Abstract
Current paradigms of carcinogenic risk suggest that genetic, hormonal, and environmental factors influence an individual’s predilection for developing metastatic breast cancer. Investigations of tumor latency and metastasis in mice have illustrated differences between inbred strains, but the possibility that mitochondrial genetic inheritance may contribute to such differences in vivo has not been directly tested. In this study, we tested this hypothesis in mitochondrial-nuclear exchange (MNX) mice we generated, where cohorts shared identical nuclear backgrounds but different mtDNA genomes on the background of the PyMT transgenic mouse model of spontaneous mammary carcinoma. In this setting, we found that primary tumor latency and metastasis segregated with mtDNA, suggesting that mtDNA influences disease progression to a far greater extent than previously appreciated. Our findings prompt further investigation into metabolic differences controlled by mitochondrial process as a basis for understanding tumor development and metastasis in individual subjects.
Keywords: Breast cancer, metastasis, mitochondrial DNA, PyMT
Introduction
Breast cancer affects 230,000 people and claims 40,000 lives annually in the United States (1). Age and ethnicity are both risk factors for breast cancer; interestingly, Caucasian women have the greatest risk of developing breast cancer, but African-American women have the highest risk of mortality, followed by Caucasian then Asian women (1). While certain nuclear genetic differences have garnered much attention regarding disease susceptibility with the BRCA1 and 2 mutations being leading examples (accounting for ~10–15% of patients), breast cancer is a complex disease with no known single cause (2–4). Furthermore, most studies investigating breast cancer susceptibility neglect to account for metastatic potential (5), which is what contributes most significantly to cancer morbidity and mortality. Consequently, a better understanding of both tumorigenesis and metastasis will complement and improve prognosis and treatment for breast cancer.
Several studies have shown differential susceptibility to tumor formation and metastatic spread in certain inbred mouse strains (3, 5–9). Hunter and colleagues were first to show metastatic susceptibility loci in a genetic screen crossing MMTV – polyoma middle T mice (FVB/N-Tg(MMTV-PyVT)634Mul/J, hereafter PyMT) on the FVB/NJ genetic background with different mice strains resulting in F1 progeny with different primary tumor latencies and metastatic potentials (8, 10, 11). Tumors from the PyMT model progress through stages that are comparable to human breast cancer, and biomarker expression in PyMT tumors is similarly consistent with those associated with poor outcome in human disease (12, 13). In work reported by Lifsted et al., F1 progeny generated from C57BL/6J or BALB/cJ mothers crossed with PyMT males were shown to be more resistant or susceptible, respectively, to advanced disease and metastases (8). Using these models, metastasis-modifier genes have been identified by comparative genomic analysis and were among the first to show inheritance of metastasis susceptibility (14, 15). However, the experimental design – male transgene mice crossed with females of other strains – left open the possibility that maternal mitochondrial DNA (mtDNA) inheritance was contributing to the observed changes in metastatic behavior.
Recent studies have reported that mtDNA mutations can significantly impact growth and metastatic potential in the tumor cell. For example, Ishikawa and colleagues performed reciprocal transfers of mtDNA from a highly and poorly metastatic mouse tumor cell lines and showed that metastatic potential segregated in a manner consistent with mtDNA background and was associated with reactive oxygen species production and complex I activity levels (16). This has led to suggestions that somatic mutations within the tumor mtDNA have an effect on oncogenic transformation. However, these studies do not address the impact of “normal” nuclear-mitochondrial genetic interactions nor do they consider that cancer development and metastatic potential can be influenced by ordinary mtDNA haplotypes (non-pathogenic mutations).
Our laboratory developed and utilized the MNX (Mitochondrial-Nuclear eXchange) mouse model in conjunction with the PyMT mammary tumor model to directly investigate the role of the mtDNA on breast cancer development (17). Through the process of nuclear transfer, MNX mice receive the intended nuclear and mitochondrial genetic components without a need for multiple backcrosses or exposures to mutagens, like ethidium bromide. Results herein provide direct support to the hypothesis that constitutional mitochondrial genetic background imparts a significant effect on breast cancer tumorigenicity and subsequent metastatic severity.
Materials and Methods
Generation of MNX mice
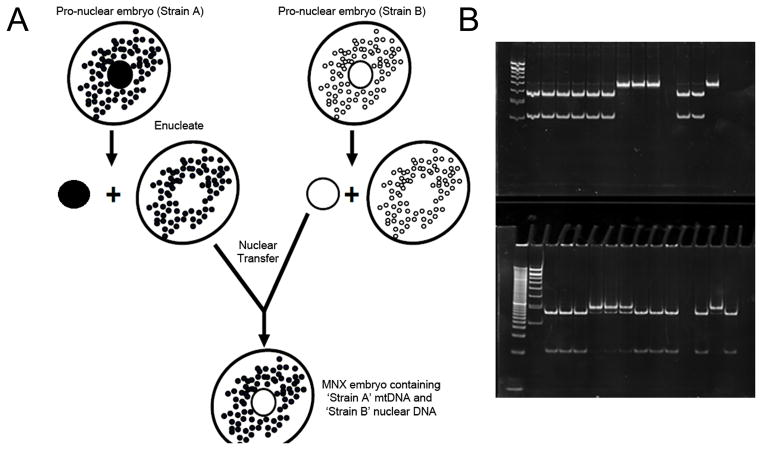
All animal studies were carried out under the approval of the University of Alabama at Birmingham (UAB) and University of Kansas Medical Center (KUMC) Institutional Animal Care and Use Committees. MNX mice were utilized in this study and generated as recently described (17). Briefly, pronuclei were removed from pronuclear embryos of FVB/NJ mice and isolated pronuclei implanted into enucleated C57BL/6J (metastasis suppressing (8)) and BALB/cJ (metastasis promoting (8)) embryos, generating reconstructed zygotes having FVB/NJ nuclei but cytoplasms (and therefore mitochondria and mtDNA) of either C57BL/6J or BALB/cJ mice (Fig. 1).
Figure 1.
(A) Schema depicting generation of the Mitochondrial-Nuclear Exchange (MNX) mouse. (B+C) Experimental lanes 1–3 correspond to tumor samples from = FVB, 4–6 = FVBn:C57mt, and 7–9 = FVBn:BALBcmt. Ear clip haplotype controls were taken from 11 = FVB, 12 = FVBn:C57mt, 134 = FVBn:BALBcmt. (B)BALB/c and FVB mtDNA were distinguished by a mutation at 9348 (valine to isoleucine) that results in the loss of a RflfI site in the BALB/c mtDNA. (C)C57BL/6 and FVB mtDNA were distinguished by a mutation at bp 9461(methionine to isoleucine) that results in the loss of a BclI site in the C57 mtDNA.
MNX mice were generated having a FVB/NJ nuclear genetic background with either a C57BL/6J mtDNA (FVBn:C57mt) or a BALB/cJ mtDNA (FVBn:BALBcmt). The FVB/NJ nuclear and mtDNA backgrounds were used as a control for these studies because this is the background of the PyMT model. All MNX mice were healthy and fertile with no overt physical or behavioral phenotypes. Wild-type FVB/NJ, C57BL/6J, and BALB/cJ animals were purchased from Jackson Laboratories (Bar Harbor, ME).
Crossing MNX with PyMT mice
The PyMT mouse spontaneous mammary tumor model was utilized for these studies. The PyMT oncogene is transcriptionally controlled by the mouse mammary tumor virus promoter (MMTV), is highly metastatic, and progresses through stages similar to human breast cancer (12). Hemizygous male PyMT mice were used as sires for breeding to control (FVB/NJ strain) and MNX mice (FVBn:C57mt and FVBn:BALBcmt) to generate hemizygous PyMT mice having FVB/NJ, C57BL/6J, or BALB/cJ mtDNA.
At weaning (~3 weeks of age) mice were genotyped for presence of the PyMT transgene as previously described (12, 18). mtDNA haplotypes were also confirmed in all female transgene-positive F1 progeny by RFLP (restriction fragment length polymorphism) analysis of PCR products screening for diagnostic mtDNA SNPs that differentiate between the FVB, C57BL/6, and BALB/c mtDNA, as previously described (17). Briefly, C57BL/6J and FVB/NJ mtDNA were distinguished by an adenine to cytosine mutation at bp 9461 that results in the loss of a BclI restriction site in the C57BL/6J mtDNA. BALB/cJ and FVB/NJ mtDNA were distinguished by a guanine to adenine mutation at bp 9348 that results in the loss of an RflfI site in the BALB/cJ mtDNA.
Tumor development and metastasis quantification
PyMT positive mice (n = 40) were palpated daily for first detection of tumor onset. Mice were aged an additional week to confirm tumor onset, and aged 40 days past initial primary tumor detection before euthanization and necropsy. Animal age and weight were recorded and tumors removed and weighed to determine total tumor burden. Lungs were resected for metastatic analysis and fixed overnight in Bouin’s fixative to facilitate metastasis quantification (19). Areas of surface metastases were determined using a Zeiss dissection microscope (Stemi 2000-C) fitted with an ocular micrometer (#474066-9901-000).
Isolation of primary mammary epithelial cells
All transgene positive female F1 progeny were sacrificed at 70 days (N: FVB/N = 5, FVBn:C57mt = 7, FVBn:BALBcmt = 7). Animals were weighed before sacrifice and tumors were removed and weighed to determine total tumor burden. The #3 and #4 mammary glands were digested in HBSS with Ca2+ and Mg2+ (GIBCO #24020-117) containing 0.1% collagenase and 0.01% pronase. Tissue in media was minced with scalpel blades, transferred to a 50 mL tube and then incubated at 37°C on a shaker platform for two hours. Samples were then transferred to a 15 mL tube and centrifuged to separate epithelial cells from fatty tissue and red blood cells. After several spins, the remaining epithelial cell fraction was resuspended in growth media (GIBCO #11039-021; phenol red-free DMEM/F12) containing 10% FBS, 20ng/mL EGF, 0.5 μg/mL hydrocortisone, 100ng/mL cholera toxin, 10 μg/mL bovine insulin, and 1% Pen-Strep) to be plated.
Bioenergetic and oxidative stress profiling
Seahorse Bioscience XF24 Calibrant pH 7.4 (1.0 mL; #100840-000, Seahorse Bioscience, North Billerica, MA) was added to each well of a 24-well plate (Seahorse Bioscience #100777-004) that was kept overnight at room temperature. Primary tumor epithelial cells were seeded into individual wells (6 × 104) cells per well) on a XF24 cell culture plate pre-warmed with unbuffered DMEM (250 μL, 37°C), leaving appropriate temperature correction wells blank (A1, B4, C3, D6). Epithelial cells were incubated in the XF24 cell culture plate in a 37°C incubator without CO2 until ready for use in order to allow them to equilibrate to the new media. Injection compound dilutions were made in appropriate running media (unbuffered DMEM) for the bioenergetic profile analysis experiment. Previously, titration experiments were run to determine each effector’s appropriate working concentration. For these studies, optimal concentrations were found to be: 10 μg/mL oligomycin, 1 μM FCCP (Trifluorocarbonylcyanide Phenylhydrazone), and 10 μM antimycin A.
Oligomycin, FCCP, antimycin A, and media were loaded into injection ports A, B, C, and D, respectively, in 75 μL volumes for each experimental well. All wells, including temperature correction wells, had control media or compounds loaded into all ports to ensure uniform injection. Once prepared, the plate containing the effectors was placed atop the plate containing the primary cells and placed into the bioanalyzer. Each oxygen consumption rate (OCR) measurement consisted of 3 minutes of mixing, 2 minutes wait time, and 3 minutes of continuous O2 level measurement. There were 3 measurement rounds per injection.
Data analysis was performed as described previously (20, 21). Briefly, baseline average, oligomycin minimum, FCCP maximum, antimycin A minimum, and reserve capacity were calculated as well as ATP linked and non-ATP linked baseline respiration and average baseline extracellular acidification (ECAR). Aconitase activity was determined by measuring the transformation of isocitrate to cis-aconitate at 240 nm in 50 mM TrisHCl (pH 7.4) containing MnCl2 and 20 mM isocitrate at 25°C. Aconitase is specifically inactivated by superoxide (O2·−) and peroxynitrite (ONOO−) and hence, decreased activity correlates with increased oxidative stress associated O2·− and ONOO−. Since ONOO− formation is related to superoxide production, decreased aconitase activity can be associated indirectly with increased O2·− generation.
Statistical analyses
Continuous parameters were compared by ANOVA for animal weight, tumor weight, primary tumor latency, and average metastasis area. In instances of non-normal distributions (e.g. visible lung metastases) a Kruskal-Wallis one-way rank test was employed. A P value <0.05 was considered statistically significant.
Results
mtDNA from C57BL/6J and BALB/cJ mouse strains were chosen to produce MNX mice because F1 hybrid progeny derived from PyMT males and females of these strains showed differential susceptibility to metastasis severity compared to offspring from PyMT male-FVB female crosses (8). In that study, F1 progeny having C57BL/6J mothers had less severe metastasis burden relative to PyMT mice from FVB/NJ mothers, while PyMT offspring with BALB/cJ females had increased metastatic burden, as determined by surface area of tumor cells in lung histologic sections (8). For our studies, FVBn:C57mt and FVBn:BALBcmt MNX and FVB/NJ control female mice were bred to PyMT males to directly test the effects of mtDNA background upon tumorigenicity and metastatic severity.
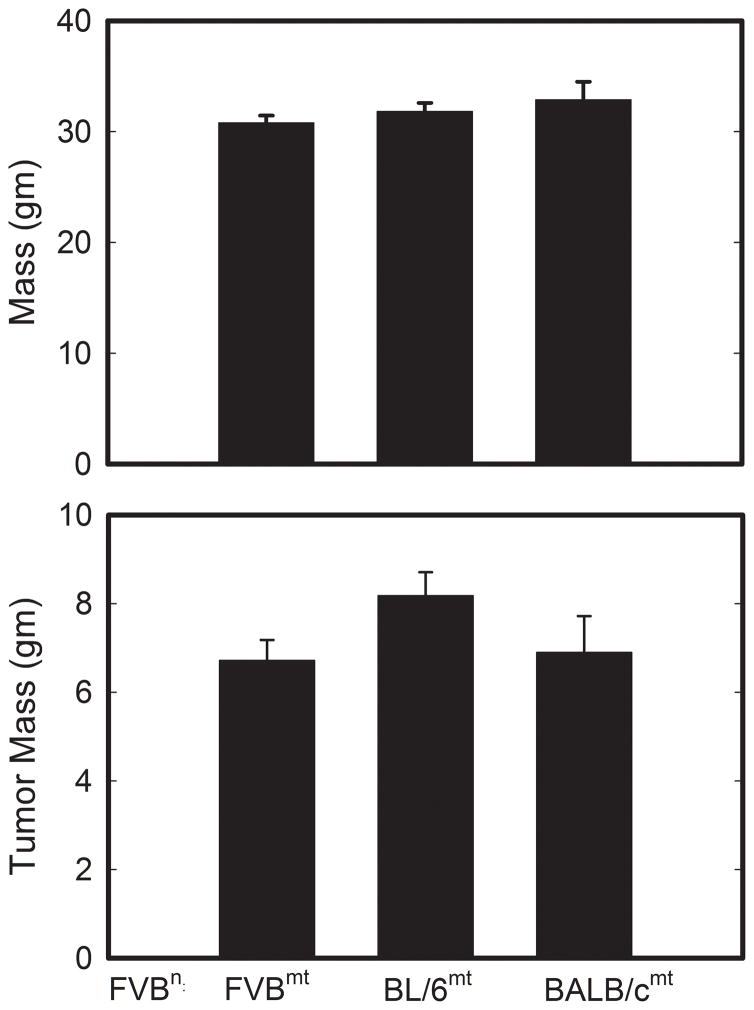
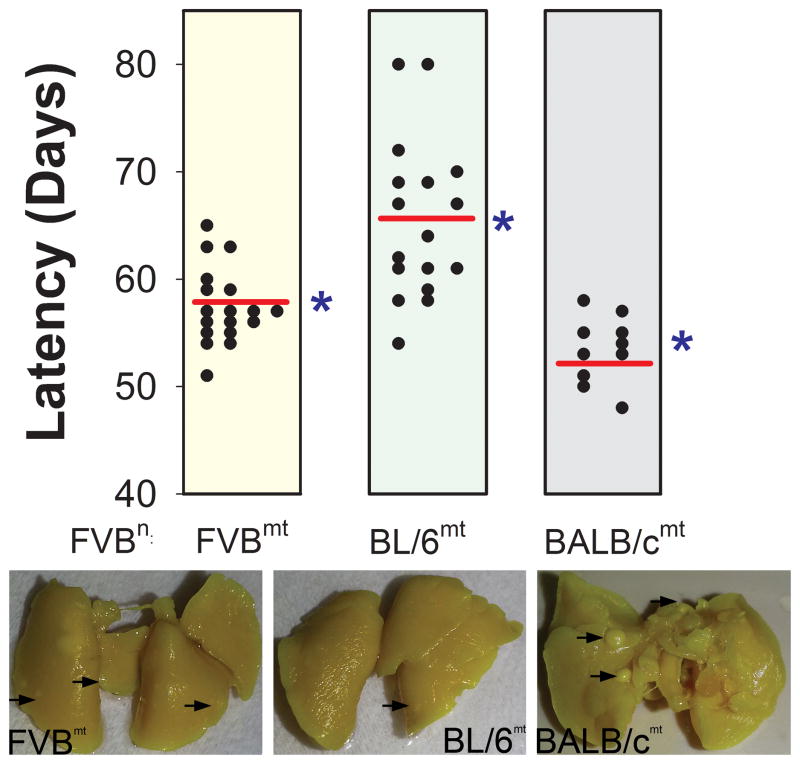
Figure 2 reveals that no significant differences in animal weight (FVBwt = 30.08 ± 0.66 g, FVBn:C57mt = 31.82 ± 0.78 g, FVBn:BALBcmt = 32.89 ± 1.62 g) or total tumor burden (FVBwt = 6.72 ± 0.46 g, FVBn:C57mt = 8.18 ± 0.53 g, FVBn:BALBcmt = 6.90 ± 0.82 g) were detected in PyMT positive F1 females 40 days subsequent to initial primary tumor detection. These data indicate that mice with all three mitochondrial genetic backgrounds had similar body weights and tumor growth rates (once tumors developed). In contrast, mitochondrial genetic background significantly affected primary tumor latency. PyMT mice with FVB/NJ, C57BL/6J, and BALB/cJ mtDNA had average latencies of 57.44 ± 0.83, 65.05 ± 1.72, and 53.40 ± 0.97 days, respectively (Figure 3). These latencies were significantly (P < 0.05) different, suggesting that mtDNA background influenced the early growth properties of PyMT mammary tumors. Interestingly, these results closely mirrored values for mean latencies observed in the matching F1 progeny reported by Lifsted et al. (FVB/NJ = 57.92 days, C57BL/6J = 64.44 days, BALB/cJ = 56.44 days) (8).
Figure 2.
(A) Animal weight and (B) total mammary tumor burden at sacrifice. Error bars indicate ± S.E.M.
Figure 3.
Primary PyMT mammary tumor latency segregates by mtDNA. Dots represent individual animals within each group, red lines represent the means, and the blue arrows indicate literature reported values for respective wild type latency. Each group has FVB nuclear DNA. (* = P < 0.05)
To determine the role of the mtDNA on metastatic severity, surface pulmonary metastases were counted and measured (19). Due to the non-normal distribution for the number of metastases per lung, the median number of metastases was utilized for statistical comparison. Whereas there were no differences in the median number of metastases between groups of PyMT mice (Table 1; FVB = 11, range: 0–83; FVBn:C57mt = 12, range: 0–62, FVBn:BALBcmt = 7, range: 1–58), a statistically significant (P <0.05) difference in the average size of individual metastases (in cm2) between groups was observed (FVB = 0.128 ± 0.012, FVBn:C57mt = 0.099 ± 0.009, and FVBn:BALBcmt = 0.181 ± 0.031; Table 1). These data suggest that, in addition to having an impact on primary tumor latency, mitochondrial genetic background influences the severity of metastatic disease. Given that the number of metastases was unchanged, the differences in average metastasis size confer an overall difference in metastatic burden. This is consistent with previous reports that PyMT F1 offspring from C57BL/6J mothers had lower metastatic burden; whereas, those from BALB/cJ mothers had more lung metastatic volume compared to PyMT offspring from FVB dams. The findings herein show that presence of the mitochondrial genome alone (i.e., no alteration of the nuclear genome) is sufficient to reproduce this finding.
Table 1.
Metastatic area, but not volume, differs in MNX mice
| MNX Strain | Metastasis area (cm−2) | Median No. Lung Metastases (Range) | |||
|---|---|---|---|---|---|
| FVBn:FVBmt | 0.128 ± 0.012 |
|
11 (0, 83) | ||
| FVBn:C57mt | 0.099 ± 0.009 | * |
|
12 (0, 62) | |
| FVBn:BALB/cmt | 0.181 ± 0.031 | * | 7 (1, 58) | ||
Numbers of pulmonary metastases are the same for each group, but they differ in the average size of individual metastases which therefore confers differences in overall metastatic burden.
P < 0.05
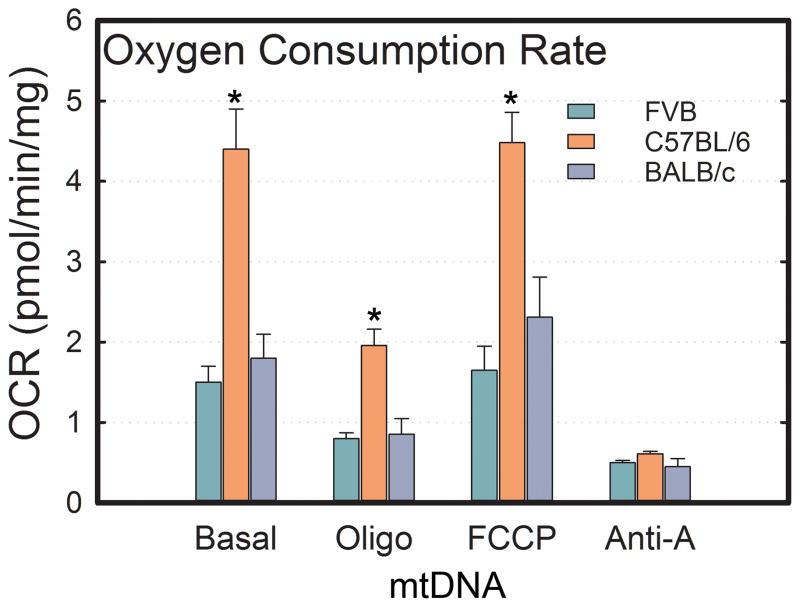
To assess whether bioenergetic profiles could be contributing to tumor behavior, epithelial cells were isolated from primary mammary tumors and seeded onto a Seahorse XF24 plate. No differences in baseline oxygen consumption rate (OCR) were observed between FVB/N and FVBn:BALBcmt cells, but FVBn:C57mt had a significantly elevated basal OCR (FVB/N = 1.50 ± 0.20, FVBn:C57mt = 4.45 ± 0.48, FVBn:BALBcmt = 1.79 ± 0.32 pmol/min/mg). This trend was consistent for OCR measurements following addition of oligomycin, FCCP, and antimycin A (Figure 4). Reserve capacity is a measure of the difference between the maximum possible OCR (following FCCP addition) and the baseline OCR. This parameter is proposed to be a measure of the ability of cells or tissue to respond to stresses by increasing their metabolic output. Due to the FVBn:C57mt tumor cells operating basally at a level near their maximum, their reserve capacity was miniscule (0.06 pmol/min/mg). FVB/N cells had an intermediate reserve at 0.18 pmol/min/mg while FVBn:BALBcmt tumor epithelial cells had a reserve capacity of 0.57 pmol/min/mg, roughly 25% of their maximum OCR.
Figure 4.
Oxygen consumption rates in epithelial cells taken from mammary tumors of 70 day old PyMT+ FVB/N, FVBn:C57mt, and FVBn:BALBcmt mice. OCR is measured under basal conditions as well as following addition of oligomycin, FCCP, and antimycin A. Each bar is the average of >10 independent measurements. Error bars indicate ± S.E.M. (* p < .05)
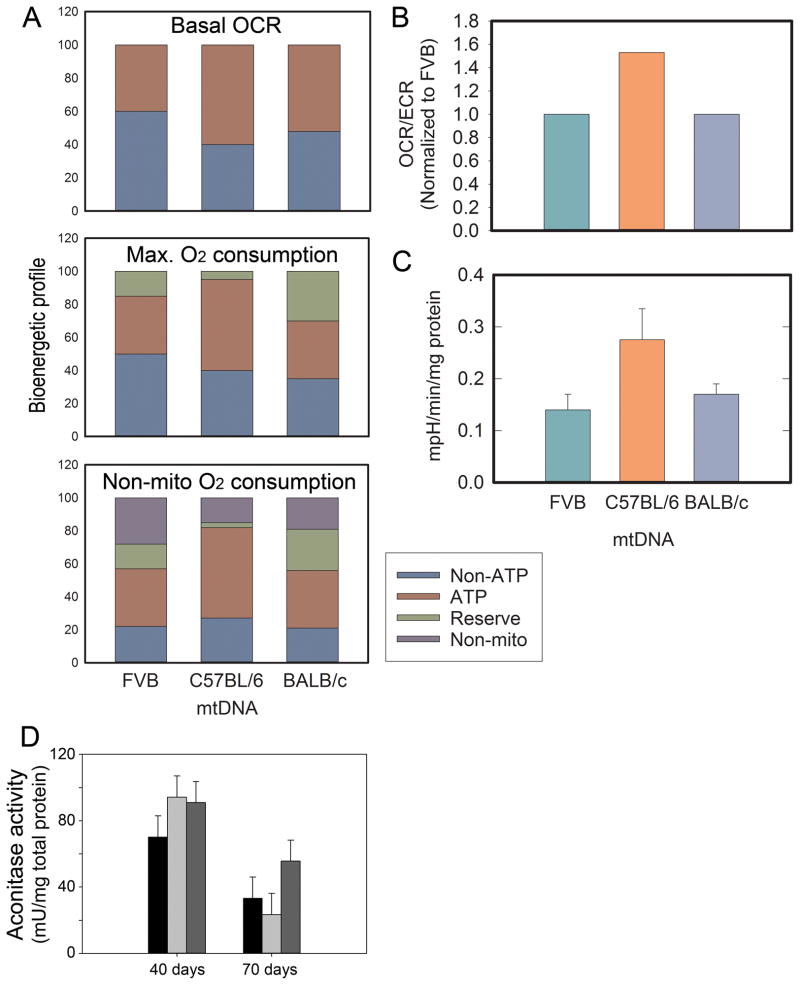
Baseline oxygen consumption levels are made up of oxygen consumption that is either linked to ATP or not linked to ATP. Along with having different total baseline OCRs, the percent contributions from ATP-linked and non ATP-linked were different for each group. The baseline OCR for FVB was 44.4% ATP-linked and 55.6% non ATP-linked, FVBn:C57mt was 56.4% ATP-linked and 43.6% non ATP-linked, and FVBn:BALBcmt was 50.8% ATP-linked and 49.2% non ATP-linked (Figure 5A). As a percentage of the maximum potential OCR, these values diverged further. ATP-linked OCR for FVB/N cells fell to 39.6% and non ATP-linked OCR fell to 49.6% with reserve capacity accounting for 10.8% of the maximum OCR. For FVBn:C57mt tumor cells, ATP-linked and non ATP-linked remained nearly the same (55.6% and 43.1%, respectively), as reserve capacity only accounted for 1.3% of the maximum attainable OCR. With the high reserve capacity observed in FVBn:BALBcmt cells (24.1%), ATP-linked OCR accounted for 38.6% and non ATP-linked OCR for 37.3% of maximum oxygen consumption. Consistent with the heightened oxygen consumption levels observed in FVBn:C57mt tumor cells, baseline extracellular acidification (ECAR) was highest in these cells as well (FVB/N = 0.14 ± 0.02, FVBn:C57mt = 0.27 ± 0.06, FVBn:BALBcmt = 0.17 ± 0.02 mpH/min/mg) (Figure 5A). Calculating OCR/ECAR determines the ratio of oxidative phosphorylation relative to glycolysis, and this value was roughly equal for FVB/N and FVBn:BALBcmt cells (10.48 and 10.51, respectively), while it was significantly higher for FVBn:C57mt cells (16.21) (Figure 5B and Figure 5C). Taken together, these bioenergetic data indicate that primary tumor cells isolated from these animals are metabolizing fundamentally differently.
Figure 5.
Oxygen utilization profiles of isolated epithelial cells from primary mammary tumors for FVB/N, FVBn:C57mt, and FVBn:BALBcmt mice. (A) The top graph displays the percentage of basal oxygen consumption (OCR) that is linked to ATP production and non-ATP production. The middle graph displays the percentage of maximal oxygen consumption that is linked to reserve capacity, ATP production, or non-ATP production. The bottom graph includes non-mitochondrial oxygen consumption as unique from non ATP-linked. (B) OCR/ECAR ratios from OCR and ECAR averages at baseline. Ratios were normalized against FVB = 1.0 (* P < .05). (C) Extracellular acidification rates in epithelial cells derived from primary mammary tumors of 70 day old PyMT+ FVB/N, FVBn:C57mt, and FVBn:BALBcmt mice. ECAR is measured under baseline conditions. Each bar represents the mean of at least 10 independent measurements. Error bars indicate ± S.E.M. (D) Mean aconitase activity of mammary epithelial tumor cells derived from FVB/N (black bar), FVBn:C57mt (light gray bar), and FVBn:BALBcmt (dark gray bar) mice. Error bars indicate ± S.E.M. from at least 3 independent samples.
To assess oxidative stress, the mammary epithelial cells, aconitase activity was assessed 40 days after detection of the first mammary tumors or in animals at 70 days of age. No significant differences between tumor cell lines were detected within either group (Figure 5D). There was a significant difference comparing the FVBn:C57mt tumor cells at the different ages. Cells at the 70 day hard cut off generally had more oxidative stress than cells isolated 40 day post-detection. However, the results must be viewed with a consideration that the 70 day hard cut-off animals were always younger than the 40 day post-detection mice. Considering that the FVBn:C57mt 40 day post-detection were the oldest of all (because they took the longest to get primary tumors), this may simply be an age-related factor. Similar measurements of citrate synthase similarly yielded no detectable differences between cells from the different crosses (unpublished data).
Discussion
Two of the major challenges facing breast cancer researchers are to expand our knowledge of what contributes to an individual’s risk for developing the disease, and to more fully understand influences on metastatic spread. Recent work investigating the roles of acquired mutations and nuclear genetic background on breast cancer susceptibility has provided important new insights; yet, the impact of the “host” mitochondrial genome has rarely been considered in such analyses. Because metabolic reprogramming occurs in cancer progression (21), it is vital to examine the genetic components responsible for providing a significant amount of cellular metabolic instructions.
This is the first study to show that constitutional mtDNA polymorphisms directly affect breast cancer tumorigenicity and metastatic spread in the PyMT model. Our studies show that in animals harboring the same nuclear DNA and subjected to the same oncogenic event (PyMT), primary tumor latency segregates with mitochondrial genetic background. Compared to progeny from control FVB females and PyMT males, offspring of FVBn:C57mt mothers had significantly delayed primary tumor latency while PyMT mice harboring the BALBcJ mtDNA had an accelerated onset. The finding that there were contrasting primary tumor latencies between groups and no differences in total tumor burdens 40 days post-onset further suggests that early stages of tumorigenesis are distinct between groups and are influenced by mtDNA background, whereupon their establishment, mtDNA background in the PyMT model does not appear to dictate primary tumor growth rate.
Metastatic efficiency of PyMT mice was modulated by mtDNA background as well. Our data show that the C57BL/6J mtDNA was associated with less severe metastatic disease while the BALB/cJ mtDNA was linked with more severe metastasis. It is possible that these differences are due to: i) the speed of primary tumor dissemination, ii) growth rates at the secondary site leading to colonization, or iii) both. It is important to emphasize that mtDNA differences were not acquired mutations in the tumor cells. Therefore, different mtDNA backgrounds influence tumor development, progression, and steps throughout the metastatic cascade, including at secondary sites. It is possible that the tumor cell’s ability to persist in circulation and eventually grow at distant sites is reflected in its capacity to metabolize efficiently while in transit or after arresting at secondary sites. Additionally, it is possible that acquired mutations may superimpose on the somatic differences to further affect tumor progression; however, additional studies will be required to discern these options.
Mitochondria have been implicated in tumor growth rates previously (25–27), and it is evident that the mitochondrial backgrounds are modifying the growth characteristics of these primary mammary tumors, presumptively through direct or indirect modification of cellular metabolic processes or oxidative stress. To explore these possibilities, mammary tumor (epithelial) cells were isolated and OCR and ECAR were measured. Heightened glycolysis is observed in most tumors and can indicate increased aggressiveness. The high ECAR seen in FVBn:C57mt tumor cells suggests elevated glycolytic activity. That these cells also have a significantly larger OCR implies that they simply have increased metabolic activity without necessarily relying more heavily on aerobic glycolysis. The similarities between FVB/N and FVBn:BALBcmt tumor cells in baseline OCR and ECAR suggest that other factors may be contributing to observed differences between their tumorigenic phenotype.
One significant difference, in particular, is reserve capacity. Reserve capacity makes up 10.8% of the total possible OCR available to FVB/N tumor cells; yet, it accounts for 24.1% of maximal OCR for FVBn:BALBcmt mammary tumor cells. Surprisingly, reserve capacity for the FVBn:C57mt tumor cells only constitutes 1.3% of their maximum potential OCR. When the reserve capacity is this low suggests that the FVBn:C57mt tumor cells have a baseline OCR nearly equal to their maximum which, counterintuitively, may indicate that they are operating inefficiently. These cells also display the highest percentage of baseline OCR that is linked to ATP production which is an often an indicator of poor mitochondrial economy, i.e., mitochondrial damage can render mitochondria less efficient to the point that they require greater oxygen utilization to meet the same metabolic requirements. Considering that reserve capacity reflects a cells ability to persist in response to stress (28), it might also provide insight into the ability for disseminated cells to complete the metastatic cascade. Interestingly, reserve capacities mirrored metastatic severity data previously reported (21). Interestingly, oxidative stress, as measured by aconitase or citrate synthase activities showed no differences when age was removed as a variable.
The metabolic changes observed could be, at least partially explained by the specific polymorphisms in the mtDNA. FVB/NJ and C57BL/6J mtDNA are different at sites encoding subunit II of complex IV (np 7778, a Asp to Tyr missense mutation), as well as subunit III of complex I (np 9461, a Met to Ile change); whereas, FVB/NJ and BALB/cJ mtDNA are different in sites encoding subunit II of complex IV (np 7778, a Asp to Tyr missense mutation) and subunit III of complex IV (np 9348, a Val to Ile missense mutation) (22). Differential complex I and complex IV activities have been recently implicated as contributing to breast cancer progression (23, 24). However, a simple explanation is not likely since polymorphisms within one electron transport complex can potentially affect other complexes or related metabolic processes (e.g. citric acid cycle) because cellular metabolism is a network of tightly coupled processes, and modification of any step might affect the entire process.
Taken together, our data suggest that modulating mitochondrial genetic background has a significant impact on tumor latency, metastasis, and disease progression. The specific causes remain to be fully elucidated; but, it is probable that they are a consequence of oncogenic mutations “co-opting” the pre-existing metabolic programming of the cell influenced by mtDNA background. To evaluate the mechanisms behind these differences, it will be necessary to further dissect the bioenergetics of PyMT control and MNX animals. Additionally, retrograde signaling to the nucleus that is altered by mitochondrial polymorphisms could be responsible for the changes in cellular behavior observed.
The concept that mtDNA polymorphisms can modify breast cancer susceptibility could be of use for risk assessment and/or prognosis before any malignant transformation has taken place. Since these are not acquired mutations, a blood sample or cheek swab could be sufficient to ascertain an individual’s risk for advanced disease as more information is found characterizing metastatic potential versus mtDNA haplotype. Petros et al. reported that germ-line mtDNA mutations contribute to prostate cancer development (29), but ours are the first functional data that directly establish a connection between mtDNA and metastasis. Both studies, however, support roles for mtDNA background as a contributor to outcomes and potential therapies. Furthermore, metabolic pathways in cancer offer a bright future for potential therapeutic targets (30). Delineating specific mitochondrial contributors to altered cancer growth kinetics will only expedite these discoveries.
Significance.
Differences in mitochondrial DNA are sufficient to fundamentally alter disease course in the PyMT mouse mammary tumor model, suggesting that functional metabolic differences direct early tumor growth and metastatic efficiency.
Acknowledgments
Grant Support:
The authors are grateful for the support of many organizations to accomplish these studies: Susan G. Komen for the Cure (SAC11037 to DRW); U.S. Army Medical Research & Material Command (W81XWH-07-1-0540d to SWB/DRW); National Cancer Institute (CA013148 to DRW/SWB; CA168524 to DRW; HL103859 to SWB; and CA134981 to DRW); National Foundation for Cancer Research (to DRW). AWB was supported by the pre-doctoral training program in cardiovascular pathophysiology (T32 HL007918); RAK was supported by NIH P30 CA13148, P30 AR048311, P30 DK074038, P30 DK05336 and P60 DK079626. We are also indebted for the support of the UAB Diabetes Research Center Bioanalytical Redox Biology Core (DK079626).
DRW is a Kansas Bioscience Authority Eminent Scholar.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Authors’ Contributions
Conception and design: S.W. Ballinger, D.R. Welch
Development of methodology: K. P. Feeley, A.W. Bray, L.W. Johnson, R.E. Kesterson,
Acquisition of data: K.P. Feeley, A.W. Bray
Analysis and interpretation of data: K.P. Feeley, S.W. Ballinger, D.R. Welch
Writing, review and/or revision of the manuscript: K.P. Feeley, S.W. Ballinger, D.R. Welch
Administrative, technical or material support: S.W. Ballinger, D.R. Welch
Study supervision: S.W. Ballinger, D.R. Welch
Reference List
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.King MC. “The race” to clone BRCA1. Science. 2014;343:1462–5. doi: 10.1126/science.1251900. [DOI] [PubMed] [Google Scholar]
- 3.Winter SF, Hunter KW. Mouse modifier genes in mammary tumorigenesis and metastasis. J Mamm Gland Biol Neopl. 2008;13:337–42. doi: 10.1007/s10911-008-9089-1. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, Sorlie T, Eisen MB, Van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Krupke DM, Begley DA, Sundberg JP, Bult CJ, Eppig JT. The mouse tumor biology database. Nature Rev Cancer. 2008;8:459–65. doi: 10.1038/nrc2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford NPS, Walker RC, Lukes L, Officewala JS, Williams RW, Hunter KW. The Diasporin Pathway: a tumor progression-related transcriptional network that predicts breast cancer survival. Clin Exptl Metastasis. 2008;25:357–69. doi: 10.1007/s10585-008-9146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter KW, Broman KW, LeVoyer T, Lukes L, Cozma D, Debies MT, et al. Predisposition to efficient mammary tumor metastatic progression is linked to the breast cancer metastasis suppressor gene Brms1. Cancer Res. 2001;61:8866–72. [PubMed] [Google Scholar]
- 8.Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, Buetow KH, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77:640–4. doi: 10.1002/(sici)1097-0215(19980812)77:4<640::aid-ijc26>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 9.Sevignani C, Calin GA, Nnadi SC, Shimizut M, Davulurit RV, Hyslop T, et al. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci. 2007;104:8017–22. doi: 10.1073/pnas.0702177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter KW. Allelic diversity in the host genetic background may be an important determinant in tumor metastatic dissemination. Cancer Lett. 2003;200:97–105. doi: 10.1016/s0304-3835(03)00420-8. [DOI] [PubMed] [Google Scholar]
- 11.Park YG, Lukes L, Yang H, Debies MT, Samant RS, Welch DR, et al. Comparative sequence analysis in eight inbred strains of the metastasis modifier QTL candidate gene Brms1. Mamm Genome. 2002;13:289–92. doi: 10.1007/s00335-001-2151-6. [DOI] [PubMed] [Google Scholar]
- 12.Lin EY, Jones JG, Li P, Zhu UY, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Molec Cell Biol. 1992;12:954–61. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park YG, Zhao XH, Lesueur F, Lowy DR, Lancaster M, Pharoah P, et al. Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat Genet. 2005;37:1055–62. doi: 10.1038/ng1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford NP, Qian X, Ziogas A, Papageorge AG, Boersma BJ, Walker RC, et al. Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLoS Genet. 2007;3:e214. doi: 10.1371/journal.pgen.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–4. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 17.Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, et al. Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem J. 2013;456:147. doi: 10.1042/BJ20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook LM, Cao X, Dowell AE, Debies MT, Edmonds MD, Beck BH, et al. Ubiquitous Brms1 expression is critical for mammary carcinoma metastasis suppression via promotion of apoptosis. Clin Exptl Metastasis. 2012 doi: 10.1007/s10585-012-9452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welch DR. Technical considerations for studying cancer metastasis in vivo. Clin Exptl Metastasis. 1997;15:272–306. doi: 10.1023/a:1018477516367. [DOI] [PubMed] [Google Scholar]
- 20.Feeley KP, Westbrook DG, Bray AW, Ballinger SW. An ex-vivo model for evaluating bioenergetics in aortic rings. Redox Biol. 2014;2C:1003–7. doi: 10.1016/j.redox.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Beck BH, Vaidya KS, Nash KT, Feeley KP, Ballinger SW, et al. Metastasis suppressor KISS1 seems to reverse the Warburg effect by enhancing mitochondrial biogenesis. Cancer Res. 2014;74:954–63. doi: 10.1158/0008-5472.CAN-13-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayona-Bafaluy MP, cin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, et al. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–55. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santidrian AF, Matsuno-Yagi A, Ritland M, Seo BB, LeBoeuf SE, Gay LJ, et al. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. JCI. 2013;123:1068–81. doi: 10.1172/JCI64264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sen S, Kawahara B, Chaudhuri G. Mitochondrial-associated nitric oxide synthase activity inhibits cytochrome c oxidase: implications for breast cancer. Free Radic Biol Med. 2013;57:210–20. doi: 10.1016/j.freeradbiomed.2012.10.545. [DOI] [PubMed] [Google Scholar]
- 25.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–96. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Ginkel PR, Sareen D, Subramanian L, Walker Q, Darjatmoko SR, Lindstrom MJ, et al. Resveratrol inhibits tumor growth of human neuroblastoma and mediates apoptosis by directly targeting mitochondria. Clin Cancer Res. 2007;13:5162–9. doi: 10.1158/1078-0432.CCR-07-0347. [DOI] [PubMed] [Google Scholar]
- 28.Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radic Biol Med. 2010;48:905–14. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci. 2005;102:719–24. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yizhak K, Le Devedec SE, Rogkoti VM, Baenke F, de Boer VC, Frezza C, et al. A computational study of the Warburg effect identifies metabolic targets inhibiting cancer migration. Mol Syst Biol. 2014;10:744. doi: 10.15252/msb.20134993. [DOI] [PMC free article] [PubMed] [Google Scholar]