ABSTRACT
G-protein-coupled receptors (GPCRs) are the largest family of receptors in many organisms, including worms, mice and humans. GPCRs are seven-transmembrane pass proteins that are activated by binding a stimulus (or ligand) in the extracellular space and then transduce that information to the inside of the cell through conformational changes. The conformational changes activate heterotrimeric G-proteins, which execute the downstream signaling pathways through the recruitment and activation of cellular enzymes. The highly specific ligand–GPCR interaction prompts an efficient cellular response, which is vital for the health of the cell and organism. In this Commentary, we review general features of GPCR signaling and then focus on the Drosophila GPCRs, which are not as well-characterized as their worm and mammalian counterparts. We discuss findings that the Drosophila odorant and gustatory receptors are not bona fide GPCRs as is the case for their mammalian counterparts. We also present here a phylogenetic analysis of the bona fide Drosophila GPCRs that suggest potential roles for several family members. Finally, we discuss recently discovered roles of GPCRs in Drosophila embryogenesis, a field we expect will uncover many previously unappreciated functions for GPCRs.
KEY WORDS: AGS, β-arrestin, GASP, G-proteins, GPCR, GPSM, GRK, RGS, Signaling
Summary: GPCR signaling is required for cells to receive input from the outside world and from neighboring cells. This Commentary reviews known GPCR families and highlights recently discovered roles for GPCRs in Drosophila embryonic development.
Introduction
How cells sense and respond to outside stimuli has been a key question in biology for well over a century. The hypothesized existence of receptors that spanned the cell membrane and were able to both sense and transduce signals seems logical today, but when this idea was first proposed, it was summarily rejected (Lefkowitz, 2013). Decades of biochemical studies have since proven that such receptors exist, and that most are G-protein-coupled receptors (GPCRs). The main feature of GPCRs is their seven transmembrane-spanning segments, which position the N-terminus of the protein on the outside of the cell and the C-terminus inside. GPCRs bind an astoundingly diverse set of ligands – proteins, small molecules, hormones, drugs, photons – usually by capturing the ligand with their N-terminus and/or with a pocket formed by the extracellular and transmembrane domains (Fig. 1). The GPCR cycle, described in more detail below, is an elegant cellular solution for sensing a specific exogenous signal, transducing it to a signaling cascade, and then terminating the signal.
Fig. 1.
The GPCR cycle. The GPCR cycle starts on the top left of the figure. In its basal state, a GPCR is free of ligand (L). Gα binds to GDP and associates with Gβγ. The heterotrimeric protein complex might associate with the receptor at this point, or remain free in the membrane as pictured. Upon ligand binding the GPCR becomes activated and undergoes a conformational change. The activated GPCR acts as a GEF for Gα. The resulting GTP-bound Gα separates from βγ, and the heterotrimeric proteins are active (*). Activated Gα can then interact with an effector (E), such as PLC or adenylate cyclase, which results in effector activation (*) and initiation of a second-messenger cascade. The GTP in Gα is then hydrolyzed to GDP through the activity of Gα and RGS proteins (not shown), leading to Gα inactivation and reassociation of the heterotrimeric protein complex. Independently, GRKs bind to and phosphorylate the GPCR. This stimulates its binding by β-arrestin (βArr), which promotes internalization of the receptor. The GPCR can then be recycled back to the cell surface without ligand, restarting the cycle.
GPCRs are widely represented in most life forms, from bacteria to fungi and animals, including all of the major model organisms. Over 800 GPCRs are encoded in the human genome and well over 700 in the zebrafish (Fredriksson and Schioth, 2005). Caenorhabditis elegans is predicted to encode over 1000 GPCRs, a particularly impressive number, as this figure accounts for over 5% of the entire worm genome. Mice also encode a large number of GPCRs (over 1300). Drosophila encodes over 200 GPCRs, whereas just over 50 are found in Dictyostelium (Prabhu and Eichinger, 2006). In contrast, yeast encode a surprisingly small number of GPCRs – three in Saccharomyces cerevisiae and nine in S. pombe. One kingdom where GPCRs might be absent is the plants, which do not require GPCRs to function as guanine-nucleotide-exchange factors (GEFs) due to the very high abundance of GTP (Urano et al., 2012). Instead, the Gα subunit of the heterotrimeric G protein complex is typically already bound to GTP, with regulation of activity at the level of GTP hydrolysis to halt signaling (Urano and Jones, 2014). Of the 56 putative plant GPCRs identified based on structural homology, only a single candidate – GCR1 – has features consistent with it functioning as a bona fide GPCR with GEF activity (Taddese et al., 2014). Whether or not GCR1 acts as a GEF remains to be determined.
GPCRs are involved in nearly every aspect of animal life, from early development and heart function to neuronal activity (Wettschureck and Offermanns, 2005). Mutations in GPCRs are linked to a number of human diseases, such as Usher syndrome, which results in variable onset deaf-blindness (Schöneberg et al., 2004). Cell migration is another process that requires GPCRs, in both beneficial and detrimental ways. The single-celled amoeba Dictyostelium uses four GPCRs, cAR1–cAR4, to detect cAMP, which triggers migration and coalescence into a multicellular organism (Prabhu and Eichinger, 2006). In both zebrafish and mice, germ cell migration is regulated by the chemokine receptor CXCR4 and its ligand stromal derived factor 1 (SDF1) (Doitsidou et al., 2002; Molyneaux, 2003). Neutrophils also migrate through activation of a GPCR (Becker et al., 1987). GPCR-regulated cell migration can also be detrimental for the organism, primarily during cancer (Lappano and Maggiolini, 2011). Overly activated GPCRs are able to trans-activate epidermal growth factor receptor (EGFR) and other receptors, which can cause unregulated growth and cell migration (Bhola and Grandis, 2008).
In this Commentary, we first cover the basics of GPCR signaling, from how the receptor is arranged in the membrane to heterotrimeric G protein activation and receptor recycling. We focus then on the previously described Drosophila GPCR family, which includes three major groups, the two chemosensory groups – odorant and gustatory – and the classical receptor group. Phylogenetic trees representing all three major groups of Drosophila GPCRs are presented. We cover recent findings that insect odorant and (likely) gustatory receptors, unlike their vertebrate counterparts, are not bona fide GPCRs, leaving the classical Drosophila GPCR family with only 116 members. Finally, we discuss the known roles of GPCRs in Drosophila development, an open and exciting field given the number of uncharacterized GPCRs encoded in the fly genome.
GPCR structure–function relationship
Because the GPCR superfamily is so diverse, there is little sequence conservation among families. Nonetheless, the superfamily does share several architectural features. The N-terminus and extracellular loops (ECLs) are responsible for ligand binding. This can involve direct binding of the ECL to the ligand [as is the case for metabotropic glutamate (mGlu) receptors] or funneling of a hydrophobic ligand into a binding pocket formed by the transmembrane domains (Venkatakrishnan et al., 2013). ECLs often contain disulfide bridges to stabilize the loops and prevent promiscuous GPCR signaling. Indeed, ligand binding does not induce a simple on-off state for GPCRs. GPCRs are dynamic proteins that fluctuate between many states (Fig. 1). Ligand binding stabilizes the GPCR into an ‘on’ position, which is further stabilized by binding of a G-protein (Kobilka, www.nobelprize.org/nobel_prizes/chemistry/laureates/2012/kobilka-lecture.html).
Bioinformatic analyses show that specific residues, such as asparagine, tryptophan and proline, cause clustering and stabilization of the transmembrane domains of the GPCR, collectively termed the GPCR barrel owing to their composite shape in the membrane (Venkatakrishnan et al., 2013). Ligand contact triggers movement of TM3 and causes conformational changes of the intracellular loops (ICLs). In the rhodopsin family, ICL2 contains an E/DRY motif near the boundary between ICL2 and TM3 (Rovati et al., 2007). Mutations within this motif can result in constitutive GPCR signaling or impaired G-protein binding. Interestingly, other than the E/DRY motif, very little is known about how GPCRs associate with specific G-proteins. Chimeras resulting from swapping of ICL3 domains between GPCRs result in a switch in their G-protein selectivity (Kobilka et al., 1988), suggesting that this domain imparts specificity for the downstream signaling pathway of the receptor.
The localization of a GPCR within a cell membrane can affect its ability to signal. Lateral movement of GPCRs within the plasma membrane is often restricted by the preferential localization of GPCRs to a specific lipid microenvironment (Allen et al., 2007). Although GPCRs are usually shown as monomers, they can form oligomers (i.e. homodimers and heterodimers) within lipid rafts and are stabilized by interactions that are mediated through their transmembrane domains (Ferré et al., 2014). Planar lipid rafts and caveolae both influence GPCR signaling by either excluding or recruiting G-proteins and their effectors. Because native GPCRs are not highly expressed in cells (McCusker et al., 2007), lipid rafts concentrate GPCRs and their associated proteins to promote receptor dimerization and signaling. Caveolar-localized GPCRs are also subject to more rapid endocytosis, which represents another way to modulate GPCR signaling (Chini and Parenti, 2004).
Signal propagation and the GPCR cycle
The presence of a ligand–receptor binding event must then be propagated and responded to by the cell itself. The effectors of GPCR activation are the heterotrimeric G-proteins Gα, Gβ and Gγ. Organisms encode several types of each G-protein, and different combinations of these proteins into heterotrimers preferentially activate different signaling pathways. Gα proteins are GTPases, which catalyzes the hydrolysis of GTP to GDP. Gα proteins are typically anchored in the membrane by N-terminal palmitoylation and can also be myristoylated (Vögler et al., 2008). Gγ proteins are isoprenylated at their C-terminal CAAX motif (Higgins and Caseys, 1994). Gβ proteins do not have any membrane-anchoring post-translational modifications. Instead, they are tightly linked to Gγ through hydrophobic interactions (Sondek et al., 1996). A heterotrimeric complex can dock to an inactivated receptor or drift in the membrane, but once it encounters a ligand-bound GPCR, downstream signaling is initiated (Fig. 1). Activated GPCRs act as GEFs and exchange GDP for GTP in the Gα subunit, which activates the protein. Upon GTP binding, Gα changes its conformation, allowing it to separate from the Gβγ dimer. The subunits are then free to interact with downstream targets. When Gα hydrolyzes GTP into GDP, it becomes inactivated, allowing Gα to reassociate with Gβγ. This process represents a full GPCR G-protein cycle.
Gα proteins are divided into four subclasses with each targeting a specific type of signaling cascade (Wettschureck and Offermanns, 2005). Gα(s), Gα(i), and Gα(o) all regulate adenylate cyclases. Gα(s) stimulates adenylate cyclase activity, whereas Gα(i) and Gα(o) are inhibitory. The third subclass, Gα(q/11) targets phospholipase C (PLC), which cleaves phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] into inositol trisphosphate (IP3) and membrane-bound diacylglycerol (DAG). Finally, Gα(12/13) activates Rho GEFs, which in turn activate Rho. Previous models have suggested that an individual GPCR interacts with only one specific type of Gα, but it is now established that GPCRs are able to activate several Gα types, albeit with a marked preference for one (Cerione et al., 1985).
Gα proteins are weak GTPases, which slows the signaling cascade because new signaling information cannot be integrated (Kleuss et al., 1994). To accelerate GTP hydrolysis, Gα proteins are targeted by the regulator of G-protein signaling (RGS) molecules (De Vries et al., 2000). RGS proteins are somewhat promiscuous for Gα proteins, and several RGS proteins have been shown to bind to specific Gβ proteins and prevent re-formation of the heterotrimeric complex (Witherow and Slepak, 2003). Conversely, activators of G-protein signaling (AGS) can act as GEFs for Gα to prolong signaling (Vögler et al., 2008). Recent work has shown that Gα can be activated by non-receptor GEFs, which themselves are activated through associations with non-GPCR signaling pathways such as the receptor tyrosine kinase (RTK) pathway (Garcia-Marcos et al., 2015). Some AGS proteins act as guanine-nucleotide-dissociation inhibitors (GDIs) (e.g. the GPR proteins, as well as proteins that contain a GoLoCo motif), causing Gα to remain in a GDP-bound state. These GDI-AGS proteins are termed G-protein signaling modulators (GPSMs) and affect the amount of free Gβγ that can signal because they also prevent re-association of the heterotrimeric protein complexes. Finally, the RZ family of RGS proteins has recently been shown to have the dual activities of promoting Gα GTP hydrolysis and inhibiting the exchange of GDP for GTP (Lin et al., 2014).
Compared to Gα (for which there are 23 genes), there are fewer Gβ and Gγ genes in mammals, five and 12 genes, respectively. This would suggest that there are as many as 60 βγ complexes, each with different preferences for specific Gα subunits (Dingus et al., 2005). Gβγ can affect a wide range of ion channels and other signaling effectors, including pathways that are also targeted by Gα, such as PLC (Lau et al., 2013). Because the Gβγ dimer has long been considered to be a less important pathway component, the exact roles different combinations of Gβγ dimers have on GPCR signaling remain to be elucidated.
Several mechanisms exist to attenuate GPCR signaling. The same conformational change in the GPCR that results in the release of the heterotrimeric complex allows accessibility for phosphorylation by G-protein-coupled receptor kinases (GRKs) (Palczewskiss et al., 1991). Gβγ is able to recruit a GRK to the GPCR, thus establishing a negative-feedback loop (Luttrell et al., 1999). GRKs usually phosphorylate GPCRs at serine or threonine residues in the ICL3 or the C-terminal tail of the activated receptor (Pitcher et al., 1998). This phosphorylation of the GPCR results in the binding of β-arrestin (Drake et al., 2006), which then recruits clathrin and its adaptor protein AP-2, to internalize the GPCR. Several different fates await the GPCR following internalization and GPCRs are divided into two classes depending upon how strongly they maintain β-arrestin binding. Class A GPCRs lose β-arrestin following internalization (Oakley et al., 2000) and can be dephosphorylated and recycled back to the cell surface. An emerging concept in the field is the ability of GPCRs to maintain signaling once they are endocytosed (Mullershausen et al., 2009). It is thought that by continuing to signal at endosomes instead of the plasma membrane, the physical distance between the GPCR and the nucleus is reduced, resulting in more efficient signaling to transcriptional pathways (Tsvetanova and von Zastrow, 2014). Returning a GPCR back to the plasma membrane completes the GPCR cycle (Fig. 1). Class B GPCRs maintain β-arrestin binding. Here, GRK-mediated phosphorylation and binding to β-arrestin can stimulate the ubiquitylation of GPCRs. Ubiquitylated receptors are then targeted to the lysozome for degradation. More recently, the so-called GPCR-associated sorting protein (GASP) family has been identified, which assists in the decision as to whether GPCRs are degraded or recycled (Bornert et al., 2013; Simonin et al., 2004). The amino acid sequence at the C-terminus of the GPCR is thought to direct the type of adaptor protein that binds the receptor, thus influencing the fate of the receptor once it is internalized (Marchese et al., 2008).
GPCR families
The rhodopsin-like family is the largest family of GPCRs in most organisms. Members of this family are well-known for the diversity of their ligands, which range from hormones and peptides to odorants and photons of light. Rhodopsin was first described as a light-sensitive compound in animals (Kühne, 1877), and was cloned over a century later (Nathans and Hogness, 1983). This finding incited the explosion of subsequent GPCR research. Because the rhodopsin-like family represents over 80% of human GPCRs (Fredriksson and Schioth, 2005), it has been intensely studied for potential therapeutic benefits. The huge number of mammalian odorant or olfactory receptors (∼700 in humans and ∼1200 in rodents) all fall into the rhodopsin-like family based on sequence homology. Olfactory GPCRs activate a Gα(olf) subunit that activates an adenylyl cyclase, leading to increases in cAMP (DeMaria and Ngai, 2010). Increased cAMP, in turn, activates cyclic-nucleotide-gated ion channels that bring in Na+ and Ca2+, thereby depolarizing the cell. Ca2+-gated Cl− channels are then activated, leading to further depolarization. Although insect odorant receptors were initially thought to function in the same way, it appears that the seven-transmembrane-pass insect odorant receptors are not bona fide GPCRs and instead function directly as ligand-gated ion channels (see below) (Ha and Smith, 2008; Leal, 2013; Wicher et al., 2008).
Another important GPCR familiar is the secretin-like family. Its main feature is its large N-terminal extracellular domain (ECD) (Watkins et al., 2012), which is crucial for the recognition and binding of ligands, typically peptides or hormones. Historically, this family was named for the intestinal hormone secretin, which, in the early 20th century, was the first hormone discovered (Bayliss and Starling, 1902). Its receptor was first described nearly 80 years later (Jensen and Gardner, 1981; Chey and Chang, 2003).
mGlu receptors bind a diverse set of ligands, such as pheromones, amino acids and Ca2+ (Chun et al., 2012). Members of this family contain a large ECD that forms a so-called Venus flytrap (VFT) module (Bessis et al., 2002). Upon ligand binding to one lobe of the VFT, the other lobe closes, introducing a conformational change that is transduced to the rest of the protein through a cysteine-rich region. mGlu receptors function as dimers that are either covalently linked by disulfide bonds or by shared ion binding. Compared to the other GPCR families, the mGlu family was discovered relatively late. Although glutamate was a known neurotransmitter, it was assumed to function solely through channels or ionotropic receptors, which themselves function as channels (Curtis and Watkins, 1965). However, metabotropic glutamate receptors function as conventional GPCRs by binding a ligand and modulating that signal through the membrane to downstream G-proteins.
The final family of GPCRs are the atypical GPCRs, which includes receptors such as Frizzled or adhesion GPCRs. Members of these families were initially thought to primarily not signal through heterotrimeric G-proteins (Tang et al., 2012). Frizzled family members contain a cysteine-rich domain in their N-terminus that binds lipoglycoproteins of the Wingless (Wnt) family (Yang-Snyder et al., 1996). Upon ligand-binding, Frizzled family members usually signal through the phosphoprotein Dishevelled (Schulte and Bryja, 2007). More recently, however, both Frizzled family members and Smoothened have been shown to also function as canonical GPCRs, with Frizzled proteins acting as GEFs for Gα(o/i) proteins (Koval and Katanaev, 2011; Nichols et al., 2013), and Smoothened acting as a GEF for Gα(i) (Shen et al., 2013). GPCRs of the related adhesion group often contain cadherin or integrin domains, and these receptors often have auto-proteolytic activity (Krasnoperov et al., 1997). Their ligands include components of the extracellular matrix, such as collagen (Luo et al., 2011).
Ligand identification
The most common way to identify the upstream and downstream components of a GPCR pathway is through heterologous expression systems, such as Xenopus oocytes, and HEK293 or CHO cells. In this way, specific GPCRs can then be exposed to individual ligands to test for functionality. Heterologous expression systems are preferentially used to identify ligands for GPCRs (a process called ‘de-orphanizing’ or ‘deorphaning’) because it reduces the possibility of endogenous proteins triggering the receptor, as most GPCRs are quite specific for ligand recognition (Caers et al., 2014). Many downstream effectors have not been singularly identified as coupling to a specific receptor and, instead, output is typically measured through changes in Ca2+ levels. Activation of Gα(q) releases Ca2+ through the downstream effector PLC, and various Ca2+ reporters can be used to track this change. Although changes in cAMP and Rho GEF activity are not as easily tracked, ligand-induced changes in cAMP levels have been used to link the neuropeptide controlling Drosophila circadium rhythms, known as pigment-dispersal factor (PDF), to an orphan secretin-related GPCR, known as PDF-R or Han (Hyun et al., 2005; Mertens et al., 2005). To circumvent challenges in linking orphan receptors to their cognate ligands, GPCRs have typically been expressed along with the promiscuous human Gα16, which readily couples with many GPCRs to cause changes in intracellular Ca2+. Although this system can identify ligand–receptor interactions, it does not identify the endogenous Gα that interacts with a GPCR. Moreover, as stated above, GPCRs are known to interact with more than one Gα, causing multiple output changes. Studies that have identified specific Gα coupling to a receptor have often been performed using physical and genetic interaction data.
Drosophila GPCRs
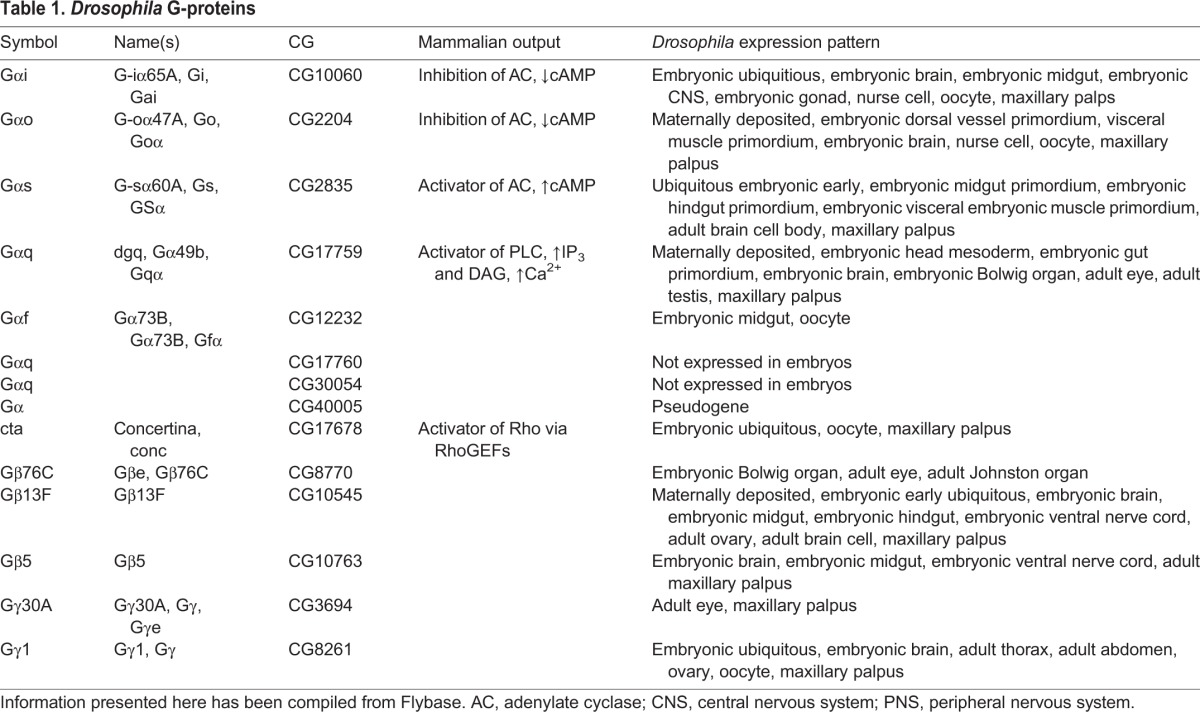
Drosophila encodes over 200 proteins originally considered to be GPCRs based on domain topology and other key structural features (Brody and Cravchik, 2000). Not surprisingly, modulators of GPCR signaling, such as GRKs, GDIs, GPCR kinases and arrestins, are also encoded in the Drosophila genome. One aspect of Drosophila GPCR signaling that makes it particularly appealing for study is the reduced number of G-proteins – nine genes encode predicted Gα proteins (six have been characterized and three are known only by their CG numbers), three genes encode Gβ proteins and two genes encode Gγ proteins (Table 1) (Katanayeva et al., 2010; Anantharaman et al., 2011). Of the three Gα subunits with only CG designations, one appears to be a pseudogene (CG40005) and the other two are not expressed in embryos (CG30054 and CG17760). Indeed, only two of the Gβ proteins (Gβ5 and Gβ13f) and a single G γ-subunit (Gγ1) are expressed during the first half of Drosophila embryogenesis. With only 13 G-protein subunits, Drosophila offers a relatively simple system for studying GPCR signaling. However, compared to worms and mammals, the fly GPCRs are not as well characterized. Nearly half of the Drosophila GPCRs are orphans, suggesting that the field is relatively wide open for new discoveries. Here, we present new insights into this field that might aid in future characterization of Drosophila GPCRs and we discuss the roles of the receptors in embryonic development, which is also a rather unstudied side of GPCR signaling.
Table 1.
Drosophila G-proteins
Insect odorant receptors function as ionotropic receptors and are not bona fide GPCRs
There are 60 odorant receptors encoded in the Drosophila genome and, based on our Clustal Omega analysis, they fall into four major clades (A–D) (supplementary material Fig. S1; Table S1). The odorant receptor A group includes 22 genes. The odorant receptor B and odorant receptor C group include 17 genes each, whereas odorant receptor D, the smallest clade, includes only four genes. Vertebrate odorant receptors are bona fide GPCRs; they are seven-transmembrane-pass proteins that are members of the large rhodopsin class of GPCRs. The vertebrate odorant receptors are coupled to Gα proteins that activate an adenylate cyclase, leading to increased levels of cAMP, which eventually leads to cell depolarization through the sequential activation of cyclic-nucleotide- and ion-gated channels. Insect odorant receptors, by contrast, do not appear to function as classical GPCRs. The first hint that they could be different was the discovery that these proteins have an inverse topology from typical GPCRs: the N-terminus is intracellular, whereas the C-terminus is extracellular (Benton et al., 2006; Lundin et al., 2007). This would suggest that the domains that normally directly interact with the Gα subunit are outside the cell. Indeed, insect odorant receptors are not metabotropic receptors. Upon ligand binding, insect odorant receptors dimerize with a co-receptor protein known as Orco, an odorant receptor found in the odorant receptor B clade. The activated heterodimer (or possibly heterotetramer) of an odorant-specific odorant receptor subunit and Orco then functions directly as a non-selective cation channel (Wicher et al., 2008), allowing for a much quicker response to perceived odorants, which has been suggested to be a particular advantage for insects that fly and need to process information quickly (Wicher, 2015). Thus, as ionotropic receptors, the Drosophila seven-transmembrane-domain olfactory receptors are not true GPCRs; they do not function as GEFs for G-proteins.
Insect gustatory receptors might also function as ionotropic receptors
Gustatory receptors are located on the antenna, labellum, wing hairs and leg hairs, and sense a diverse array of compounds including sugars, bitter compounds, amino acids and sex pheromones. The 60 identified Drosophila gustatory receptors separate into three major clades A, B and C based on a Clustal Omega analysis of all members (supplementary material Fig. S2; Table S2). The large A clade, which includes 45 of the known gustatory receptors, can be further divided into two subordinate clades, I and II. Clade A-I includes the eight related sweet-taste receptors (Gr5a, Gr61a, Gr64a Gr64b, Gr64c, Gr64d, Gr64e and Gr64f) and the CO2 receptors (Gr21a and Gr63a), which form a functional heterodimer. Gustatory neurons that respond to sweet taste are also known to express γ-aminobutyric acid (GABA) receptors, and the loss of these receptors influences the perception of sugar (Chu et al., 2014). The GABA receptors are proposed to be important for increasing the dynamic range of sweet perception and for suppressing output when bitter tastes are also present. The two receptors for a female mating pheromone (Gr68a and Gr32a) cluster together in the small C clade. Unlike the gustatory receptors of vertebrates, which, upon ligand binding, undergo conformational changes that initiate G-protein signaling to activate specific downstream enzymes, the Gr proteins of Drosophila and other insects might function as ligand-gated ion channels. Based entirely on sequence analyses, the gustatory receptors of Drosophila and other insects, like the odorant receptors, appear to be inserted into the plasma membrane with a reverse topology relative to that of the vertebrate gustatory receptors, which are bona fide GPCRs (Kent and Robertson, 2009). Indeed, many Gr proteins are predicted to have more than seven transmembrane domains, further separating them from classical GPCRs. Thus, the Drosophila gustatory receptors might also function as ionotropic receptors, although direct experimental evidence for this is lacking. Overall, it appears that insects sense large parts of their chemical worlds by linking ligand binding directly to the opening of ion channels rather than using G-proteins to indirectly produce second messengers, which then open ion channels (Silbering and Benton, 2010).
Phylogenetic analysis of classical Drosophila GPCRs
Having removed the odorant and gustatory receptors from the list of GPCRs, we are left with 116 presumed GPCRs encoded by the Drosophila genome. Using Clustal Omega with five combined iterations, we created a phylogenetic tree using these sequences (Fig. 2, supplementary material Table S3). We termed this GPCR set the classical GPCRs. The classical data set comprises 116 proteins, whose sequences were obtained from Flybase (www.flybase.org); for proteins with more than one isoform, only the first isoform listed was used. Historically, Drosophila GPCRs have been classified into four separate families: rhodopsin-like, secretin-like, mGlu-like and atypical, which included the Frizzled group. Based on our analysis, which includes only the Drosophila seven-transmembrane-pass proteins that are likely to function as bona fide GPCRs, we see a very different tree. The classical GPCRs separate into three major clades, A, B and C. The large clade A includes many members of the classical rhodopsin-like family but also includes all of the other previously categorized families: secretin-like, mGlu-like, and Frizzled and atypical. Clade B includes the receptors for the biogenic amines, receptors that are bound and activated by molecules such as serotonin, dopamine, adrenaline and epinephrine. This clade includes three proteins identified only by CG number (CG18208, CG12796 and CG13579) that, based on this classification, might also bind biogenic amines. Clade C includes a subset of rhodopsin-like receptors that bind small peptides that often function as neuropeptides, including the allostatin, tachykinin and pyrokinin receptors; we refer to this clade as peptide A.
Fig. 2.
Rooted phylogenetic tree of classical Drosophila GPCRs. Full-length sequences of GPCRs obtained from Flybase (www.flybase.org) were used to construct the tree. The names correspond with the colored clades. Colors also correspond to those used in supplementary material Table S3.
Within clade A, the remaining rhodopsin-like receptors fall into five different groups: the peptide B (eight proteins) and peptide C (three proteins) groups bind different flavors of small peptides. Two proteins with only CG numbers share a common branch with the peptide B group of proteins, indicating that they might have related ligands. The glycoprotein group (four proteins) includes a single uncharacterized CG protein (CG34411), Rk, and the Lgr1 and Lgr2 proteins; these receptors have large leucine-rich repeat domains. The opsins fall into a separate group that includes all seven of the known rhodopsin vision receptors. Finally, two CG proteins (CG30340 and CG13995) cluster with the ‘orphan’ receptors, receptors for which the ligands remain completely unknown. Also included in the large A clade are GPCRs known as the ‘glutamate’ or ‘Class C’ family; these include two glutamate and three GABA receptors. The Frizzled and Smoothened receptors, which have been shown to function as bona fide GPCRs with GEF activity (Riobo et al., 2006; Koval and Katanaev, 2011; Shen et al., 2013), fall into a single cluster, which was previously known as the ‘Frizzled’ or ‘Class F’ GPCRs. The Frizzled proteins are closely grouped with the cell adhesion, cysteine-rich and secretin-like proteins. The Methuselah-like proteins, which have been described as being secretin-like, share a common upstream branch, but this branch is also shared with the glutamate or Class C group. Only two of the 16 Methuselah-like proteins have been characterized, one is involved in aging and another involved in gastrulation (see below). Two additional groups in clade A fall outside the previously categorized GPCRs, including one with two proteins, Boss and CG32547, and another with six proteins, including Pog and five genes that are only known by their CG numbers.
Further supporting our phylogenetic analysis is the finding that genes of known related functions cluster into single groups. For example, all of the visual (rhodopsin) receptors cluster into a single group, as do all Methuselah-like proteins. Likewise, neurotransmitter receptors, including those for dopamine, octopamine, histamine, serotonin and acetylcholine, group together into the biogenic amine clade.
GPCRs in Drosophila development
Many of the characterized Drosophila GPCRs regulate adult behavior, but considerably less is known about the role of GPCRs in development. Before cellularization occurs, embryonic development in Drosophila is largely dictated by the action of diffusible gradients. Because the early embryo is essentially one cell with many nuclei, cell-to-cell communication is not necessary. However, as development progresses, the activities of newly formed tissues and organs must be coordinated to form a viable organism. Although many GPCRs are expressed during embryonic stages, very little is known with regard to their roles in specific developmental processes. Interestingly, only six Gα, two Gβ (Gβ5 and Gβ13f) and one Gγ (Gγ1) genes are expressed in early embryos (Table 1). Here, we discuss the only known roles of GPCRs in Drosophila development.
One of the first major events of embryonic development is gastrulation. Concertina (a Gα) and Fog (a secreted protein) were known to be necessary for this process by the early 1990s, but the GPCR that linked these two molecules remained undiscovered for over 20 years (Parks and Wieschaus, 1991; Costa et al., 1994). In addition, the GEF Ric8a was also identified as a crucial component for gastrulation (Peters and Rogers, 2013), further supporting the idea that a GPCR was involved in this process. Recently, Mist has been identified as a GPCR that coordinates these events and triggers the initial cell shape changes (Fig. 3A; Manning et al., 2013). The GPCR kinase Gprk2 acts downstream of Fog signaling and limits the cell-shape changes to ventral mesodermal cells (Fuse et al., 2013), potentially by limiting Mist activity.
Fig. 3.

GPCRs in Drosophila development. (A) The GPCR Mist binds to its ligand Fog in the early embryo; this activates the Gα Concertina (Cta) and its downstream effector Rho1 to mediate apical constriction of mesodermal cells during gastrulation. (B) Tre1 has two roles in the developing embryo. In germ cells (teal), Tre1 activates Gβ13f and Gγ1 to relocalize Rho1 and E-cadherin (ECad) to the rear of the migrating cells. In neuroblast stem cells (green), Tre1 interacts with Gα(o), which binds to Pins. Pins interacts with proteins that orient the mitotic spindle in such a way that correct cell fates are established. (C) Moody activates Gα(i), Gα(o), Gβ13f, and Gγ1 to alter the accumulation of actin in the surface glia, and affecting the organization of septate junction proteins. (D) An unknown GPCR is activated in cardial cells, which activates Gα(o), Gβ13f, and Gγ1. These proteins influence the localization of septate junction proteins in cardial cells, which interact in trans with septate junction proteins on pericardial cells, thereby facilitating adhesion between these two types of cells to ensure proper cardiogenesis. PNS, peripheral nervous system; CNS, central nervous system.
The role of the atypical GPCRs of the Frizzled family in establishing segment polarity through the Wnt signaling pathway has been well characterized. Frizzled family members can also act as canonical GPCRs by signaling through Gα(o) (Katanaev et al., 2005), which in turn recruits Rab5, an important regulator of endocytosis, to the plasma membrane (Purvanov et al., 2010). Endocytosis and morphogenesis are tightly linked, so this link between Gα(o) and Rab5 has potentially important implications for embryonic development. Frizzled and other proteins involved in planar polarity have also proven roles in tracheal development (Chung et al., 2009; Warrington et al., 2013), but a specific role of Frizzled in GPCR signaling within the context of this developmental process has not been examined.
As in other organisms, embryonic germ cell migration is also directed by GPCR signaling in the fly. Loss of Tre1 severely affects germ cell migration in the embryo (Fig. 3B; Kunwar et al., 2003). Tre1 was named for its phenotype: instead of migrating to the gonad, the germ cells remain trapped in the endoderm. The arginine in the DRY motif is necessary for Tre1 function (Kamps et al., 2010). Tre1 has been shown to influence the distribution of Rho1 and E-cadherin (known as Shotgun in Drosophila) in the germ cells, although the mechanism for this interaction remains unknown (Kunwar et al., 2008). Tre1 also influences embryonic neuroblast stem cell divisions (Yoshiura et al., 2012). Through interactions with its partners Gα(o) and Pins (a GDI protein), Tre1 has been shown to play a key role in the oriented cell divisions that establish neuroblast cell polarity and cell fate. Despite these known roles for Tre1 in controlling individual cell behavior, Tre1 remains an orphan receptor as its ligand is unknown.
There are also some insights into the role of Drosophila GPCRs in cell adhesion and boundary integrity. The Gα(o)–Gβ13f–Gγ1 complex mediates cardiomyocyte adhesion, and loss of Gγ1 causes mislocalization of septate junction proteins in the embryonic heart (Fig. 3D; Yi et al., 2008). Septate junctions are also compromised in moody mutants, which functions in the late embryonic surface glia to form the blood–brain barrier that insulates the nerve cord (Fig. 3C; Schwabe et al., 2005). Gα(i), Gα(o) and Loco likely function in the same pathway, which is thought to directly affect the actin organization required for septate junction assembly. Like Tre1, Moody is also an orphan receptor, and these proteins belong to the same group within clade A in our phylogenetic analysis. As many Drosophila GPCRs remain uncharacterized, exciting work remains to fully understand their potential roles in developmental processes.
Conclusions
In conclusion, GPCR signaling is vital to life. GPCRs are able to translate an outside stimulus into a cellular response on a millisecond timescale, thus connecting the behavior of a cell to its outer environment. GPCRs are involved in many key developmental events, but they also participate in more refined events, such as discerning the difference between closely related neuropeptides. Ligands for GPCRs are immensely diverse, and many GPCRs remain orphans. Through phylogenetic analysis of the bona fide GPCRs encoded by the Drosophila genome, we hope to add new insights into the activities of this important family. Heterologous expression systems have historically been used to identify GPCR ligands. However, this method has limitations. Most notably, this method ignores spatiotemporal information, an aspect that is vital in living organisms. Most ligand–receptor pairs have been identified in adult organisms as responding to behavior inputs such as odorants or mating pheromones. More nuanced analysis is needed to understand the role of GPCRs in developmental processes, where both components are genetically encoded and must be expressed at the right time and place to interact with one another. Bioinformatic analysis can point to potential shared or related ligands in clades where well-characterized and uncharacterized receptors cluster together. Our analysis suggests that there are roles for several uncharacterized Drosophila GPCRs, which will hopefully spur further research about the exact ligand, pathway and expression profile for each of these genes.
Acknowledgements
We would like to thank members of the Andrew laboratory for helpful comments and suggestions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Our own work in this area has been funded by the Ruth l. Kirschstein national individual research award [grant number 5F31DE022233] and the National Institutes of Health [grant number RO1 DE013899]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.175158/-/DC1
References
- Allen J. A., Halverson-Tamboli R. A. and Rasenick M. M. (2007). Lipid raft microdomains and neurotransmitter signalling. Nat. Rev. Neurosci. 8, 128-140. 10.1038/nrn2059 [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Abhiman S., de Souza R. F. and Aravind L. (2011). Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene 475, 63-78. 10.1016/j.gene.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss W. M. and Starling E. H. (1902). The mechanism of pancreatic secretion. J. Physiol. 28, 325-353. 10.1113/jphysiol.1902.sp000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E. L. Kanaho Y. and Kermode J. C. (1987). Nature and functioning of the pertussis toxin-sensitive G protein of neutorphils. Biomed. Pharmacother. 41, 289-297. [PubMed] [Google Scholar]
- Benton R., Sachse S., Michnick S. W. and Vosshall L. B. (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4, e20 10.1371/journal.pbio.0040020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessis A.-S., Rondard P., Gaven F., Brabet I., Triballeau N., Prezeau L., Acher F. and Pin J.-P. (2002). Closure of the Venus flytrap module of mGlu8 receptor and the activation process: insights from mutations converting antagonists into agonists. Proc. Natl. Acad. Sci. USA 99, 11097-11102. 10.1073/pnas.162138699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhola N. E. and Grandis J. R. (2008). Crosstalk between G-protein-coupled receptors and epidermal growth factor receptor in cancer. Front. Biosci. 13, 1857-1865. 10.2741/2805 [DOI] [PubMed] [Google Scholar]
- Bornert O., Møller T. C., Boeuf J., Candusso M.-P., Wagner R., Martinez K. L. and Simonin F. (2013). Identification of a novel protein-protein interaction motif mediating interaction of GPCR-associated sorting proteins with G protein-coupled receptors. PLoS ONE 8, e56336 10.1371/journal.pone.0056336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T. and Cravchik A. (2000). Drosophila melanogaster G protein – coupled receptors. J. Cell Biol. 150, F83-F88. 10.1083/jcb.150.2.F83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caers J., Peymen K., Suetens N., Temmerman L., Janssen T., Schoofs L. and Beets I. (2014). Characterizatino of G protein-coupled receptors by a fluorescence-based calcium mobilization assay. J. Vis. Exp. 89, e51516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerione R. A., Staniszewski C., Benovic J. L., Lefkowitz R. J., Caron M. G., Gierschik P. Somers R. Spiegel A. M., Codina J. and Birnbaumer L. (1985). Specificity of the functional interactions of the beta-adrenergic receptor and rhodopsin with guanine nucleotide regulatory proteins reconstituted in phospholipid vesicles. J. Biol. Chem. 260, 1493-1500. [PubMed] [Google Scholar]
- Chey W. Y. and Chang T. M. (2003). Neural control of the release and action of secretin. J. Physiol. Pharmacol. 54, 105-112. 10.1007/BF01315215 [DOI] [PubMed] [Google Scholar]
- Chini B. and Parenti M. (2004). G-protein coupled receptors in lipid rafts and caveolae: how, when, and why do they go there? J. Mol. Endocrinol. 32, 325-338. 10.1677/jme.0.0320325 [DOI] [PubMed] [Google Scholar]
- Chu B., Chui V., Mann K. and Gordon M. D. (2014). Presynaptic gain control drives sweet and bitter taste integration in Drosophila. Curr. Biol. 24, 1978-1984. 10.1016/j.cub.2014.07.020 [DOI] [PubMed] [Google Scholar]
- Chun L., Zhang W.-H. and Liu J.-F. (2012). Structure and ligand recognition of class C GPCRs. Acta Pharmacol. Sin. 33, 312-323. 10.1038/aps.2011.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Vining M. S., Bradley P. L., Chan C.-C., Wharton K. A. Jr and Andrew D. J. (2009). Serrano (Sano) functions with the planar cell polarity genes to control tracheal tube length. PLoS Genet. 5, e1000746 10.1371/journal.pgen.1000746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Wilson E. T. and Wieschaus E. (1994). A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell 76, 1075-1089. 10.1016/0092-8674(94)90384-0 [DOI] [PubMed] [Google Scholar]
- Curtis D. R. and Watkins J. C. (1965). The pharmacology of amino acids related to gamma-aminobutyric acid. Pharmacol. Rev. 17, 347-391. [PubMed] [Google Scholar]
- De Vries L., Zheng B., Fischer T., Elenko E. and Farquhar M. G. (2000). The regulator of G protein signaling family. Annu. Rev. Pharmacol. Toxicol. 40, 235-271. 10.1146/annurev.pharmtox.40.1.235 [DOI] [PubMed] [Google Scholar]
- DeMaria S. and Ngai J. (2010). The cell biology of smell. J. Cell Biol. 191, 443-452. 10.1083/jcb.201008163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingus J., Wells C. A., Campbell L., Cleator J. H., Robinson K. and Hildebrandt J. D. (2005). G protein βγ dimer formation: Gβ and Gγ differentially determine efficiency of in vitro dimer formation. Biochemistry 44, 11882-11890. 10.1021/bi0504254 [DOI] [PubMed] [Google Scholar]
- Doitsidou M., Reichman-Fried M., Stebler J., Köprunner M., Dörries J., Meyer D. Esguerra C. V., Leung T. and Raz E. (2002). Guidance of primordial germ cell migration by the chemokine SDF-1. Cell 111, 647-659. 10.1016/S0092-8674(02)01135-2 [DOI] [PubMed] [Google Scholar]
- Drake M. T., Shenoy S. K. and Lefkowitz R. J. (2006). Trafficking of G protein-coupled receptors. Circ. Res. 99, 570-582. 10.1161/01.RES.0000242563.47507.ce [DOI] [PubMed] [Google Scholar]
- Ferré S., Casadó V., Devi L. A., Filizola M., Jockers R., Lohse M. J., Milligan G. Pin J.-P. and Guitart X. (2014). G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol. Rev. 66, 413-434. 10.1124/pr.113.008052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R. and Schioth H. B. (2005). The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol. Pharmacol. 67, 1414-1425. 10.1124/mol.104.009001 [DOI] [PubMed] [Google Scholar]
- Fuse N., Yu F. and Hirose S. (2013). Gprk2 adjusts Fog signaling to organize cell movements in Drosophila gastrulation. Development 140, 4246-4255. 10.1242/dev.093625 [DOI] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ghosh P. and Farquhar M. G. (2015). GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J. Biol. Chem. 290, 6697-6704. 10.1074/jbc.R114.613414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. S. and Smith D. P. (2008). Insect odorant receptors: channeling scent. Cell 5, 761-763. 10.1016/j.cell.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Higgins J. B. and Caseys P. J. (1994). In vitro processing of recombinant G protein γ subunits. J. Biol. Chem. 269, 9067-9073. [PubMed] [Google Scholar]
- Hyun S., Lee Y., Hong S.-T., Bang S., Paik D., Kang J., Shin J., Lee J., Jeon K., Hwang S. et al. (2005). Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron 48, 267-278. 10.1016/j.neuron.2005.08.025 [DOI] [PubMed] [Google Scholar]
- Jensen R. T. and Gardner J. D. (1981). Identification and characterization of receptors for secretagogues on pancreatic acinar cells. Fed. Proc. 40, 2486-2496. [PubMed] [Google Scholar]
- Kamps A. R., Pruitt M. M., Herriges J. C. and Coffman C. R. (2010). An evolutionarily conserved arginine is essential for Tre1 G protein-coupled receptor function during germ cell migration in Drosophila melanogaster. PLoS ONE 5, e11839 10.1371/journal.pone.0011839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katanaev V. L., Ponzielli R., Sémériva M. and Tomlinson A. (2005). Trimeric G protein-dependent frizzled signaling in Drosophila. Cell 120, 111-122. 10.1016/j.cell.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Katanayeva N., Kopein D., Portmann R., Hess D. and Katanaev V. L. (2010). Competing activities of heterotrimeric G proteins in Drosophila wing maturation. PLoS ONE 5, e12331 10.1371/journal.pone.0012331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent L. B. and Robertson H. M. (2009). Evolution of the sugar receptors in insects. BMC Evol. Biol. 9, 41 10.1186/1471-2148-9-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleuss C., Raw A. S., Lee E., Sprang S. R. and Gilman A. G. (1994). Mechanism of GTP hydrolysis by G-protein alpha subunits. Proc. Natl. Acad. Sci. USA 91, 9828-9831. 10.1073/pnas.91.21.9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobilka B. K., Kobilka T. S., Daniel K., Regan J. W., Caron M. G. and Lefkowitz R. J. (1988). Chimeric alpha 2-, beta 2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science 240, 1310-1316. 10.1126/science.2836950 [DOI] [PubMed] [Google Scholar]
- Koval A. and Katanaev V. L. (2011). Wnt3a stimulation elicits G-protein-coupled receptor properties of mammalian Frizzled proteins. Biochem. J. 433, 435-440. 10.1042/BJ20101878 [DOI] [PubMed] [Google Scholar]
- Krasnoperov V. G., Bittner M. A., Beavis R., Kuang Y., Salnikow K. V., Chepurny O. G., Little A. R., Plotnikov A. N., Wu D., Holz R. W. et al. (1997). α-latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron 18, 925-937. 10.1016/S0896-6273(00)80332-3 [DOI] [PubMed] [Google Scholar]
- Kühne W. (1877). Zur Photochemie der Netzhaut. Untersuch. Physiol. Instit. Univ. Heidelberg 1, 1-14. [Google Scholar]
- Kunwar P. S., Starz-Gaiano M., Bainton R. J., Heberlein U. and Lehmann R. (2003). Tre1, a G protein-coupled receptor, directs transepithelial migration of Drosophila germ cells. PLoS Biol. 1, e80 10.1371/journal.pbio.0000080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar P. S., Sano H., Renault A. D., Barbosa V., Fuse N. and Lehmann R. (2008). Tre1 GPCR initiates germ cell transepithelial migration by regulating Drosophila melanogaster E-cadherin. J. Cell Biol. 183, 157-168. 10.1083/jcb.200807049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappano R. and Maggiolini M. (2011). G protein-coupled receptors: novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 10, 47-60. 10.1038/nrd3320 [DOI] [PubMed] [Google Scholar]
- Lau W. W. I., Chan A. S. L., Poon L. S. W., Zhu J. and Wong Y. H. (2013). Gβγ-mediated activation of protein kinase D exhibits subunit specificity and requires Gβγ-responsive phospholipase C β isoforms. Cell Commun. Signal. 11, 1-17. 10.1186/1478-811X-11-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373-391. 10.1146/annurev-ento-120811-153635 [DOI] [PubMed] [Google Scholar]
- Lefkowitz R. J. (2013). A brief history of G-protein coupled receptors (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 52, 6366-6378. 10.1002/anie.201301924 [DOI] [PubMed] [Google Scholar]
- Lin C., Koval A., Tishchenko S., Gabdulkhakov A., Tin U., Solis G. P. and Katanaev V. L. (2014). Double suppression of the Gα protein activity by RGS proteins. Mol. Cell 53, 663-671. 10.1016/j.molcel.2014.01.014 [DOI] [PubMed] [Google Scholar]
- Lundin C., Käll L., Kreher S. A., Kapp K., Sonnhammer E. L., Carlson J. R., von Heijne G. and Nilsson I. (2007). Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 581, 5601-5604. 10.1016/j.febslet.2007.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Jeong S.-J., Jin Z., Strokes N., Li S. and Piao X. (2011). G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. USA 108, 12925-12930. 10.1073/pnas.1104821108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell L. M., Ferguson S. S. G., Daaka Y., Miller W. E., Maudsley S., Della Rocca G. J., Lin F.-T., Kawakatsu H., Owada K., Luttrell D. K. et al. (1999). Β-arrestin-dependent formation of the β2 adrenergic receptor-Src protein kinase complexes. Science 283, 655-661. 10.1126/science.283.5402.655 [DOI] [PubMed] [Google Scholar]
- Manning A. J., Peters K. A., Peifer M. and Rogers S. L. (2013). Regulation of epithelial morphogenesis by the G protein-coupled receptor mist and its ligand fog. Sci. Signal. 6, ra98 10.1126/scisignal.2004427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A., and Paing M. M., Temple B. R. S. and Trejo J. (2008). G protein-coupled receptor sorting to endosomes and lysosomes. Annu. Rev. Pharmacol. Toxicol. 48, 601-629. 10.1146/annurev.pharmtox.48.113006.094646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker E. C., Bane S. E., O'Malley M. A. and Robinson A. S. (2007). Heterologous GPCR expression: a bottleneck to obtaining crystal structures. Biotechnol. Prog. 23, 540-547. 10.1021/bp060349b [DOI] [PubMed] [Google Scholar]
- Mertens I., Vandingenen A., Johnson E. C., Shafer O. T., Li W., Trigg J. S., De Loof A., Schoofs L. and Taghert P. H. (2005). PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 48, 213-219. 10.1016/j.neuron.2005.09.009 [DOI] [PubMed] [Google Scholar]
- Molyneaux K. A. (2003). The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development 130, 4279-4286. 10.1242/dev.00640 [DOI] [PubMed] [Google Scholar]
- Mullershausen F., Zecri F., Cetin C., Billich A., Guerini D. and Seuwen K. (2009). Persistent signaling induced by FTY720-phosphate is mediated by internalized S1P1 receptors. Nat. Chem. Biol. 5, 428-434. 10.1038/nchembio.173 [DOI] [PubMed] [Google Scholar]
- Nathans J. and Hogness D. S. (1983). Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell 34, 807-814. 10.1016/0092-8674(83)90537-8 [DOI] [PubMed] [Google Scholar]
- Nichols A. S., Floyd D. H., Bruinsma S. P., Narzinski K. and Baranski T. J. (2013). Frizzled receptors signal through G proteins. Cell Signal. 25, 1468-1475. 10.1016/j.cellsig.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley R. H., Laporte S. A., Holt J. A., Caron M. G. and Barak L. S. (2000). Differential affinities of visual arrestin, beta arrestin1, and beta arrestin2 for G protein-coupled receptors delineate two major classes of receptors. J. Biol. Chem. 275, 17201-17210. 10.1074/jbc.M910348199 [DOI] [PubMed] [Google Scholar]
- Palczewskiss K., Buczylkost J., Kaplans M. W., Polanss A. S. and Crabb J. W. (1991). Mechanism of rhodopsin kinase activation. J. Biol. Chem. 266, 12949-12955. [PubMed] [Google Scholar]
- Parks S. and Wieschaus E. (1991). The Drosophila gastrulation gene concertina encodes a Ga-like protein. Cell 64, 447-458. 10.1016/0092-8674(91)90652-F [DOI] [PubMed] [Google Scholar]
- Peters K. A. and Rogers S. L. (2013). Drosophila Ric-8 interacts with the Ga12/13 subunit, Concertina, during activation of the Folded gastrulation pathway. Mol. Biol. Cell 24, 3460-3471. 10.1091/mbc.E12-11-0813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher J. A., Freedman N. J. and Lefkowitz R. J. (1998). G protein-coupled receptor kinases. Annu. Rev. Biochem. 67, 653-692. 10.1146/annurev.biochem.67.1.653 [DOI] [PubMed] [Google Scholar]
- Prabhu Y. and Eichinger L. (2006). The Dictyostelium repertoire of seven transmembrane domain receptors. Eur. J. Cell Biol. 85, 937-946. 10.1016/j.ejcb.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Purvanov V., Koval A. and Katanaev V. L. (2010). A direct and functional interaction between go and rab5 during g protein coupled receptor signaling. Sci. Signal. 3, ra65 10.1126/scisignal.2000877 [DOI] [PubMed] [Google Scholar]
- Riobo N. A., Lu K. and Emerson C. P. Jr (2006). Hedgehog signal transduction: signal integration and crosstalk in development and cancer. Cell Cycle 5, 1612-1615. 10.4161/cc.5.15.3130 [DOI] [PubMed] [Google Scholar]
- Rovati G. E., Capra V. and Neubig R. R. (2007). The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol. Pharmacol. 71, 959-964. 10.1124/mol.106.029470 [DOI] [PubMed] [Google Scholar]
- Schöneberg T., Schulz A., Biebermann H., Hermsdorf T., Römpler H. and Sangkuhl K. (2004). Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol. Ther. 104, 173-206. 10.1016/j.pharmthera.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Schulte G. and Bryja V. (2007). The Frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol. Sci. 28, 518-525. 10.1016/j.tips.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Schwabe T., Bainton R. J., Fetter R. D., Heberlein U. and Gaul U. (2005). GPCR signaling is required for blood-brain barrier formation in Drosophila. Cell 123, 133-144. 10.1016/j.cell.2005.08.037 [DOI] [PubMed] [Google Scholar]
- Shen F., Cheng L., Douglas A. E., Riobo N. A. and Manning D. R. (2013). Smoothened is a fully competent activator of the heterotrimeric G protein Gi. Mol. Pharmacol. 83, 691-697. 10.1124/mol.112.082511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering A. F. and Benton R. (2010). Ionotropic and metabotropic mechanisms in chemoreception: ‘chance or design’? EMBO Rep. 11, 173-179. 10.1038/embor.2010.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin F., Karcher P., Boeuf J. J.-M., Matifas A. and Kieffer B. L. (2004). Identification of a novel family of G protein-coupled receptor associated sorting proteins. J. Neurochem. 89, 766-775. 10.1111/j.1471-4159.2004.02411.x [DOI] [PubMed] [Google Scholar]
- Sondek J., Bohm A., Lambright D. G., Hamm H. E. and Sigler P. B. (1996). Crystal structure of a Gα protein βγ dimer at 2.1A resolution. Nature 379, 369-374. 10.1038/379369a0 [DOI] [PubMed] [Google Scholar]
- Taddese B., Upton G. J. G., Bailey G. R., Jordan S. R. D., Abdulla N. Y., Reeves P. J. and Reynolds C. A. (2014). Do plants contain g protein-coupled receptors? Plant Physiol. 164, 287-307. 10.1104/pp.113.228874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.-L., Wang Y., Li D.-L., Luo J. and Liu M.-Y. (2012). Orphan G protein-coupled receptors (GPCRs): biological functions and potential drug targets. Acta Pharmacol. Sin. 33, 363-371. 10.1038/aps.2011.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova N. G. and von Zastrow M. (2014). Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat. Chem. Biol. 10, 1061-1065. 10.1038/nchembio.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D. and Jones A. M. (2014). Heterotrimeric G protein-coupled signaling in plants. Annu. Rev. Plant Biol. 65, 365-384. 10.1146/annurev-arplant-050213-040133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Jones J. C., Wang H., Matthews M., Bradford W., Bennetzen J. L. and Jones A. M. (2012). G protein activation without a GEF in the plant kingdom. PLoS Genet. 8, e1002756 10.1371/journal.pgen.1002756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A. J., Deupi X., Lebon G., Tate C. G., Schertler G. F. and Babu M. M. (2013). Molecular signatures of G-protein-coupled receptors. Nature 494, 185-194. 10.1038/nature11896 [DOI] [PubMed] [Google Scholar]
- Vögler O., Barceló J. M., Ribas C. and Escribá P. V. (2008). Membrane interactions of G proteins and other related proteins. Biochim. Biophys. Acta 1778, 1640-1652. 10.1016/j.bbamem.2008.03.008 [DOI] [PubMed] [Google Scholar]
- Warrington S. J., Strutt H. and Strutt D. (2013). The Frizzled-dependent planar polarity pathway locally promotes E-Cadherin turnover via recruitment of RhoGEF2. Development 140, 1045-1054. 10.1242/dev.088724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins H. A., Au M. and Hay D. L. (2012). The structure of secretin family GPCR peptide ligands: implications for receptor pharmacology and drug development. Drug Discov. Today 17, 1006-1014. 10.1016/j.drudis.2012.05.005 [DOI] [PubMed] [Google Scholar]
- Wettschureck N. and Offermanns S. (2005). Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159-1204. 10.1152/physrev.00003.2005 [DOI] [PubMed] [Google Scholar]
- Wicher D. (2015). Olfactory signaling in insects. Prog. Mol. Biol. Transl. Sci. 130, 37-54. 10.1016/bs.pmbts.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Wicher D., Schäfer R., Bauernfeind R., Stensmyr M. C., Heller R., Heinemann S. H. and Hansson B. S. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007-1011. 10.1038/nature06861 [DOI] [PubMed] [Google Scholar]
- Witherow D. S. and Slepak V. Z. (2003). A novel kind of G protein heterodimer: the G beta5-RGS complex. Receptors Channels 9, 205-212. 10.1080/10606820308239 [DOI] [PubMed] [Google Scholar]
- Yang-Snyder J., Miller J. R., Brown J. D., Lai C.-J. and Moon R. T. (1996). A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr. Biol. 6, 1302-1306. 10.1016/S0960-9822(02)70716-1 [DOI] [PubMed] [Google Scholar]
- Yi P., Johnson A. N., Han Z., Wu J. and Olson E. N. (2008). Heterotrimeric G proteins regulate a non-canonical function of septate junction proteins to maintain cardiac integrity in Drosophila. Dev. Cell. 15, 704-713. 10.1016/j.devcel.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura S., Ohta N. and Matsuzaki F. (2012). Tre1 GPCR signaling orients stem cell divisions in the Drosophila central nervous system. Dev. Cell 22, 79-91. 10.1016/j.devcel.2011.10.027 [DOI] [PubMed] [Google Scholar]




