DOI] [PubMed] [Google Scholar]
- 32.Tsatsanis C., Androulidaki A., Dermitzaki E. Urocortin 1 and urocortin 2 induce macrophage apoptosis via CRFR2. FEBS Lett. 2005;579:4259–4264. doi: 10.1016/j.febslet.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 33.Kageyama K., Hanada K., Nigawara T. Urocortin induces interleukin-6 gene expression via cyclooxygenase-2 activity in aortic smooth muscle cells. Endocrinology. 2006;147:4454–4462. doi: 10.1210/en.2006-0008. [DOI] [PubMed] [Google Scholar]
- 34.Jarnicki A., Putoczki T., Ernst M. Stat3: linking inflammation to epithelial cancer—more than a “gut” feeling? Cell Div. 2010;5:14. doi: 10.1186/1747-1028-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spano J.P., Milano G., Rixe C. JAK/STAT signalling pathway in colorectal cancer: a new biological target with therapeutic implications. Eur J Cancer. 2006;42:2668–2670. doi: 10.1016/j.ejca.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Waldner M.J., Foersch S., Neurath M.F. Interleukin-6—a key regulator of colorectal cancer development. Int J Biol Sci. 2012;8:1248–1253. doi: 10.7150/ijbs.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bromberg J.F., Wrzeszczynska M.H., Devgan G. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 38.Masuda M., Suzui M., Yasumatu R. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–3355. [PubMed] [Google Scholar]
- 39.Bowman T., Broome M.A., Sinibaldi D. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci USA. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bollrath J., Phesse T.J., von Burstin V.A. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Kanda N., Seno H., Konda Y. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23:4921–4929. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 42.Chen C.L., Cen L., Kohout J. Signal transducer and activator of transcription 3 activation is associated with bladder cancer cell growth and survival. Mol Cancer. 2008;7:78. doi: 10.1186/1476-4598-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Connor D.S., Grossman D., Plescia J. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci USA. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu G., Wright K.L., Ma Y. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25:7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joyce T., Cantarella D., Isella C. A molecular signature for Epithelial to Mesenchymal transition in a human colon cancer cell system is revealed by large-scale microarray analysis. Clin Exp Metastasis. 2009;26:569–587. doi: 10.1007/s10585-009-9256-9. [DOI] [PubMed] [Google Scholar]
- 46.Xiong H., Hong J., Du W. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J Biol Chem. 2012;287:5819–5832. doi: 10.1074/jbc.M111.295964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin L., Li C., Li R. Corticotropin-releasing hormone receptors mediate apoptosis via cytosolic calcium-dependent phospholipase A(2) and migration in prostate cancer cell RM-1. J Mol Endocrinol. 2014;52:255–267. doi: 10.1530/JME-13-0270. [DOI] [PubMed] [Google Scholar]
- 48.Zhu C., Sun Z., Li C. Urocortin affects migration of hepatic cancer cell lines via differential regulation of cPLA2 and iPLA2. Cell Signal. 2014;26:1125–1134. doi: 10.1016/j.cellsig.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Colomiere M., Ward A.C., Riley C. Cross talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Br J Cancer. 2009;100:134–144. doi: 10.1038/sj.bjc.6604794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yadav A., Kumar B., Datta J. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res. 2011;9:1658–1667. doi: 10.1158/1541-7786.MCR-11-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei D., Le X., Zheng L. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–329. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]