Abstract
Phosphatidylinositol 4-phosphate 5-kinases (PIP5Ks) regulate a variety of cellular processes including signaling through G protein-coupled receptors (GPCRs), endocytosis, exocytosis, and cell migration. These lipid kinases synthesize phosphatidylinositol 4,5-bisphosphate (PIP2) from phosphatidylinositol 4-phosphate [PI(4)P]. Since small molecule inhibitors of these lipid kinases did not exist, molecular and genetic approaches were predominantly used to study PIP5K1 regulation of these cellular processes. Moreover, standard radioisotope-based lipid kinase assays cannot be easily adapted for high-throughput screening. Here, we report a novel high-throughput microfluidic mobility shift assay to identify inhibitors of PIP5K1C. This assay utilizes fluorescently labeled phosphatidylinositol 4-phosphate as the substrate and recombinant human PIP5K1C. Our assay exhibited high reproducibility, had a calculated ATP Km of 15 µM, performed with z’ values >0.7, and was used to screen a kinase-focused library of ~4,700 compounds. From this screen, we identified several potent inhibitors of PIP5K1C, including UNC3230, a compound that we recently found can reduce nociceptive sensitization in animal models of chronic pain. This novel assay will allow continued drug discovery efforts for PIP5K1C and can be easily adapted to screen additional lipid kinases.
Keywords: Lipid kinase, PIP5K1C, PIP2
INTRODUCTION
Phosphatidylinositol (PI) lipids tightly regulate many cellular processes including cell migration, vesicle trafficking, endocytosis, surface receptor signaling, cell proliferation, and cell survival—all of which underlie normal physiology and disease mechanisms.1, 2, 3 The role of phosphatidylinositol 3-kinases (PI3Ks) in cancer proliferation and survival has been extensively characterized explaining why the vast majority of lipid kinase drug discovery efforts over the past 20 years (starting with the discovery of the first PI3K inhibitor, wortmannin) have been directed towards PI3Ks.4 There has been relatively little effort on identifying inhibitors of other lipid kinase family members, likely due to a lack of conclusive evidence for a direct link between specific diseases and individual lipids and/or lipid kinases. Recent studies connecting additional lipid kinases to disease-relevant cellular processes include sphingosine 1-kinase in cancer progression5, 6, phosphatidylinositol 5-phosphate 4-kinase (PIP4K2B) in insulin signaling7, 8, and phosphatidylinositol 4-phosphate 5 kinase (PIP5K1C) in nociceptive sensitization.9 There is thus growing interest in using high-throughput screening (HTS) assays to identify inhibitors of these lipid kinases.8, 10 Here, we describe a HTS assay developed during our studies that examined the role of PIP5K1C in nociceptive signaling and sensitization.9
Type I PIP5 kinases (PIP5K1s) and type II PIP4 kinases (PIP4K2s) synthesize the predominant lipid second messenger, PI(4,5)P2, from phosphatidylinositol monophosphates (PIPs), PI(4)P and PI(5)P, respectively. Although there have been two recent publications reporting the development of independent high-throughput assays to identify inhibitors of PIP4K2A11 and PIP4K2B8 there were no known inhibitors of any of the three mammalian PIP5K1 isoforms (alpha, beta, or gamma). Our studies using genetic Pip5k1c knockout mice demonstrated the need for a pharmacological inhibitor that could be used to complement our genetic studies by acutely inhibiting PIP5K1C and as a tool for target validation.9, 12
When we began our studies, available assays to monitor PIP5K1-dependent PIP2 synthesis required the use of lengthy lipid extraction protocols, radiolabeled ATP, and/or thin layer chromatography to separate substrate and product, none of which were amenable to a HTS assay.10, 13, 14 Here, we overcame these limitations by developing a HTS assay using fluorescently conjugated PI(4)P, the natural substrate for PIP5K1C, and full length recombinant PIP5K1C.
MATERIALS AND METHODS
Materials
Fluorescein conjugated phosphatidylinositol 4-phosphate [PI(4)P, 9000655] and fluorescein conjugated phosphatidylinositol 4,5-bisphosphate (PIP2, 10010388) were purchased from Cayman Chemical and reconstituted in 100% dimethylsulfoxide (DMSO) to 1.5 mM. N-terminal His6-tagged full length (90 kDa) recombinant human PIP5K1C was purchased from Millipore (14–845M). ProfilerPro separation buffer (760367) and coating-reagent 8 (CR-8; 760278) were purchased from PerkinElmer. PIP5K1C enzyme was used at a final concentration of 3 nM in assay buffer (Supplemental Table 1) containing 0.01% BSA, 1 mM DTT, 0.5× protease inhibitors (Roche mini complete tablets), and 0.5× phosphatase inhibitors. PI(4)P substrate solution was used at a final concentration of 1 µM in assay buffer containing 0.05% DMSO and 15 µM ATP (Km for PIP5K1C determined using the PerkinElmer LabChip assay).
LOPAC Library
The library of pharmacologically active compounds (LOPAC) was purchased from Sigma and was used as an assay validation library. The 1,280 compounds were supplied as 1 µL samples (10 mM) in 384-well polypropylene microplates (Grenier). On the day of screening, plates were thawed and diluted (1:100) to 0.1 mM (10× the final assay concentration) with assay buffer (Supplemental Table 1) in the 384-well plate. A Multidrop Combi Reagent Dispenser (ThermoScientific) was used to add 100 µL of 1% DMSO to columns 1, 2, 23, and 24 which did not contain compound and served as control columns. A MultiMek NSX-1536 assay workstation system fitted with a 384-well head (Nanoscreen, Charleston, SC) was used to transfer 2 µL of each sample into 384-well ShallowWell Nunc assay plates (ThermoScientific, 267459). After enzyme and substrate addition (18 µL total), the final concentration of compound is 10 µM in a final DMSO concentration of ~0.125%.
Kinase-focused Library
The 4,727 compound kinase-focused library was prepared and generously provided by the UNC Center for Integrative Chemical Biology and Drug Discovery (CICBDD).15 On the day of screening, plates were prepared as described for the LOPAC library.
Screening
A Multidrop Combi Reagent Dispenser (ThermoScientific) was used for the addition of all reagents to assay plates. First, 10 µL of 90 mM EDTA (in assay buffer) was added to each well in columns 1 and 2 and served as positive control reactions (100% inhibition). Nine microliters of 2× enzyme solution was added to each well of the entire plate. Plates were incubated at room temperature for 10 minutes then 9 µL of 2× substrate solution was added to each well of the entire plate. Assay plates were incubated in the dark for 40 minutes at room temperature. Ten microliters of 90 mM EDTA (in assay buffer) was then added to columns 3–24 to stop the reactions. Fluorescently conjsuugated substrate, PI(4)P, and product, PIP2, were detected using the LabChip EZ Reader II microfluidic mobility shift assay (MSA) platform from PerkinElmer and ProfilerPro separation buffer containing 1.5% CR-8.
For dose response curves, compounds were plated as 3-fold serial dilutions starting with a top concentration of 10 mM. The lowest concentration prepared in the 10-point dose response was 0.0005 mM. Dose response compound plates were prepared using a Tecan Genesis 200 (Research Triangle Park, NC). Dose response plates were heat-sealed and stored at −20°C until day of use. On the day of use, plates were prepared as described the LOPAC library (see above). The final top assay concentration was 10 µM and the lowest assay concentration was 0.005 µM.
Data analysis
Screening data was analyzed using Screenable software (Screenable Solutions, Chapel Hill, NC). Screenable was used to calculate the mean of the positive and negative controls (positive controls in columns 1 and 2 and negative controls in columns 23 and 24), the percent inhibition (with respect to on-plate controls) for each reaction and the common assay performance measure, z’, for each plate. Where p is the positive control (+90 mM EDTA; 100% inhibition) and n is the negative control (no compound addition; no inhibition).16 A z’>0.5 was considered acceptable for the plate to be included in the overall data analysis. The LabChip software calculated percent conversion for each reaction.
Compounds from the kinase library were considered hits if they inhibited PIP5K1C at ≥80%. The 80% threshold was determined as greater than 3 standard deviations from the mean percent inhibition for the entire screen. Mean inhibition was 14% with a standard deviation of 21%. Dose response curves were calculated using Screenable Software by converting the % conversion to % inhibition with respect to on-plate controls and using a 3 or 4-parameter curve fit.
RESULTS
Assay design and development
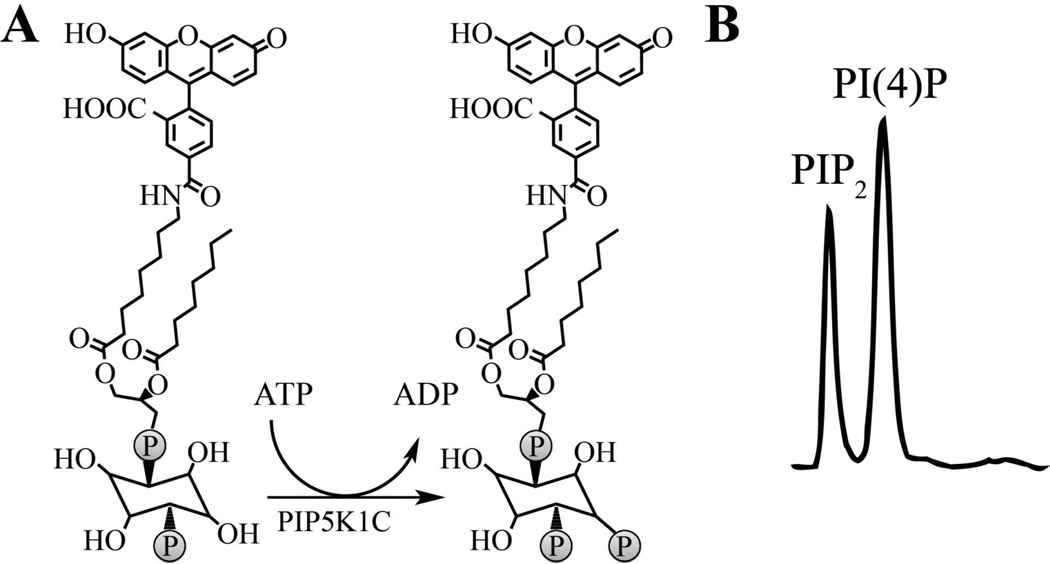
Our initial goal was to develop an assay that was compatible with high-throughput techniques. To avoid lengthy lipid extraction and preparation protocols required when using eukaryotic cell lysates, we used recombinant human PIP5K1C and a fluorescein conjugated PI(4)P substrate in an in vitro biochemical reaction (Figure 1A). Although there are no reported assays that monitor PIP2 from any of the three natural substrates, there was a recent assay developed that used the PerkinElmer microfluidic MSA LabChip platform to monitor the conversion of PIP2 to phosphatidylinositol 3,4,5-triphosphate (PIP3) by PI3K.17 Using the reported PI3K assay buffer and the PerkinElmer LabChip microfluidic MSA platform, we observed baseline separation of 1 µM fluorescein-conjugated PIP2 and 1 µM fluorescein conjugated PI(4)P using ProfilerPro separation buffer with 1.5% coating reagent 8 (CR-8) (Figure 1B). Notably, we found that the presence of sodium cholate in the assay buffer is imperative to achieve separation and prevent aggregation of lipids. The proprietary CR-8 promotes run-to-run separation consistency, prevents aggregation of substrate and kinase, and reduces adsorption of substrate and kinase to the microchannel surfaces.18 Baseline separation of the substrate and product indicated that the LabChip MSA platform would be suitable for continued assay development.
Figure 1. Recombinant PIP5K1C generates fluorescein conjugated PIP2 from fluorescein conjugated PI(4)P.
(A) Schematic representation of the PIP5K1C reaction using fluorescein conjugated PI(4)P as the substrate. (B) Separation of the fluorescein conjugated substrate, PI(4)P, and fluorescein conjugated product (PIP2) using the PerkinElmer LabChip EZ Reader II and microfluidic mobility shift platform.
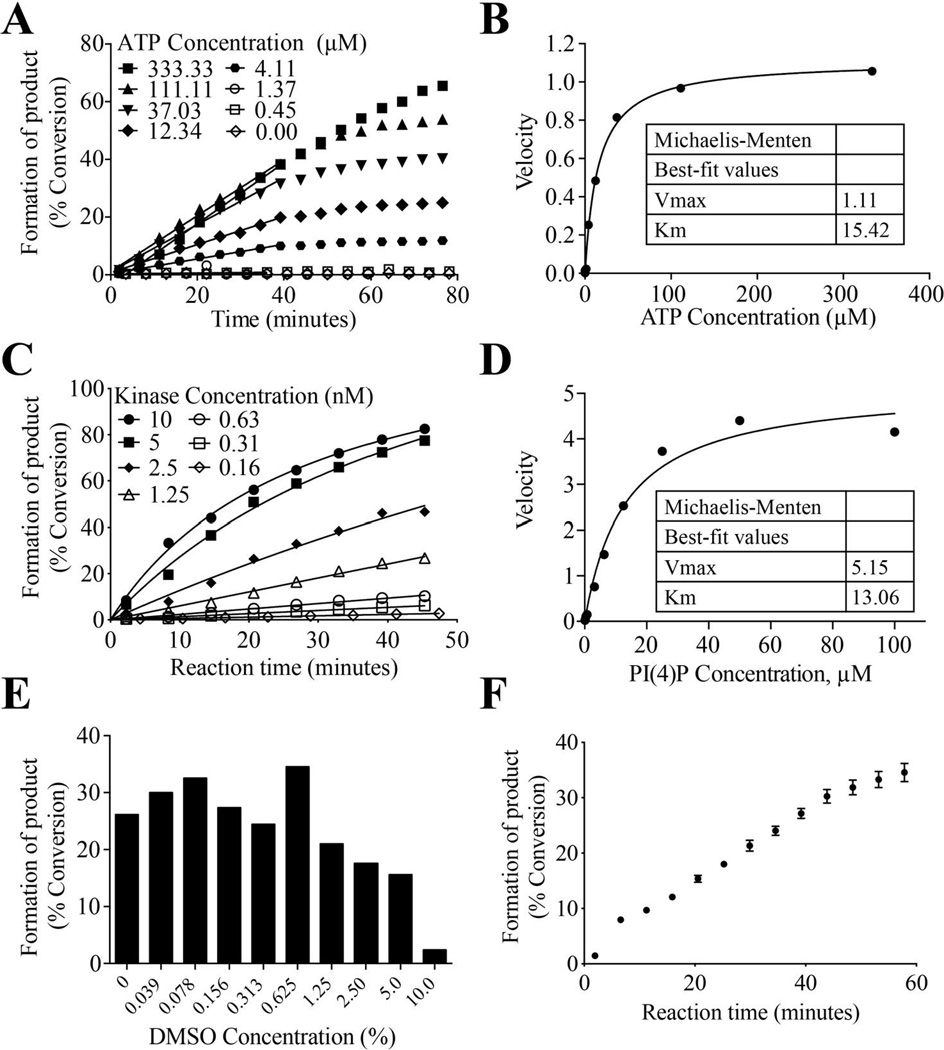
Fluorescein conjugated PI(4)P (1 µM) was incubated with 100 nM N-terminal His6-tagged full length (90 kDa) recombinant human PIP5K1C and varying concentrations of adenosine triphosphate (ATP; three fold serial dilution starting at 1000 µM; 11 concentrations total) to determine the ATP Km. The LabChip MSA platform was used to monitor the reactions kinetically (every 5 minutes for ~80 minutes). Michaelis-Menten analysis, using initial rate velocities, revealed an ATP Km of 15 µM (Figure 2A and 2B). Next, reactions containing 1 µM PI(4)P, 15 µM ATP, and varying concentrations (two-fold serial dilution starting at 10 nM; 7 concentrations total) of PIP5K1C were read kinetically (every 6 minutes for 45 minutes) to determine the concentration of enzyme that would give ~30% conversion of substrate to product at an end-point within the linear range of the reaction (Figure 2C). A final concentration of 3 nM PIP5K1C and a 40-minute incubation time were chosen. Kinase titrations were performed for every vial of enzyme purchased over the course of assay development and screening to account for slight differences in kinase activity from lot-to-lot. Varying concentrations of PI(4)P (two fold serial dilution starting at 100 µM; 11 concentrations total) were incubated with 3 nM PIP5K1C and 15 µM ATP and reactions were read kinetically (every 4 minutes for 45 minutes) using the LabChip platform to calculate substrate Km. Percent conversion of the reactions was converted to pmol product produced to determine initial rate velocities. Michaelis-Menten analysis, using these initial rate velocities, revealed a substrate Km of 13 µM (Figure 2D).
Figure 2. Assay development to determine kinase concentration, ATP and substrate Km, and DMSO sensitivity.
(A) Progression curves for PIP5K1C reactions with varying concentrations of ATP. (B) Michaelis-Menten analysis of initial rates in (A) to determine ATP Km for PIP5K1C. (C) Progression curves with varying concentrations of PIP5K1C to determine the optimal kinase concentration to obtain ~30% conversion after 40-minute incubation. (D) Michaelis-Menten analysis of progression curves for PI5K1C reactions with varying concentrations of substrate to determine substrate Km (E) Percent conversion of PIP5K1C reactions at various DMSO concentrations after 40-minute incubation to determine DMSO sensitivity of the reaction. (F) Final progression curve for reactions containing all optimized parameters. Mean ± SEM. n=12 reactions.
Given that all UNC compound libraries are dissolved in 100% DMSO, reactions containing 1 µM PI(4)P, 15 µM ATP, 3 nM PIP5K1C and varying concentrations of DMSO (two-fold serial dilutions starting at 10%; 9 concentrations total) were monitored using the LabChip MSA platform to determine DMSO tolerance (Figure 2E). There was decreased activity in reactions containing >1% DMSO indicating the final concentration of DMSO within the reaction must be lower than 1%. Compounds are prepared as 10 mM stocks in 100% DMSO. Compounds are then diluted in the compound plate to 0.1 mM with assay buffer (DMSO concentration of 1%). Compounds are then transferred to the assay plate where the final concentration after enzyme and substrate addition is 10 µM in ~0.125% DMSO (0.1% from compound addition and 0.125% from substrate solution) which is well within the DMSO tolerability of the reaction. Finalized reactions containing 1 µM PI(4), 15 µM ATP, 3 nM PIP5K1C, and 0.15% DMSO result in 30% conversion of substrate to product following a 40 minute incubation (Figure 2F), an ideal percent conversion (within the linear range of the reaction) for detecting inhibitors.
Next, we transitioned this assay into a HTS format beginning with automation validation using a Multidrop Combi Reagent Dispenser for delivery of the enzyme and substrate solutions. We initially observed inconsistent delivery of the lipid substrate and kinase to the plate, likely because of lipid adsorption to the silicone tubing of the Multidrop delivery cassette.19 To overcome adsorption problems, a final concentration of 0.01% BSA and 0.05% DMSO were included in the enzyme and substrate solutions, respectively, to serve as carriers and mitigate adsorption.20 There was slight reduction in PIP5K1C activity upon addition of 0.01% BSA to the enzyme solution (23% conversion in the presence of BSA compared to 30% conversion in the absence of BSA). Regardless, reactions in the presence of BSA remained in the linear range of detection and displayed the desired 20–40% conversion of substrate to product after a 40 minute incubation, indicating no adjustments were needed to the previously determined PIP5K1C concentration of 3 nM. To further minimize adsorption problems, the silicone lines of the Multidrop delivery cassette were primed with 5 mL of 0.2% BSA prior to kinase dispensing and 5 mL of 1% DMSO prior to substrate dispensing. Addition of BSA and DMSO to the enzyme and substrate solutions, respectively, allowed for consistent delivery of both solutions to multiple plates (data not shown). By incorporating these changes, all plates had coefficients of variation less than 10% (CVs >20% were observed prior to making these changes) and no discernible, systematic patterns, thus completing automation validation.
Assay validation
Following automation validation, the assay was revalidated in an HTS format using the Nanoscreen MultiMek to transfer 1% DMSO (to mimic compound delivery) to each assay plate followed by Multidrop delivery of the enzyme and substrate solutions. During this stage we confirmed the plate layout for the assay in which positive controls (+90 mM EDTA in assay buffer; 100% inhibition) would occupy columns 1 and 2 while negative controls (no compound; no inhibition) would occupy columns 23 and 24; all plates were run in this manner for HTS validation. HTS validation was carried out over three days, 2 plates per day. HTS validation revealed excellent assay performance with z’ values above 0.9 on all three days (Figure 3A). We then used the library of pharmacologically active compounds (LOPAC, Sigma Chemical) as a validation library to examine reproducibility of our assay. The 1,280 compounds were run in duplicate over two days (4 plates per day). The assay was deemed excellent with respect to z’ which ranged from 0.7 to 0.9 (Figure 3A) for all plates. The data fitted with a linear regression line revealed an r2 and slope of 0.966, indicating a highly reproducible assay (Figure 3B).
Figure 3. Assay validation using the LOPAC library.
(A) Assay performance measurement (z’) for 6 plates (over 3 days) of HTS validation and 4 plates of the LOPAC library in duplicate (2 days). (B) Duplicate runs (each on a different axis) of the LOPAC library (1,280 compounds) to assess reproducibility of the assay.
Screening
A kinase-focused library of 4,727 compounds that was designed and made available by the UNC Center for Integrative Chemical Biology and Drug Discovery (CICBDD) was screened at 10 µM in 0.1% DMSO.15 Results from the kinase-focused library were found to follow a normal distribution (Figure 4A and 4B). All 16 plates of the kinase-focused library had z’ values greater than 0.5 with an average z’ of 0.862 (Figure 4C). Compounds that exhibited inhibition greater than three standard deviations from the mean value were considered active (>80% inhibition; mean of 14% and standard deviation of 21%). There were 22 compounds that exhibited >80% inhibition of PIP5K1C and were considered active (hit rate of 0.42%; Supplemental Table 2). Primary hits were subjected to a set of structure- and property-based filters in order to remove compounds whose physical properties would prevent them to penetrate through the cell membrane or induce substantial cellular toxicity. We made use of a softened version of the Lipinski rule21 (2+ violations of Number of H-bond donors < 6, Number of H-bond acceptors < 12, Molecular Weight between 200 and 600, ALogP < 5.5) and REOS.22 Likewise, we removed compounds featuring at least one reactive or toxicity-causing substructure. After filtering, 20 compounds remained, all of which were classified as singletons (3 or more compounds with similar structures would have been considered a cluster) (Supplemental Table 2).
Figure 4. HTS of 4,727 compound kinase-focused library.
(A) PIP5K1C reactions (expressed as % inhibition determined by on-plate controls) for each well of the assay including all positive (yellow) and negative controls (red). (B) Distribution of compound activity for the kinase-focused library. The number of compounds with a given activity level (% inhibition) are plotted in a frequency histogram. (C) z’ values for each of the 16 plates of the kinase-focused library.
Hit confirmation and follow up of select inhibitors
All available active compounds were re-tested in triplicate in a 10-point dose response assay to confirm activity and provide potency information (Supplemental Table 2). Compound 20 was unavailable at the time of potency testing (Supplemental Table 2). Potency values were obtained for 18 of the 19 compounds tested, revealing an overall 90% confirmation rate for our active compounds. In addition to active compounds from the kinase-focused library, there were 6 active compounds (inhibition >80%; after drug-likeness filtering) in the LOPAC library; however, the LOPAC library was utilized as a validation library and potency information was not obtained for these compounds (Supplemental Table 3).
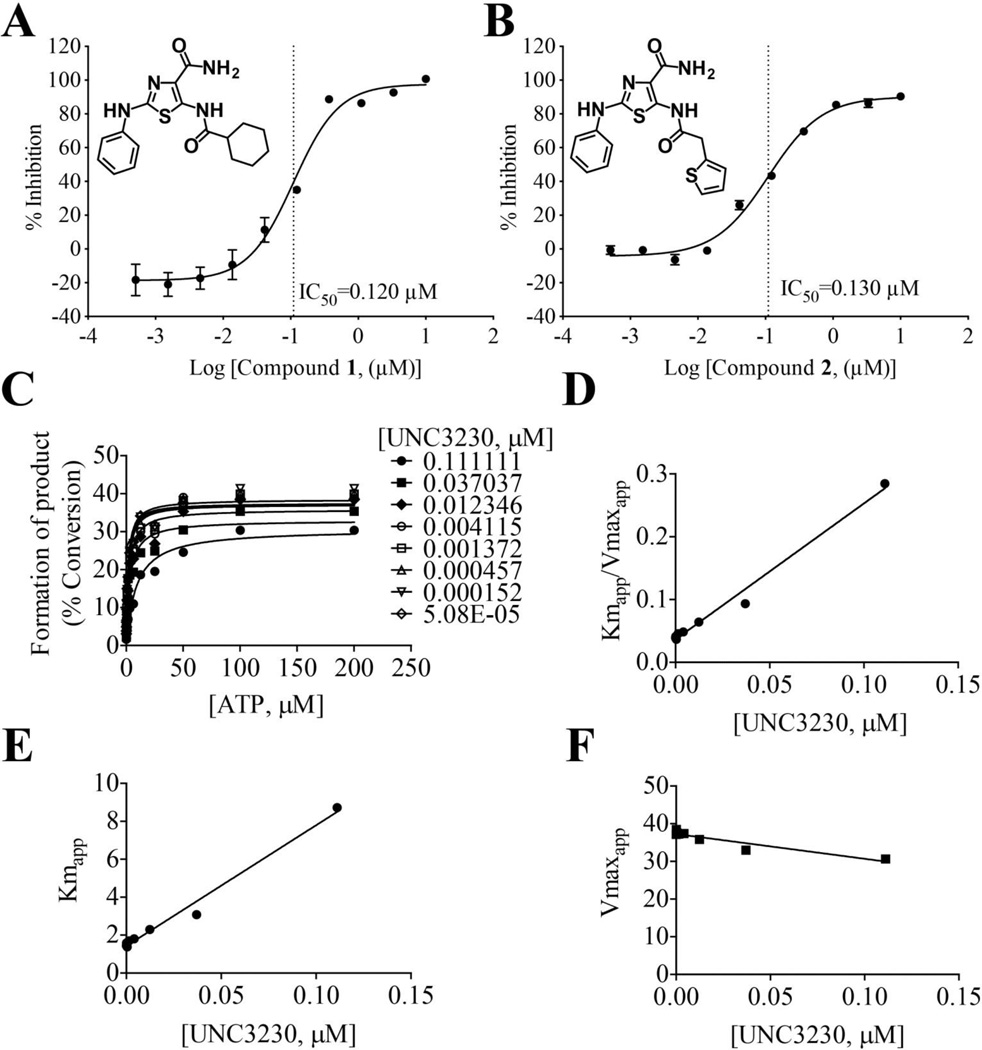
Of the 18 active kinase-focused library compounds, compound 1 (UNC3230) and 2 had identical thiazole carboxamide core structures and were the most potent inhibitors of PIP5K1C (IC50 of 0.120 and 0.130 µM, respectively; Figure 5 and Supplemental Table 2). Further optimization of the assay (incubation of kinase with compound for 20 minutes prior to the addition of kinase), using a new lot of kinase and a tighter dose response curve (10 point; 1 µM highest concentration) gave a reproducible potency value of ~40 nM for UNC3230.9 There were 3 additional compounds with IC50 values less than 1 µM and a total of 7 compounds with IC50 values less than 5 µM. Selectivity and in vivo characterization of UNC3230 was completed as described.9 Briefly, UNC3230 was highly selective for PIP5K1C and PIP4K2C (also generates PIP2 albeit with a different substrate, phosphatidylinositol 5-phosphate), reduced PIP2 in DRG neurons, and attenuated pain hypersensitivity when injected intrathecally in rodent models of chronic pain. Selectivity screening reported in Wright et al. was performed by DiscoveRx using an orthogonal competitive binding assay that was not reliant on fluorescently labeled substrate providing independent confirmation that UNC3230 is directly inhibiting PIP5K1C phosphorylation of PI(4)P.9
Figure 5. The two most potent inhibitors of PIP5K1C have the same core structure and UNC3230 exhibits ATP-competitive mode of inhibition.
(A) Compound 1 (UNC3230) and (B) Compound 2 (UNC2828) have the same thiazole carboxamide core structure and IC50 values of 120 and 130 nM, respectively. Mean ± SD. n=3 reactions per data point. (C) Progression curves for ATP competition studies with PIP5K1C and various concentrations of UNC3230. (D) Plot of Kmapp/Vmaxapp to determine inhibitory constant, Ki. (E) Plot of Kmapp and (F) Vmaxapp against increasing concentrations of UNC3230 to determine mode of inhibition.
In additional follow up studies, the mode of inhibition of our most potent compound, UNC3230, was evaluated in two independent ATP competition studies using our mobility shift assay (Figure 5C–F). These studies demonstrated that UNC3230 was competitive against ATP with an average Ki of 23 nM (Figure 5C–F). The slight decrease in Vmaxapp is not statistically significant, thus we conclude that UNC3230 is an ATP-competitive inhibitor of PIP5K1C.
DISCUSSION
Previous studies of cellular process that are regulated by type I PIP5Ks have been limited to genetic and molecular biology techniques, including the use of genetic knockout mice and targeted RNA interference, due to the lack of available pharmacological inhibitors.3, 13, 14, 23 The use of genetic knockout mice is time- and resource-intensive and limits studies to irreversible deletion of the enzyme, thwarting information on acute inhibition of the enzyme.24 Although PIP5K1 is thought to regulate cellular processes via its enzymatic activity (production of PIP2), PIP5K1s contain several protein binding motifs and hence might also exert some degree of regulation independent of catalytic activity.25, 26, 27 With the use of our inhibitors, studies of acute inhibition of PIP5K1C are now possible. Additionally, these inhibitors could provide a necessary tool to differentiate between scaffolding and/or enzymatic regulation of each PIP5K1C-dependent cellular process25, 27, 28, assuming inhibitor binding reduces only the catalytic activity and does not change the protein conformation substantially. To make this assumption, additional studies are needed on inhibitor binding to the protein, which include acquisition of a crystal structure. As noted previously, our assay identified UNC3230 that was used successfully as a complementary pharmacological approach to validate PIP5K1C regulation of nociceptive signaling that was shown to be dependent on PIP5K1C catalytic activity using genetic-based experiments.9
Furthermore, current techniques in the field of PIP5K1C-regulated cellular processes depend on overexpression of phosphatases and kinases that transiently eliminate or synthesize PIP2, respectively.3, 29, 30 These techniques perturb the complex, interconnected phosphatidylinositol signaling cascade at several critical enzymatic points in a single experiment. The identified inhibitors provide an attractive alternative to reduce PIP2 by specifically inhibiting PIP5K1C and PIP5K2C without disturbing other critical enzymes in the cascade.
Unlike the PIP4K2s, there are no crystal structures of any of the PIP5K1s making structure-guided drug design impractical11; however, standard structure-activity relationship (SAR) studies are feasible and required. As we previously reported9, UNC3230 has limited aqueous solubility, highlighting the need for medicinal chemistry optimization to increase solubility while maintaining potency and selectivity. The assay described herein will be used for high-throughput evaluation of analogs derived from the identified thiazole carboxamide core structure that would be more feasible for further preclinical studies.
Importantly, our HTS assay can be extended to the study of other lipid kinases that regulate cellular signaling and disease physiology. The only limitation to extending our assay to other lipid kinases is the availability of fluorescently conjugated substrates and recombinant proteins. However, many fluorescently labeled lipid substrates are available for purchase and recombinant proteins can be expressed and purified from bacculovirus or bacterial systems. Furthermore, this new assay can be expanded to screen larger libraries to provide additional scaffolds for continued development of PIP5K1C inhibitors for analgesic drug development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chatura Jayakody for compound management, Melissa Porter and Michael Stashko for help with assay development, and Drs. Stephen Frye and Jian Jin for medicinal chemistry expertise and guidance. This work was supported by grants to M.J.Z. from NINDS (R01NS081127, R01NS067688).
REFERENCES
- 1.Delage E, Puyaubert J, Zachowski A, et al. Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: convergences and divergences among eukaryotic kingdoms. Prog Lipid Res. 2013;52(1):1–14. doi: 10.1016/j.plipres.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9(2):162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 3.Balla T, Szentpetery Z, Kim YJ. Phosphoinositide signaling: new tools and insights. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes D. PI3K pathway inhibitors approach junction. Nat Rev Drug Discov. 2011;10(8):563–564. doi: 10.1038/nrd3527. [DOI] [PubMed] [Google Scholar]
- 5.Alshaker H, Sauer L, Monteil D, et al. Therapeutic potential of targeting SK1 in human cancers. Adv Cancer Res. 2013;117:143–200. doi: 10.1016/B978-0-12-394274-6.00006-6. [DOI] [PubMed] [Google Scholar]
- 6.Nagahashi M, Hait NC, Maceyka M, et al. Sphingosine-1-phosphate in chronic intestinal inflammation and cancer. Adv Biol Regul. 2014;54:112–120. doi: 10.1016/j.jbior.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamia KA, Peroni OD, Kim YB, et al. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta−/− mice. Mol Cell Biol. 2004;24(11):5080–5087. doi: 10.1128/MCB.24.11.5080-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voss MD, Czechtizky W, Li Z, et al. Discovery and pharmacological characterization of a novel small molecule inhibitor of phosphatidylinositol-5-phosphate 4-kinase, type II, beta. Biochem Biophys Res Commun. 2014;449(3):327–331. doi: 10.1016/j.bbrc.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 9.Wright BD, Loo L, Street SE, et al. The lipid kinase PIP5K1C regulates pain signaling and sensitization. Neuron. 2014;82(4):836–847. doi: 10.1016/j.neuron.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lima S, Milstien S, Spiegel S. A real-time high-throughput fluorescence assay for sphingosine kinases. J Lipid Res. 2014;55(7):1525–1530. doi: 10.1194/jlr.D048132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis MI, Sasaki AT, Shen M, et al. A homogeneous, high-throughput assay for phosphatidylinositol 5-phosphate 4-kinase with a novel, rapid substrate preparation. PLoS One. 2013;8(1):e54127. doi: 10.1371/journal.pone.0054127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frye SV. The art of the chemical probe. Nat Chem Biol. 2010;6(3):159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 13.Volpicelli-Daley LA, Lucast L, Gong LW, et al. Phosphatidylinositol-4-phosphate 5-kinases and phosphatidylinositol 4,5-bisphosphate synthesis in the brain. J Biol Chem. 2010;285(37):28708–28714. doi: 10.1074/jbc.M110.132191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenk MR, Pellegrini L, Klenchin VA, et al. PIP kinase Igamma is the major PI(4,5)P(2) synthesizing enzyme at the synapse. Neuron. 2001;32(1):79–88. doi: 10.1016/s0896-6273(01)00456-1. [DOI] [PubMed] [Google Scholar]
- 15.Hutti JE, Porter MA, Cheely AW, et al. Development of a high-throughput assay for identifying inhibitors of TBK1 and IKKepsilon. PLoS One. 2012;7(7):e41494. doi: 10.1371/journal.pone.0041494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 17.Sciences CL. LabChip Mobility-Shift Assay: Phosphatidylinositol-3 Kinase PI3Ka. LC3000-AP-212. 2008:1–4. LC3000-AP-212 (LC3000-AP-212) [Google Scholar]
- 18.Lin S, Fischl AS, Bi X, et al. Separation of phospholipids in microfluidic chip device: application to high-throughput screening assays for lipid-modifying enzymes. Anal Biochem. 2003;314(1):97–107. doi: 10.1016/s0003-2697(02)00616-4. [DOI] [PubMed] [Google Scholar]
- 19.Lee KY. Loss of lipid to plastic tubing. J Lipid Res. 1971;12(5):635–636. [PubMed] [Google Scholar]
- 20.Copeland RA. Mechanistic considerations in high-throughput screening. Anal Biochem. 2003;320(1):1–12. doi: 10.1016/s0003-2697(03)00346-4. [DOI] [PubMed] [Google Scholar]
- 21.Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2000;44(1):235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 22.Walters WP, Murcko MA. Prediction of 'drug-likeness'. Adv Drug Deliv Rev. 2002;54(3):255–271. doi: 10.1016/s0169-409x(02)00003-0. [DOI] [PubMed] [Google Scholar]
- 23.Di Paolo G, Moskowitz HS, Gipson K, et al. Impaired PtdIns(4,5)P2 synthesis in nerve terminals produces defects in synaptic vesicle trafficking. Nature. 2004;431(7007):415–422. doi: 10.1038/nature02896. [DOI] [PubMed] [Google Scholar]
- 24.Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3(12):739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bairstow SF, Ling K, Su X, et al. Type Igamma661 phosphatidylinositol phosphate kinase directly interacts with AP2 and regulates endocytosis. J Biol Chem. 2006;281(29):20632–20642. doi: 10.1074/jbc.M601465200. [DOI] [PubMed] [Google Scholar]
- 26.Mao YS, Yamaga M, Zhu X, et al. Essential and unique roles of PIP5K-gamma and -alpha in Fcgamma receptor-mediated phagocytosis. J Cell Biol. 2009;184(2):281–296. doi: 10.1083/jcb.200806121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao YS, Yin HL. Regulation of the actin cytoskeleton by phosphatidylinositol 4-phosphate 5 kinases. Pflugers Arch. 2007;455(1):5–18. doi: 10.1007/s00424-007-0286-3. [DOI] [PubMed] [Google Scholar]
- 28.Noda Y, Niwa S, Homma N, et al. Phosphatidylinositol 4-phosphate 5-kinase alpha (PIPKalpha) regulates neuronal microtubule depolymerase kinesin, KIF2A and suppresses elongation of axon branches. Proc Natl Acad Sci U S A. 2012;109(5):1725–1730. doi: 10.1073/pnas.1107808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond GR, Fischer MJ, Anderson KE, et al. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337(6095):727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toth DJ, Toth JT, Gulyas G, et al. Acute depletion of plasma membrane phosphatidylinositol 4,5-bisphosphate impairs specific steps in endocytosis of the G-protein-coupled receptor. J Cell Sci. 2012;125(Pt 9):2185–2197. doi: 10.1242/jcs.097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.